1. Introduction
The need for improved tools in clinical histopathology has been repeatedly emphasized. Commonly cited arguments include an increasing need for accuracy, a lack of specialized histopathologists, subjectivity, and a lack of reproducibility [
1,
2]. Furthermore, the situation is expected to worsen due to increasing cancer incidence and more screening campaigns [
3]. Fortunately, deep neural networks are expected to significantly change the study of cancer tissue in histopathology [
1]. In fact, deep learning (DL) in computational histopathology promises various benefits. These include time and cost savings, lower error rates, better accessibility, and the learning of more accurate representations [
2]. Furthermore, it can improve clinical decision making by providing access to high-level information [
4]. It also plays an important role in mining big data [
5]. As a result, the yearly number of research papers on DL-based histopathology is steadily increasing [
3,
6].
However, DL models must achieve a sufficient level of accuracy to be approved for clinical practice, while digital histopathology poses several challenges for the application of DL, including large image size, insufficient data annotation, varying magnification levels of the microscope, staining artifacts, and color variation [
2]. Transfer learning, including the fine-tuning of pre-trained models, is a standard technical solution for insufficient annotation. Given limited training data, fine-tuning can significantly improve the performance compared to training from scratch [
7]. Kornblith et al. [
8] also found that, on average, fine-tuning achieved a 17-fold increase in the training speed compared to training from scratch. This was confirmed by He et al. [
9]. In histopathology, fine-tuning appears to be generally superior to using off-the-shelf features [
10,
11].
The exact procedure for fine-tuning, however, is still an active field of research. Early studies attempted to systematically evaluate the relationship between model parameters and transferability in convolutional neural networks (CNNs) [
12]. Recently, several studies have tried to systematically investigate the impact of pre-training [
13,
14], algorithms [
15,
16], and parameters [
8,
17]. Raghu et al. [
18] reported findings on model size in the medical domain. A recent literature review for computer vision (CV) with further information can be found in the dissertation by Plested [
19], among others.
Especially the technical approach to transfer learning in histopathology leaves room for improvement. The most straightforward algorithm consists of copying the pre-trained weights of the initial layers, reinitializing the subsequent layers, and then training them together. This algorithm dates back to the early 2010s [
7,
20,
21]. At present, researchers using pre-trained DL models for digital histopathology can choose from various recently proposed algorithms from CV research [
22,
23,
24]. Some initially successful applications of advanced transfer learning techniques in the medical domain can be found with respect to radiological images [
25,
26,
27,
28] or lung sound analysis [
29]. However, the wider adoption of these techniques in medicine has not occurred to date. For example, “task adaptation,” which transfers a pre-trained model into a supervised target domain [
30], has not been widely explored for histopathology. In general, Sauter et al. [
31] recently noticed a lack of transfer of state-of-the-art DL methodology into the histopathology of malignant melanoma.
On the other hand, choosing a transfer learning algorithm for histopathology can be challenging. It is known that no machine learning (ML) algorithm is generally superior in every conceivable use case [
32]. More specifically, Zhang et al. [
33] noted that, in addition to other factors, the benefit of transfer learning depends on the algorithm due to assumptions made and specific application scenarios. Accordingly, the question arises of whether transfer learning algorithms that perform well on datasets like ImageNet are also suitable for histopathology. In addition, there is the related question of how much each algorithm benefits from more labeled training data. From a practical point of view, gaining only a small increase in accuracy would perhaps speak against the labor- and cost-intensive effort of labeling additional training data by pathology experts [
2]. Systematic evaluations of other technical aspects, such as pre-training [
34,
35] and data augmentation [
36], exist. However, there is—to our knowledge—no systematic comparison of task adaptation strategies applied to histopathological data.
The present study intends to advance the development of DL models for histopathology via improved transfer learning techniques from CV. Based on our findings, follow-up research can enhance their models addressing specific clinical questions and achieve accuracy levels sufficient for clinical approval. This will hopefully help to realize the potential benefits of DL in clinical patient care. Therefore, in our study, we would like to know which task adaptation technique(s) should be used when developing DL models for medical use cases to achieve the best possible accuracy. Consequently, our research question is as follows: which task adaptation technique achieves the highest accuracy (in terms of AUC) for classification in digital histopathology tasks?
