A Genetic Circuit Design for Targeted Viral RNA Degradation
Abstract
:1. Introduction
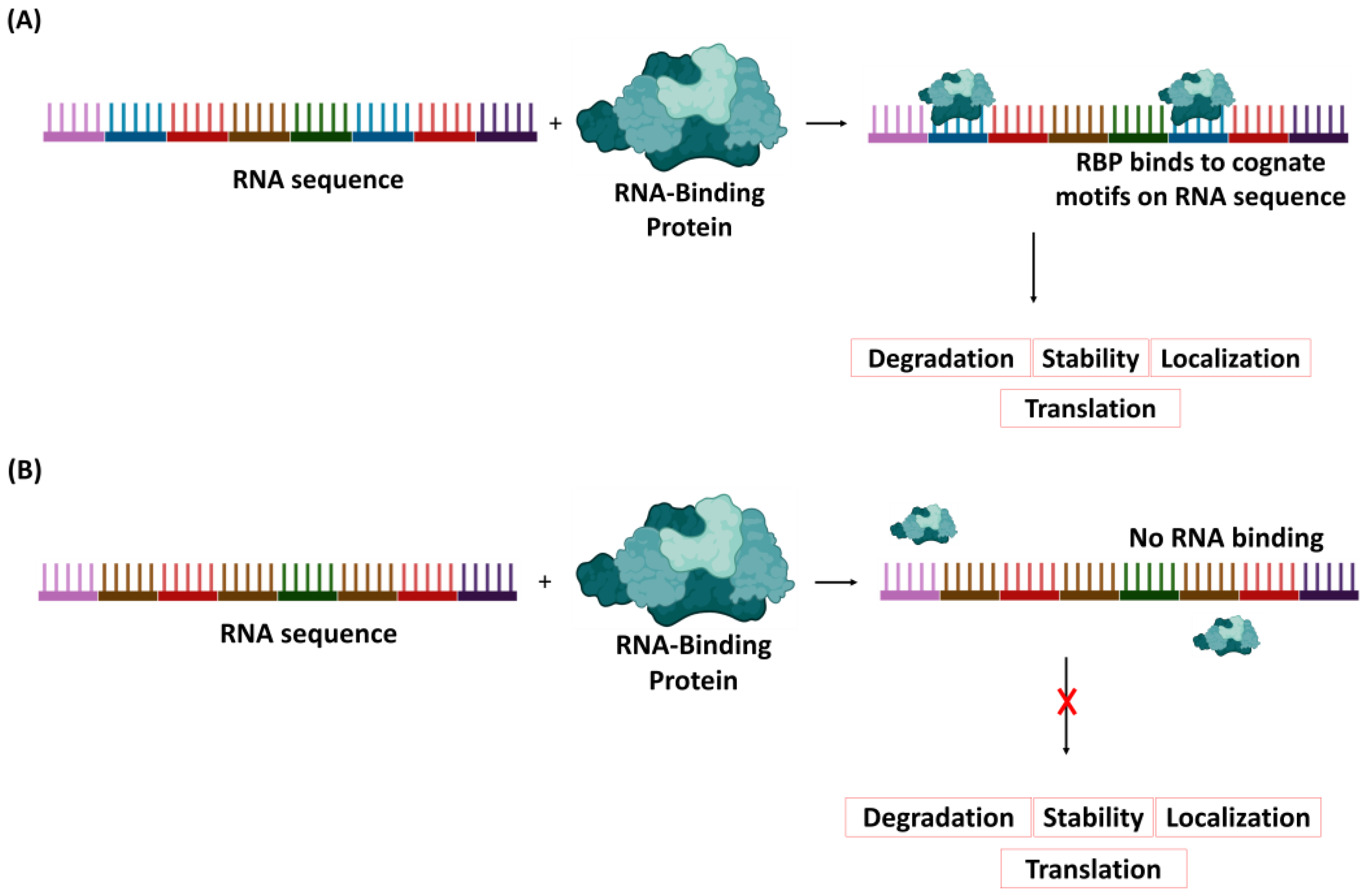
2. Process of Viral RNA Degradation
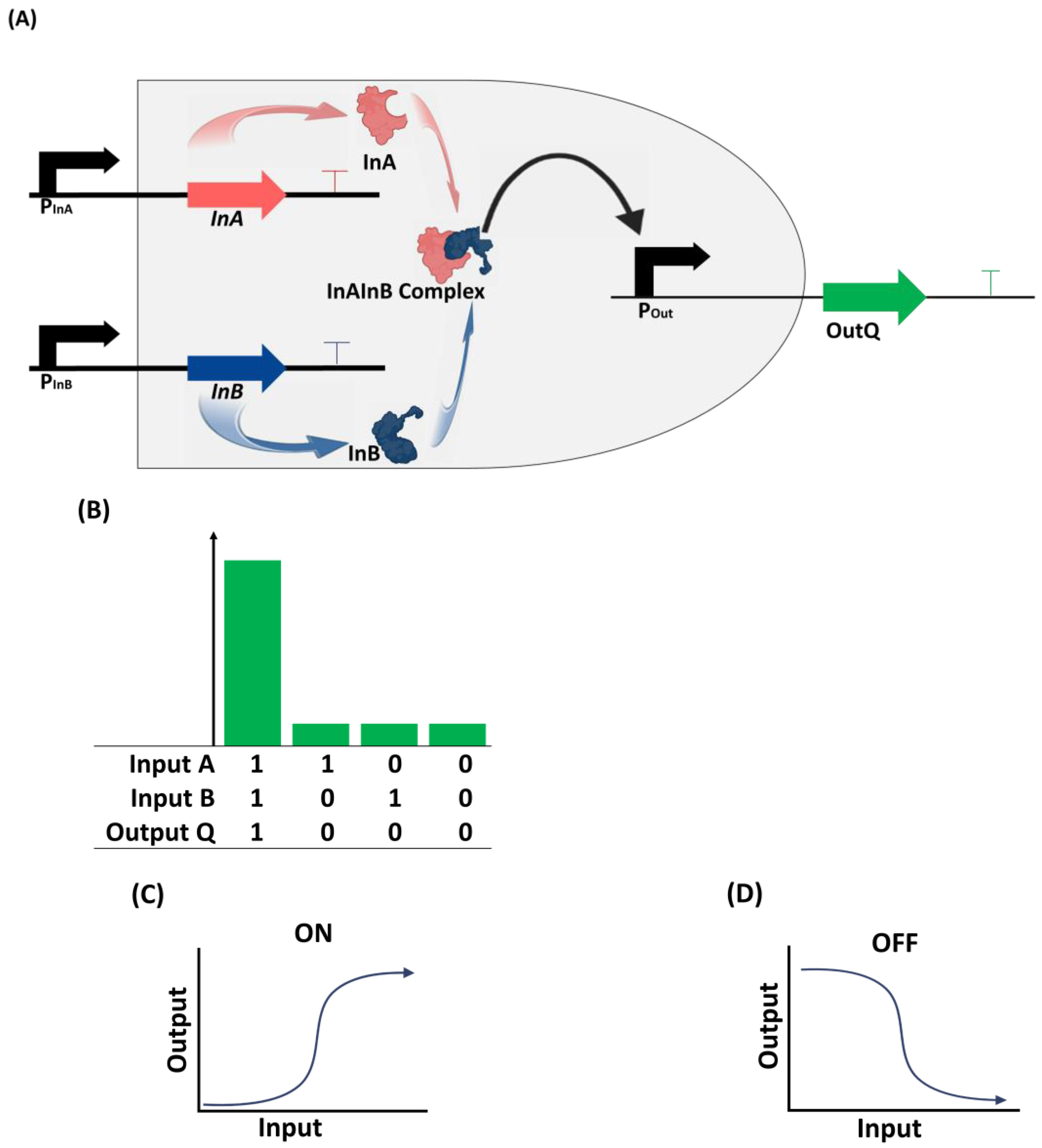
3. Design of Genetic Circuits
4. Perspectives and Conclusions
- (1)
- Would the RBP of interest be upregulated during viral infection? This can be addressed by proteomic analysis of the infected cells. In addition, techniques such as quantitative proteomics, UV protein–RNA crosslinking, and the oligo(dT) selection of polyadenylated (poly(A)) RNA can be used to study expression patterns [86,87,88].