Our study has several unique contributions, which can be summarized as follows:
It is the first systematic evaluation of up-to-date task adaptation techniques for classification tasks in digital histopathology.
We comprehensively carried out evaluations based on five histopathological classification tasks (mitotic figure detection, tumor metastasis detection, tumor-infiltrating lymphocytes detection, colorectal cancer tissue-type classification, and skin cancer tissue-type classification) and eight datasets. Our evaluations include external testing and statistical validation.
We show that the standard fine-tuning procedure can be outperformed by more advanced task adaptation techniques depending on the task at hand.
Furthermore, the impact of dataset size is investigated. We show that the Co-Tuning technique can offer further improvements in large-scale settings.
3. Methodology
This section describes the methodology of our study with respect to task adaptation and its various techniques. As shown in
Section 2, task adaptation comprises several closely related research areas as well as a variety of algorithms. Therefore, in this section, we begin with a precise delimitation of our object of investigation based on various criteria. Secondly, we describe an appropriate experimental design to answer our research question for the previously delimited algorithms. Thirdly, our research question itself does not imply a specific DL pipeline architecture. To collect empirical results, however, we must restrict ourselves to a single representative architecture. We therefore define and describe the exemplary DL CNN pipeline used in this study. Finally, we lay out the details regarding its hardware and the software implementation.
3.1. Transfer Learning Techniques
Our study defined a specific application scenario regarding the availability of labeled data: we assumed that there was a small amount of labeled data available. While the amount was enough for supervised model training, it was not sufficient for “training from scratch”. Therefore, we subsequently disregarded the application areas of data efficiency, domain adaptation, and the related areas of few-shot/zero-shot learning. Furthermore, we were interested in training a model with high performance. Thus, we were not interested in runtime performance and disregarded parameter efficiency as well. Our focus was, therefore, on task adaptation, especially catastrophic forgetting and negative transfer. We further narrowed down the use case scenario. We assumed that there was only one model in the source domain (no model zoo) and that the time-consuming pre-training was already carried out (no model selection). The focus was on the target domain; thus, accuracy in the source domain was irrelevant (no continual learning).
The task adaptation techniques to be included in our study were selected according to several criteria. One constraint was to keep the total computation time of our experimental setup feasible with respect to our available computational resources. Model training in DL takes significant time and resources. More specifically, we had to train
m ×
n ×
k models, where
m is the number of techniques to be evaluated,
n is the number of tasks, and
k is the number of training repetitions [
71]. Another motivation was to restrict the findings to procedures that are relevant to practical applications.
For the reasons mentioned above, we defined the following theoretical and practical inclusion criteria:
We required the code availability of a PyTorch/TensorFlow/Keras implementation.
The task adaptation technique was designed for/was suitable to image classification models.
The task adaptation technique was compatible with the ConvNeXt architecture.
The task adaptation technique was designed for high accuracy/AUC (catastrophic forgetting and negative transfer).
The original publication was published in a peer-reviewed journal/conference and holds a good ranking in the SJR (Q1) or CORE ranking (A*, A, B).
Furthermore, the following restrictions were used to reduce the computational complexity of our evaluation:
We dropped L2-PGM and MARS-SP, as MARS-PGM seems to be favored by Gouk et al. [
52].
We dropped LwF in favor of Co-Tuning as the approaches are similar, and You et al. [
22] found that “Co-Tuning fits better for transfer learning than LwF because Co-Tuning explicitly models the category relationship” [
22].
We finally included DELTA, L
2-SP, and MARS-PGM from distance regularization; Bi-Tuning and BSS from feature space regularization; MultiTune and SpotTune from layer routing; and Co-Tuning from shared domains. As a baseline, we chose to train a model using the standard vanilla fine-tuning approach [
7].