- (2)
- If the RBP is upregulated, would the designed short non-coding RNA bind to the protein, and if it does, would the complex induce the expression of an RNA-degrading enzyme? Although the protein could be abundant in the cell and could bind to other RNAs as part of its cellular function, the careful design of the ncRNA with a motif specific to the protein is expected to enable the RNA to form a complex with some of the proteins, thereby triggering the expression of the RNA-degrading enzyme.
- (3)
- How would the degrading enzyme recognize the RNA? This can be investigated by using specific methods such as the use of guide RNA to direct the enzyme to its target RNA cut site.
- (4)
- How would we regulate gene expression in the circuit? The ncRNA is an important device in the circuit. The template provides a recombinase ON/OFF switch for the expression of the ncRNA by flipping its coding gene in the opposite direction. The recombinase can also be used to flip either the promoter or the terminator to control ncRNA expression.
Author Contributions
Funding
Conflicts of Interest
References
- Santomauro, D.F.; Herrera, A.M.M.; Shadid, J.; Zheng, P.; Ashbaugh, C.; Pigott, D.M.; Abbafati, C.; Adolph, C.; Amlag, J.O.; Aravkin, A.Y. Global Prevalence and Burden of Depressive and Anxiety Disorders in 204 Countries and Territories in 2020 Due to the COVID-19 Pandemic. Lancet 2021, 398, 1700–1712. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, W. Fast-Spreading SARS-CoV-2 Variants: Challenges to and New Design Strategies of COVID-19 Vaccines. Signal Transduct. Target. Ther. 2021, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.; Mohr, I. Viral Subversion of the Host Protein Synthesis Machinery. Nat. Rev. Microbiol. 2011, 9, 860–875. [Google Scholar] [CrossRef] [PubMed]
- Jaafar, Z.A.; Kieft, J.S. Viral RNA Structure-Based Strategies to Manipulate Translation. Nat. Rev. Microbiol. 2019, 17, 110–123. [Google Scholar] [CrossRef]
- Heck, A.M.; Wilusz, J. The Interplay between the RNA Decay and Translation Machinery in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032839. [Google Scholar] [CrossRef]
- Nagy, P.D.; Pogany, J. The Dependence of Viral RNA Replication on Co-Opted Host Factors. Nat. Rev. Microbiol. 2012, 10, 137–149. [Google Scholar] [CrossRef]
- Brophy, J.A.; Voigt, C.A. Principles of Genetic Circuit Design. Nat. Methods 2014, 11, 508–520. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. The Many Pathways of RNA Degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.; Kellner, M.J.; Regev, A. RNA Targeting with CRISPR–Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA Editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Kushawah, G.; Hernandez-Huertas, L.; Del Prado, J.A.-N.; Martinez-Morales, J.R.; DeVore, M.L.; Hassan, H.; Moreno-Sanchez, I.; Tomas-Gallardo, L.; Diaz-Moscoso, A.; Monges, D.E. CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos. Dev. Cell 2020, 54, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 2020, 181, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, E.L.; Vanover, D.; Bawage, S.S.; Tiwari, P.M.; Rotolo, L.; Beyersdorf, J.; Peck, H.E.; Bruno, N.C.; Hincapie, R.; Michel, F. Treatment of Influenza and SARS-CoV-2 Infections via mRNA-Encoded Cas13a in Rodents. Nat. Biotechnol. 2021, 39, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Moreno, M.; Järvelin, A.I.; Castello, A. Unconventional RNA-binding Proteins Step into the Virus–Host Battlefront. WIREs RNA 2018, 9, e1498. [Google Scholar] [CrossRef]