3.2. Experimental Design
In our experimental design, we limited ourselves to a subset of histopathological classification tasks and their corresponding public datasets. Then, we distinguished between two aspects: comparing task adaptation algorithms and comparing training dataset sizes, each with respect to their achieved accuracy. The respective evaluation procedures follow the latest best practices for the comparison of ML algorithms to ensure generalizable findings [
71,
72].
3.2.1. Classification Tasks and Datasets
The study focused on histopathology in general and therefore addressed exemplary clinical tasks based on previously published datasets. To cover a wider variety of application scenarios in histopathology, we included five classification tasks from multiple cancer types. We also collected multiple public image datasets for each task. The following describes the classification tasks and the corresponding datasets in detail.
Mitotic Figure Detection. Our first classification task was partly adopted from Tellez et al.’s study [
36]. In this case, the task was to detect mitotic figures in the center of image patches from breast cancer. The predicted variable is binary, namely, “mitotic figure present” vs. “no mitotic figure present” in the center. As our first dataset, we used the “breast cancer histopathological annotation and diagnosis dataset” (BreCaHAD) [
73]. Hematoxylin and eosin (H&E)-stained images with a size of 1360 × 1024 pixels were collected at the University of Calgary. Our second dataset comes from the “TUmor Proliferation Assessment Challenge” in 2016 (TUPAC16) [
74]. More specifically, the “mitosis detection dataset” consists of H&E-stained square images of different sizes. It was gathered from three pathology centers in The Netherlands: the University Medical Center in Utrecht, the Symbiant Pathology Expert Center in Alkmaar, and the Symbiant Pathology Expert Center in Zaandam. The images from the TUPAC16 and the BreCaHAD datasets had a spatial resolution of 0.25 µm/pixel.
Tumor Metastasis Detection. The second classification task was partly adopted from Tellez et al.’s study [
36] as well. The purpose of the classifier was to detect patches containing metastatic tumor cells of breast cancer. Therefore, the binary classes were “metastatic tumor cells present” and “others”. Both the training and test data came from the “CAncer MEtastases in LYmph nOdes challeNge” in 2017 (CAMELYON17) [
75]. The annotated data included H&E-stained whole-slide images (WSIs) from the Radboud University Medical Center (Radboudumc), the Utrecht University Medical Center, the Rijnstate Hospital, the Canisius-Wilhelmina Hospital, and the LabPON in The Netherlands. Model training and external testing were performed on the data from the first, second, third, and fourth centers. Image data from all centers in the CAMELYON17 dataset had a spatial resolution of 0.23–0.25 μm/pixel.
Tumor-infiltrating Lymphocytes (TIL) Detection. The third task was to classify TIL-positive/-negative image patches. In this case, TIL-positive meant that at least two TILs were present in the image [
76,
77,
78]. The dataset included H&E-stained square images with 100 × 100 pixels from 22 cancer types in “The Cancer Genome Atlas” (TCGA) program. We excluded four cancer types (CESC, LUSC, READ, and STAD) as they were annotated using DL. This resulted in a dataset of 18 remaining TCGA projects (ACC, BRCA, COAD, ESCA, HNSC, KIRC, LIHC, LUAD, MESO, OV, PAAD, PRAD, SARC, SKCM, TGCT, THYM, UCEC, and UVM). We obtained two independent, non-overlapping mitosis datasets by randomly dividing cancer types into two splits. The spatial resolution of all image patches in TCGA-TIL was 0.50 µm/pixel.
Colorectal Cancer Tissue-type Classification. Again, based on the study by Tellez et al. [
36], the goal of colorectal cancer (CRC) tissue type classification was to distinguish among six different CRC tissue classes. The classes for classification were “tumor epithelium,” “simple stroma,” “immune cells,” “debris and mucus,” “normal mucosal glands,” and “adipose tissue”. As the first dataset, CRC-5000 by Kather et al. [
79] was used. They provided 5000 patches (625 per class) of archived H&E-stained WSIs from the University Medical Center Mannheim. Each patch had a resolution of 150 × 150 pixels. As the second dataset, the colon cancer dataset (DRCO) from the DROID project was used [
80]. DRCO provided H&E-stained WSIs with annotation masks. To align the classes for CRC-5000 and DRCO, we dropped the “complex stroma” and “background” classes from CRC-5000. The severe imbalance of some classes made it difficult to divide the classes into training and test splits without data leakage. Thus, we excluded two “immune cells” and “debris and mucus” classes for the task “DRCO Small”. The spatial resolution of CRC-5000 was 0.50 µm/pixel, and DRCO used a resolution of 0.45–0.50 µm/pixel.
Skin Cancer Tissue-type Classification. For skin cancer classification, the goal of the classifier was to distinguish among eight different skin tissue types. The tissue classes were “skin appendage” (including hair follicles and sweat glands), “inflammation,” “hypodermis,” “dermis,” “epidermis,” “basal cell carcinoma“ (BCC), “squamous cell carcinoma” (SCC), and “intraepidermal carcinoma” (IEC). We used a dataset that we subsequently referred to as “Queensland” as our first dataset. The dataset was published by Thomas et al. [
81] and provided by MyLab Pathology in Australia. It included segmented H&E-stained WSIs of BCC, SCC, and IEC. As our second dataset, the skin cancer dataset (DRSK) from the DROID project was used [
80]. DRSK provided H&E-stained WSIs with annotation masks. To align the classes of Queensland and DRSK, we dropped the “keratin” and “background” classes with respect to Queensland. Furthermore, “reticular dermis” and “papillary dermis” were combined into “dermis”. The classes “sweat glands” and “hair follicles” were combined into “skin appendage structure”. The severe imbalance of some classes made dividing these classes into training and test splits difficult without causing data leakage. Thus, we dropped three classes, BCC, SCC, and IEC, for the task “DRSK Small”. The spatial resolution of Queensland was 0.67 µm/pixel, while the images in DRSK had a resolution of 0.45–0.50 µm/pixel.
3.2.2. Comparison of the Task Adaptation Techniques
The experimental design for comparing task adaptation techniques was based on the statistical comparison of multiple ML algorithms across multiple datasets, as described by Benavoli et al. [
71]. Generally, for the comparison of
m techniques on
n tasks, model training is repeated
k times for each combination of
m and
n. The mean values of the
k repetitions form the set of observations per algorithm. These means are then statistically compared. For our case with
m = 9 techniques,
n = 12 tasks, and
k = 5 repetitions, this resulted in a set of 9 × 12 × 5 = 540 models in total.
Our experimental design was robust. Multiple tasks were used to evaluate the classifier (see
Table 2). We carried out stratified splitting with respect to the training and validation datasets at the case level to avoid patient-specific data leakage. For a more realistic task adaptation scenario, only a fraction of all data was randomly sampled for training and validation. The proportions were chosen so that the smaller class sizes were roughly in the lower three-digit range, similarly to other experiments on task adaptation [
22,
24,
53]. However, achieving equal dataset sizes among the different tasks was challenging due to varying class imbalances. Training and subsequent testing were repeated five times. We kept the stratified random samples constant across the evaluated algorithms [
72]. For every task, we tested them on independent external test datasets. For computational efficiency, a stratified random subset with a maximum size of 100,000 patches was chosen for all datasets.
The experimental design includes solid statistical validation. The area under the curve (AUC) was chosen as the final measure. It is a standard metric for comparing classification algorithms [
72]. For each combination of tasks and algorithms, the mean and standard deviation of the AUC values were calculated and reported in the Results Section. We used statistical tests to verify the generalizability of our results beyond the tasks in our experimental setup [
82]. The classical null hypothesis significance testing (NHST) procedure was recently criticized for being inappropriate for ML algorithm comparisons. We therefore used Bayesian hypothesis testing instead [
71].