- Guo, X.; Ma, J.; Sun, J.; Gao, G. The Zinc-Finger Antiviral Protein Recruits the RNA Processing Exosome to Degrade the Target mRNA. Proc. Natl. Acad. Sci. USA 2007, 104, 151–156. [Google Scholar] [CrossRef]
- Meagher, J.L.; Takata, M.; Gonçalves-Carneiro, D.; Keane, S.C.; Rebendenne, A.; Ong, H.; Orr, V.K.; MacDonald, M.R.; Stuckey, J.A.; Bieniasz, P.D.; et al. Structure of the Zinc-Finger Antiviral Protein in Complex with RNA Reveals a Mechanism for Selective Targeting of CG-Rich Viral Sequences. Proc. Natl. Acad. Sci. USA 2019, 116, 24303–24309. [Google Scholar] [CrossRef]
- Yang, E.; Nguyen, L.P.; Wisherop, C.A.; Kan, R.L.; Li, M.M. The Role of ZAP and TRIM25 RNA Binding in Restricting Viral Translation. Front. Cell. Infect. Microbiol. 2022, 12, 886929. [Google Scholar] [CrossRef]
- Cheng, A.A.; Lu, T.K. Synthetic Biology: An Emerging Engineering Discipline. Annu. Rev. Biomed. Eng. 2012, 14, 155–178. [Google Scholar] [CrossRef]
- Goold, H.D.; Wright, P.; Hailstones, D. Emerging Opportunities for Synthetic Biology in Agriculture. Genes 2018, 9, 341. [Google Scholar] [CrossRef]
- Clomburg, J.M.; Crumbley, A.M.; Gonzalez, R. Industrial Biomanufacturing: The Future of Chemical Production. Science 2017, 355, aag0804. [Google Scholar] [CrossRef]
- Nguyen, P.Q.; Huang, X.; Collins, D.S.; Collins, J.J.; Lu, T. Harnessing Synthetic Biology to Enhance Ocean Health. Trends Biotechnol. 2023, 41, 860–874. [Google Scholar] [CrossRef] [PubMed]
- Colloms, S.D.; Merrick, C.A.; Olorunniji, F.J.; Stark, W.M.; Smith, M.C.; Osbourn, A.; Keasling, J.D.; Rosser, S.J. Rapid Metabolic Pathway Assembly and Modification Using Serine Integrase Site-Specific Recombination. Nucleic Acids Res. 2014, 42, e23. [Google Scholar] [CrossRef] [PubMed]
- Awan, A.R.; Blount, B.A.; Bell, D.J.; Shaw, W.M.; Ho, J.C.; McKiernan, R.M.; Ellis, T. Biosynthesis of the Antibiotic Nonribosomal Peptide Penicillin in Baker’s Yeast. Nat. Commun. 2017, 8, 15202. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Barahona, M.; Buck, M. A Modular Cell-Based Biosensor Using Engineered Genetic Logic Circuits to Detect and Integrate Multiple Environmental Signals. Biosens. Bioelectron. 2013, 40, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Volpetti, F.; Petrova, E.; French, C.; Maerkl, S.J.; Wang, B. Cascaded Amplifying Circuits Enable Ultrasensitive Cellular Sensors for Toxic Metals. Nat. Chem. Biol. 2019, 15, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic Acid Detection with CRISPR Nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Gerber, P.P.; Donde, M.J.; Matheson, N.J.; Taylor, A.I. XNAzymes Targeting the SARS-CoV-2 Genome Inhibit Viral Infection. Nat. Commun. 2022, 13, 6716. [Google Scholar] [CrossRef]
- Moon, S.L.; Anderson, J.R.; Kumagai, Y.; Wilusz, C.J.; Akira, S.; Khromykh, A.A.; Wilusz, J. A Noncoding RNA Produced by Arthropod-Borne Flaviviruses Inhibits the Cellular Exoribonuclease XRN1 and Alters Host mRNA Stability. RNA 2012, 18, 2029–2040. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Mok, B.W.-Y.; Wang, P.; Kuo, R.-L.; Chen, H.; Shih, S.-R. Cellular 5′-3′ mRNA Exoribonuclease XRN1 Inhibits Interferon Beta Activation and Facilitates Influenza A Virus Replication. mBio 2021, 12, e00945-21. [Google Scholar] [CrossRef]
- Roby, J.A.; Pijlman, G.P.; Wilusz, J.; Khromykh, A.A. Noncoding Subgenomic Flavivirus RNA: Multiple Functions in West Nile Virus Pathogenesis and Modulation of Host Responses. Viruses 2014, 6, 404–427. [Google Scholar] [CrossRef]
- Steckelberg, A.-L.; Akiyama, B.M.; Costantino, D.A.; Sit, T.L.; Nix, J.C.; Kieft, J.S. A Folded Viral Noncoding RNA Blocks Host Cell Exoribonucleases through a Conformationally Dynamic RNA Structure. Proc. Natl. Acad. Sci. USA 2018, 115, 6404–6409. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Sun, R.; Chen, Z.; Yao, Y.; Zuo, X.; Chen, C.; Fang, X. Pseudoknot Length Modulates the Folding, Conformational Dynamics, and Robustness of Xrn1 Resistance of Flaviviral xrRNAs. Nat. Commun. 2021, 12, 6417. [Google Scholar] [CrossRef] [PubMed]
- Steckelberg, A.-L.; Vicens, Q.; Kieft, J.S. Exoribonuclease-Resistant RNAs Exist within Both Coding and Noncoding Subgenomic RNAs. mBio 2018, 9, e02461-18. [Google Scholar] [CrossRef]
- Jones, R.A.; Steckelberg, A.-L.; Vicens, Q.; Szucs, M.J.; Akiyama, B.M.; Kieft, J.S. Different Tertiary Interactions Create the Same Important 3D Features in a Distinct Flavivirus xrRNA. RNA 2021, 27, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Woodside, M.T. Mechanical Strength of RNA Knot in Zika Virus Protects against Cellular Defenses. Nat. Chem. Biol. 2021, 17, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Nagy, P.D. Diverse Roles of Host RNA Binding Proteins in RNA Virus Replication. RNA Biol. 2011, 8, 305–315. [Google Scholar] [CrossRef]
- Gebauer, F.; Schwarzl, T.; Valcárcel, J.; Hentze, M.W. RNA-Binding Proteins in Human Genetic Disease. Nat. Rev. Genet. 2021, 22, 185–198. [Google Scholar] [CrossRef]
- Kelaini, S.; Chan, C.; Cornelius, V.A.; Margariti, A. RNA-Binding Proteins Hold Key Roles in Function, Dysfunction, and Disease. Biology 2021, 10, 366. [Google Scholar] [CrossRef]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-Binding Proteins and Post-Transcriptional Gene Regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR Target Circular RNAs Uncovers Suppression of PABPN1 Translation by CircPABPN1. RNA Biol. 2017, 14, 361–369. [Google Scholar] [CrossRef]
- Charlesworth, A.; Meijer, H.A.; De Moor, C.H. Specificity Factors in Cytoplasmic Polyadenylation. WIREs RNA 2013, 4, 437–461. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.S.; von Lindern, M. RNA Binding Proteins and Regulation of mRNA Translation in Erythropoiesis. Front. Physiol. 2018, 9, 910. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G.; Lorsch, J.R. The Mechanism of Eukaryotic Translation Initiation: New Insights and Challenges. Cold Spring Harb. Perspect. Biol. 2012, 4, a011544. [Google Scholar] [CrossRef] [PubMed]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-Binding Proteins: Modular Design for Efficient Function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A Brave New World of RNA-Binding Proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef]
- Ferrarini, M.G.; Lal, A.; Rebollo, R.; Gruber, A.J.; Guarracino, A.; Gonzalez, I.M.; Floyd, T.; de Oliveira, D.S.; Shanklin, J.; Beausoleil, E. Genome-Wide Bioinformatic Analyses Predict Key Host and Viral Factors in SARS-CoV-2 Pathogenesis. Commun. Biol. 2021, 4, 590. [Google Scholar] [CrossRef]