Bayesian testing is preferable because it avoids several shortcomings of classical NHST [
71]. With respect to NHST, decisions are made based on the probability of observing an effect when the actual mean difference of two classifiers,
A and
B, is assumed to be 0 (
H0 hypothesis). Bayesian testing, instead, can directly estimate the probability of
A being better than
B (and vice versa) based on some observations. Furthermore, NHST is based on the unrealistic assumption that two algorithms can potentially have equal performance. Thus, trivial effect sizes can become significant by increasing the number of observations. In Bayesian testing, one can define a region of practical equivalence (ROPE) to take this problem into account beforehand. NHST does not allow drawing conclusions from non-significant results. However, Bayesian testing can show the equivalence of two algorithms based on the ROPE.
In addition to Bayesian testing, we additionally studied effect sizes [
83]. Given means
mA and
mB and the pooled standard deviation,
σpooled, of two normally distributed populations
A and
B, Cohen’s
d is calculated as follows [
84]:
We omitted effect sizes in the pairwise comparison based on multiple datasets in
Section 4.1. In this case, the denominator in Equation (1) reflects the classification tasks’ difficulty and not the model’s performance dispersion. As a nonparametric variant of Cohen’s d for populations without a normal distribution, we adopted Akinshin’s gamma from the Python package “Autorank” [
85].
3.2.3. Comparison of Training Dataset Sizes
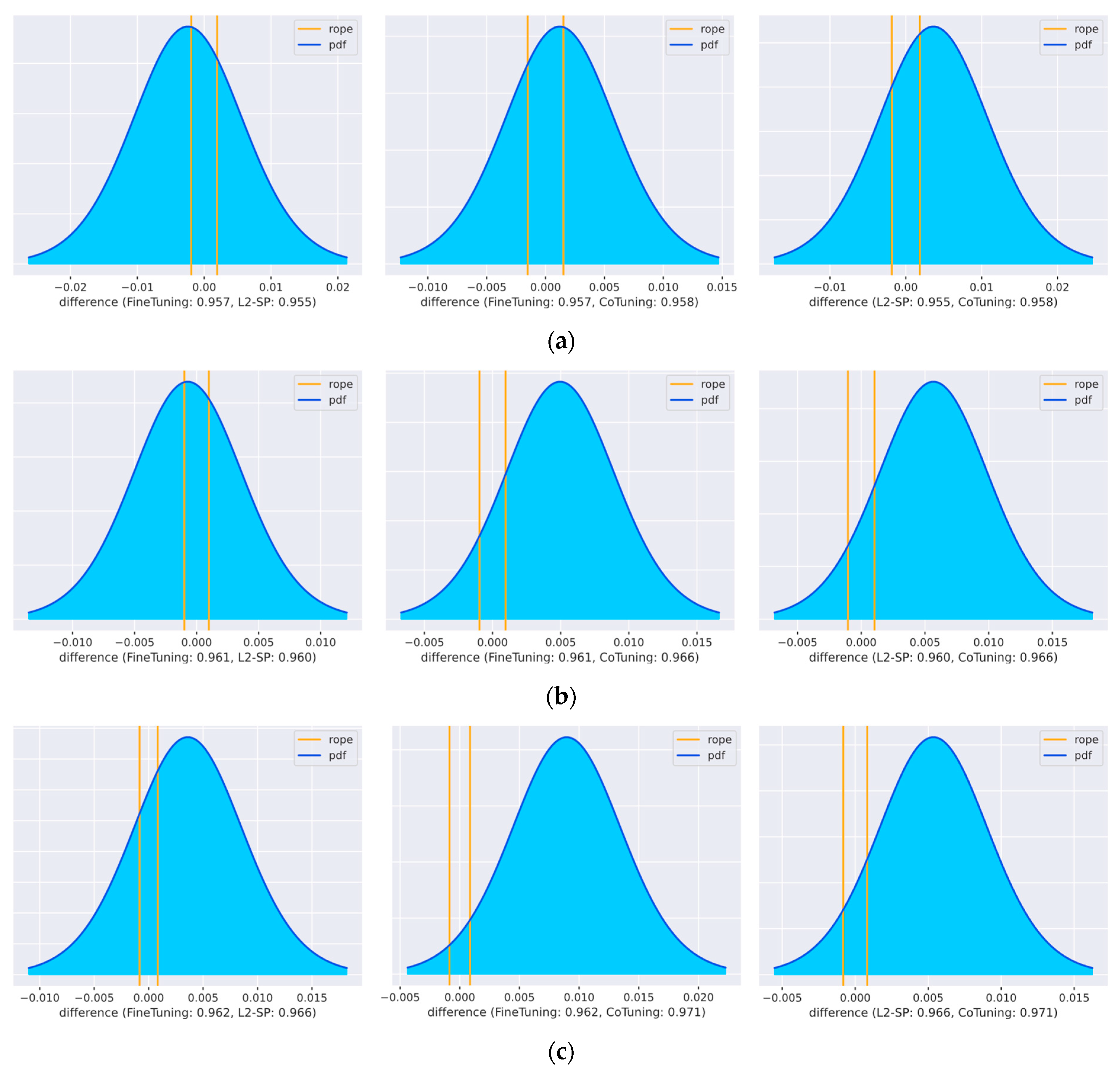
We were also interested in how the performance of the techniques under investigation would behave with a larger amount of data. The performance gain of vanilla fine-tuning is known to become saturated with the increase in the dataset size [
8]. Some recent studies on advanced task adaptation techniques carried out comparisons with respect to the training datasets’ size. Some techniques performed better in a large-scale setting with more than 1000 images per class, while others did not [
22,
54,
56]. To evaluate the impact of dataset sizes in histopathology, we chose to re-evaluate some of the task adaptation techniques using additional experiments.
The experimental design for comparing multiple dataset sizes was based on the statistical comparison of two algorithms using one dataset, as described by Benavoli et al. [
71]. In this case, we compared an algorithm trained on a small training split vs. trained on a larger training split of the same dataset. Therefore, for each combination of
m techniques and
n training splits,
k repetitions of the model training were performed. We chose three techniques (L
2-SP, fine-tuning, and Co-Tuning) from our set of task adaptation algorithms. We re-used dataset #10 from the colorectal cancer tissue-type classification (see
Table 2). Three variants of the training split were generated using a varying subsampling factor: “Base,” “Large,” and “XLarge” (see
Table 3). Base is equivalent to the size shown in
Table 2. For every combination of split size and learning technique, we trained
k = 20 models. With
m = 3 techniques,
n = 3 training split variants, and
k = 20 repetitions, this resulted in a set of 3 × 3 × 20 = 180 models in total. Again, the AUC was chosen as our metric for comparisons. We verified the results using the Bayesian correlated
t-test. We further report effect sizes in the Results Section.
3.3. Exemplary Image Classification Pipeline
A DL pipeline for image classification typically includes multiple sequential components. Each component comes with its own set of design choices. Here, we limited ourselves to a single exemplary DL pipeline, used for all experiments in this study. The core building blocks are typically image preprocessing and the neural network itself. We first started with the definition of our image preprocessing. We then described the state-of-the-art CNN architecture. Finally, we addressed the hyperparameters of our pipeline.
3.3.1. Image Preprocessing
Neural networks for image processing typically operate on smaller image patches. Therefore, we used square image patches with a default edge length of 128 pixels. The spatial resolution of the training and testing images were identical for all tasks, except for the skin cancer tissue-type classification. To account for the different spatial resolutions in the latter case, we used patch sizes with an edge length of 128 × (0.67/0.5) = 171 pixels for the DRSK dataset. Subsequently, all patches were scaled uniformly to 224 × 224 pixels, the default input image size of ConvNeXt.
Inequalities in class distribution must be considered when training representation learning algorithms. An imbalance of varying magnitudes is present in all datasets, except for CRC5000. Oversampled minority classes were therefore used during training time. This prevents the classifier from becoming biased towards one or more specific class(es). For validation and testing, we kept the original class distribution of the datasets.
When training neural networks for image processing, increasing the training set size using various image augmentations is common. We used random horizontal flipping. Following the advice of Tellez et al. [
36], we further used HSV color augmentation. As commonly carried out in PyTorch, the mean and standard deviation were normalized to [0.485, 0.456, 0.406] and [0.229, 0.224, 0.225] in the RGB color space, respectively.