- Ficarelli, M.; Wilson, H.; Pedro Galão, R.; Mazzon, M.; Antzin-Anduetza, I.; Marsh, M.; Neil, S.J.; Swanson, C.M. KHNYN Is Essential for the Zinc Finger Antiviral Protein (ZAP) to Restrict HIV-1 Containing Clustered CpG Dinucleotides. eLife 2019, 8, e46767. [Google Scholar] [CrossRef]
- Nchioua, R.; Kmiec, D.; Müller, J.A.; Conzelmann, C.; Groß, R.; Swanson, C.M.; Neil, S.J.D.; Stenger, S.; Sauter, D.; Münch, J.; et al. SARS-CoV-2 Is Restricted by Zinc Finger Antiviral Protein despite Preadaptation to the Low-CpG Environment in Humans. mBio 2020, 11, e01930-20. [Google Scholar] [CrossRef]
- Zimmer, M.M.; Kibe, A.; Rand, U.; Pekarek, L.; Ye, L.; Buck, S.; Smyth, R.P.; Cicin-Sain, L.; Caliskan, N. The Short Isoform of the Host Antiviral Protein ZAP Acts as an Inhibitor of SARS-CoV-2 Programmed Ribosomal Frameshifting. Nat. Commun. 2021, 12, 7193. [Google Scholar] [CrossRef]
- Kases, K.; Schubert, E.; Hajikhezri, Z.; Larsson, M.; Devi, P.; Darweesh, M.; Andersson, L.; Akusjärvi, G.; Punga, T.; Younis, S. The RNA-Binding Protein ZC3H11A Interacts with the Nuclear Poly (A)-Binding Protein PABPN1 and Alters Polyadenylation of Viral Transcripts. J. Biol. Chem. 2023, 299, 104959. [Google Scholar] [CrossRef]
- Girardi, E.; Pfeffer, S.; Baumert, T.F.; Majzoub, K. Roadblocks and Fast Tracks: How RNA Binding Proteins Affect the Viral RNA Journey in the Cell. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 111, pp. 86–100. [Google Scholar]
- Laudenbach, B.T.; Krey, K.; Emslander, Q.; Andersen, L.L.; Reim, A.; Scaturro, P.; Mundigl, S.; Dächert, C.; Manske, K.; Moser, M. NUDT2 Initiates Viral RNA Degradation by Removal of 5′-Phosphates. Nat. Commun. 2021, 12, 6918. [Google Scholar] [CrossRef] [PubMed]
- Charley, P.A.; Wilusz, J. Standing Your Ground to Exoribonucleases: Function of Flavivirus Long Non-Coding RNAs. Virus Res. 2016, 212, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.W.; Bass, B.L. The Role of RNA Editing by ADARs in RNAi. Mol. Cell 2002, 10, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Kallunki, T.; Barisic, M.; Jäättelä, M.; Liu, B. How to Choose the Right Inducible Gene Expression System for Mammalian Studies? Cells 2019, 8, 796. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Saito, H. Caliciviral Protein-Based Artificial Translational Activator for Mammalian Gene Circuits with RNA-Only Delivery. Nat. Commun. 2020, 11, 1297. [Google Scholar] [CrossRef]
- Tang, T.-C.; An, B.; Huang, Y.; Vasikaran, S.; Wang, Y.; Jiang, X.; Lu, T.K.; Zhong, C. Materials Design by Synthetic Biology. Nat. Rev. Mater. 2021, 6, 332–350. [Google Scholar] [CrossRef]
- Wu, C.; Wan, S.; Hou, W.; Zhang, L.; Xu, J.; Cui, C.; Wang, Y.; Hu, J.; Tan, W. A Survey of Advancements in Nucleic Acid-Based Logic Gates and Computing for Applications in Biotechnology and Biomedicine. Chem. Commun. 2015, 51, 3723–3734. [Google Scholar] [CrossRef]
- Sedlmayer, F.; Aubel, D.; Fussenegger, M. Synthetic Gene Circuits for the Detection, Elimination and Prevention of Disease. Nat. Biomed. Eng. 2018, 2, 399–415. [Google Scholar] [CrossRef]
- Church, G.M.; Elowitz, M.B.; Smolke, C.D.; Voigt, C.A.; Weiss, R. Realizing the Potential of Synthetic Biology. Nat. Rev. Mol. Cell Biol. 2014, 15, 289–294. [Google Scholar] [CrossRef]
- Slusarczyk, A.L.; Lin, A.; Weiss, R. Foundations for the Design and Implementation of Synthetic Genetic Circuits. Nat. Rev. Genet. 2012, 13, 406–420. [Google Scholar] [CrossRef]