3.3.2. CNN Architecture
Since the development of AlexNet in 2012 [
86], neural networks based on convolution have been widely used as a standard approach for CV. Research has constantly improved network architecture around this fundamental principle, including architectural milestones like VGG [
87], ResNet [
88], Xception [
89], or EfficientNet [
90]. Recently, transformer-based architectures from natural language processing (NLP) challenged the use of convolution operations [
91,
92,
93,
94]. Other than CNNs, they are based on multi-head self-attention.
Whether to use convolution- or attention-based architectures for CV remains an open research question. Liu et al. [
95] found that transformers gain importance compared to CNNs mainly because of their better scaling behavior. However, recent studies showed that CNNs can still achieve an equivalent or better performance than transformers after optimizing several design choices. One of these options is larger kernel sizes [
95,
96,
97,
98,
99]. This raises the question of whether the recent performance of transformer models is really a result of multi-head self-attention. At the same time, the established convolution approach was designed around efficiency [
95]. The new design principle of choosing large kernels also seems to push the networks to shift towards a shape bias [
97]. This effect resolves a previous issue of pre-trained CNNs being biased due to texture [
100].
For this study, we chose a state-of-the-art CNN architecture. As described above, the newest architectures achieved cutting-edge performance for image classification. Furthermore, most task adaptation techniques were explicitly designed for CNNs. Therefore, we selected ConvNeXt [
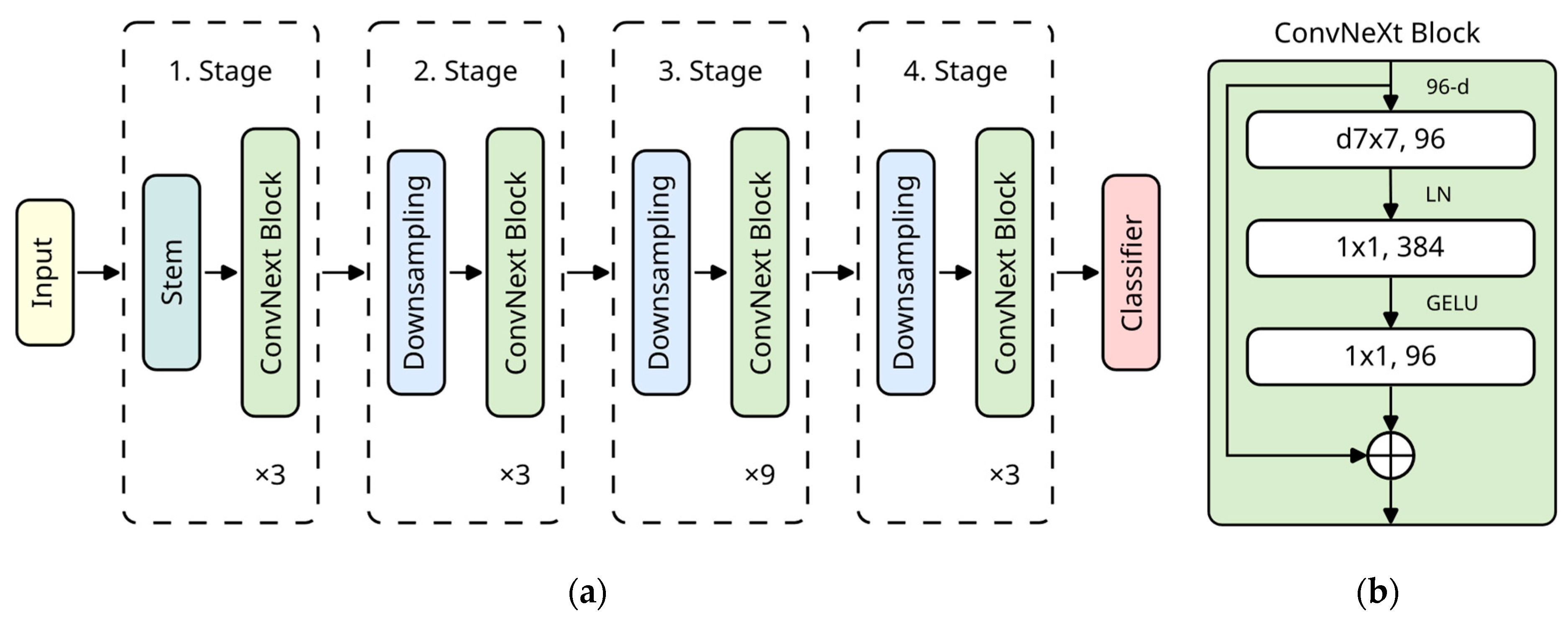
95] to represent novel CNN architectures. We chose the smallest one, “ConvNeXt Tiny”, as we assumed it had enough capacity for our tasks while keeping training time manageable. The building blocks are shown in
Figure 3. The pre-trained ImageNet weights originate from PyTorch [
101]. We transferred these weights to all used models. Furthermore, we ensured that all implementations (both in PyTorch and Keras) produced the same model results (excluding rounding errors) so that different model implementations do not distort the results.