- Xia, P.-F.; Ling, H.; Foo, J.L.; Chang, M.W. Synthetic Genetic Circuits for Programmable Biological Functionalities. Biotechnol. Adv. 2019, 37, 107393. [Google Scholar] [CrossRef]
- Voigt, C.A. Genetic Parts to Program Bacteria. Curr. Opin. Biotechnol. 2006, 17, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Canton, B.; Labno, A.; Endy, D. Refinement and Standardization of Synthetic Biological Parts and Devices. Nat. Biotechnol. 2008, 26, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Purcell, O.; Lu, T.K. Synthetic Analog and Digital Circuits for Cellular Computation and Memory. Curr. Opin. Biotechnol. 2014, 29, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Roquet, N.; Lu, T.K. Digital and Analog Gene Circuits for Biotechnology. Biotechnol. J. 2014, 9, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Buck, M. Customizing Cell Signaling Using Engineered Genetic Logic Circuits. Trends Microbiol. 2012, 20, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a Genetic Toggle Switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Jusiak, B.; Cleto, S.; Perez-Piñera, P.; Lu, T.K. Engineering Synthetic Gene Circuits in Living Cells with CRISPR Technology. Trends Biotechnol. 2016, 34, 535–547. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhan, H.; Liao, X.; Fang, L.; Liu, Y.; Xie, H.; Yang, K.; Gao, Q.; Ding, M.; Cai, Z.; et al. A Revolutionary Tool: CRISPR Technology Plays an Important Role in Construction of Intelligentized Gene Circuits. Cell Prolif. 2019, 52, e12552. [Google Scholar] [CrossRef]
- Chiu, T.-Y.; Jiang, J.-H.R. Logic Synthesis of Recombinase-Based Genetic Circuits. Sci. Rep. 2017, 7, 12873. [Google Scholar] [CrossRef]
- Olorunniji, F.J.; Merrick, C.; Rosser, S.J.; Smith, M.C.M.; Stark, W.M.; Colloms, S.D. Multipart DNA Assembly Using Site-Specific Recombinases from the Large Serine Integrase Family. In Site-Specific Recombinases; Eroshenko, N., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1642, pp. 303–323. [Google Scholar] [CrossRef]
- Olorunniji, F.J.; Lawson-Williams, M.; McPherson, A.L.; Paget, J.E.; Stark, W.M.; Rosser, S.J. Control of ϕC31 Integrase-Mediated Site-Specific Recombination by Protein Trans-Splicing. Nucleic Acids Res. 2019, 47, 11452–11460. [Google Scholar] [CrossRef] [PubMed]
- Akboğa, D.; Saltepe, B.; Bozkurt, E.U.; Şeker, U.Ö.Ş. A Recombinase-Based Genetic Circuit for Heavy Metal Monitoring. Biosensors 2022, 12, 122. [Google Scholar] [CrossRef] [PubMed]
- Wroblewska, L.; Kitada, T.; Endo, K.; Siciliano, V.; Stillo, B.; Saito, H.; Weiss, R. Mammalian Synthetic Circuits with RNA Binding Proteins for RNA-Only Delivery. Nat. Biotechnol. 2015, 33, 839–841. [Google Scholar] [CrossRef] [PubMed]
- Brophy, J.A.N.; Magallon, K.J.; Duan, L.; Zhong, V.; Ramachandran, P.; Kniazev, K.; Dinneny, J.R. Synthetic Genetic Circuits as a Means of Reprogramming Plant Roots. Science 2022, 377, 747–751. [Google Scholar] [CrossRef]
- Santos-Moreno, J.; Tasiudi, E.; Stelling, J.; Schaerli, Y. Multistable and Dynamic CRISPRi-Based Synthetic Circuits. Nat. Commun. 2020, 11, 2746. [Google Scholar] [CrossRef]
- McCarty, N.S.; Graham, A.E.; Studená, L.; Ledesma-Amaro, R. Multiplexed CRISPR Technologies for Gene Editing and Transcriptional Regulation. Nat. Commun. 2020, 11, 1281. [Google Scholar] [CrossRef]
- Costello, A.; Badran, A.H. Synthetic Biological Circuits within an Orthogonal Central Dogma. Trends Biotechnol. 2021, 39, 59–71. [Google Scholar] [CrossRef]
- Santos-Moreno, J.; Schaerli, Y. CRISPR-Based Gene Expression Control for Synthetic Gene Circuits. Biochem. Soc. Trans. 2020, 48, 1979–1993. [Google Scholar] [CrossRef]
- Buecherl, L.; Myers, C.J. Engineering Genetic Circuits: Advancements in Genetic Design Automation Tools and Standards for Synthetic Biology. Curr. Opin. Microbiol. 2022, 68, 102155. [Google Scholar] [CrossRef]
- Chakraborty, D.; Rengaswamy, R.; Raman, K. Designing Biological Circuits: From Principles to Applications. ACS Synth. Biol. 2022, 11, 1377–1388. [Google Scholar] [CrossRef]
- Pardi, M.L.; Wu, J.; Kawasaki, S.; Saito, H. Synthetic RNA-Based Post-Transcriptional Expression Control Methods and Genetic Circuits. Adv. Drug Deliv. Rev. 2022, 184, 114196. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Wang, X.; Gao, Y.; Zhu, J.; Liu, S.; Gao, G.; Gao, P. Molecular Mechanism of RNA Recognition by Zinc-Finger Antiviral Protein. Cell Rep. 2020, 30, 46–52. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Garcia-Moreno, M.; Noerenberg, M.; Ni, S.; Järvelin, A.I.; González-Almela, E.; Lenz, C.E.; Bach-Pages, M.; Cox, V.; Avolio, R.; Davis, T. System-Wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection. Mol. Cell 2019, 74, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perri, J.I.; Noerenberg, M.; Kamel, W.; Lenz, C.E.; Mohammed, S.; Hentze, M.W.; Castello, A. Global Analysis of RNA-Binding Protein Dynamics by Comparative and Enhanced RNA Interactome Capture. Nat. Protoc. 2021, 16, 27–60. [Google Scholar] [CrossRef] [PubMed]
- Sysoev, V.O.; Fischer, B.; Frese, C.K.; Gupta, I.; Krijgsveld, J.; Hentze, M.W.; Castello, A.; Ephrussi, A. Global Changes of the RNA-Bound Proteome during the Maternal-to-Zygotic Transition in Drosophila. Nat. Commun. 2016, 7, 12128. [Google Scholar] [CrossRef]
- Lecca, P.; Ihekwaba-Ndibe, A.E. Dynamic modelling of DNA repair pathway at the molecular level: A new perspective. Front. Mol. Biosci. 2022, 9, 878148. [Google Scholar] [CrossRef]
- Ihekwaba, A.E.; Mura, I.; Walshaw, J.; Peck, M.W.; Barker, G.C. An integrative approach to the computational modelling of the gene regulatory network controlling Clostridium botulinum type A1 toxin production. PLoS Comput. Biol. 2016, 12, e1005205. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bello, A.J.; Popoola, A.; Okpuzor, J.; Ihekwaba-Ndibe, A.E.; Olorunniji, F.J. A Genetic Circuit Design for Targeted Viral RNA Degradation. Bioengineering 2024, 11, 22. https://doi.org/10.3390/bioengineering11010022
Bello AJ, Popoola A, Okpuzor J, Ihekwaba-Ndibe AE, Olorunniji FJ. A Genetic Circuit Design for Targeted Viral RNA Degradation. Bioengineering. 2024; 11(1):22. https://doi.org/10.3390/bioengineering11010022
Chicago/Turabian StyleBello, Adebayo J., Abdulgafar Popoola, Joy Okpuzor, Adaoha E. Ihekwaba-Ndibe, and Femi J. Olorunniji. 2024. "A Genetic Circuit Design for Targeted Viral RNA Degradation" Bioengineering 11, no. 1: 22. https://doi.org/10.3390/bioengineering11010022
APA StyleBello, A. J., Popoola, A., Okpuzor, J., Ihekwaba-Ndibe, A. E., & Olorunniji, F. J. (2024). A Genetic Circuit Design for Targeted Viral RNA Degradation. Bioengineering, 11(1), 22. https://doi.org/10.3390/bioengineering11010022