3.3.3. Hyperparameters
For our experiments, we preferred the stochastic gradient descent (SGD) optimizer over Adam, another common choice, based on two reasons. First, SGD generalizes better for image classification [
102]. The performance of Adam seems to depend on the hyperparameter optimization of decay rates
β1 and
β2. Second, Adam was only used in the study performed by Gouk et al. [
52]. The authors of the other studies used SGD. From a practical perspective, this results in less implementation effort when using SGD as well.
Figure 3.
(
a) Overall architecture of ConvNeXt Tiny [
95]. The macro-level architecture with a ratio of 1:1:3:1 was adopted from Swin Transformers [
103]. (
b) Detailed structure of the ConvNeXt block [
95]. The micro-level architecture is based on the inverted bottleneck design [
104]. Inspired by transformer architectures, the depthwise convolution uses a kernel size of 7 × 7. For computational efficiency, it is computed before the expansion of channels. The Gaussian error linear unit (GELU) [
105] and layer normalization [
106] were also adopted from recent transformer-based architectures [
95]. Adapted from Jiang et al. [
107], © 2023, Frontiers Media S.A., CC BY 4.0 DEED.
Figure 3.
(
a) Overall architecture of ConvNeXt Tiny [
95]. The macro-level architecture with a ratio of 1:1:3:1 was adopted from Swin Transformers [
103]. (
b) Detailed structure of the ConvNeXt block [
95]. The micro-level architecture is based on the inverted bottleneck design [
104]. Inspired by transformer architectures, the depthwise convolution uses a kernel size of 7 × 7. For computational efficiency, it is computed before the expansion of channels. The Gaussian error linear unit (GELU) [
105] and layer normalization [
106] were also adopted from recent transformer-based architectures [
95]. Adapted from Jiang et al. [
107], © 2023, Frontiers Media S.A., CC BY 4.0 DEED.
Broadly consistent parameters for SGD were used across the algorithms. In line with recent recommendations on fine-tuning in dissimilar source and target domains [
17], we used SGD with an initial learning rate of 0.01 and a momentum of 0.9 and activated Nesterov. We adopted step decay from the majority of the evaluated studies [
7,
23,
24,
52,
53,
54,
61]. We consistently set a milestone at the 10th epoch with a gamma value of 0.1. We used a lower learning rate for the pre-trained layers in the fine-tuning setting. We also adopted lower learning rates for the policy network of SpotTune and the last block in MultiTune from the original studies [
23,
61]. To avoid exploding gradients, we applied gradient value clipping with a value of 1.0. We chose to use a batch size of 64. During training, iterations per epoch were fixed to 128. Models were trained with early stopping, patience of 15, and a maximum number of 100 epochs.
Various hyperparameters must be specified for the algorithms under study. However, conducting hyperparameter optimization to identify optimal parameters for each task on each dataset was not practical due to the substantial computation time required. Instead, we utilized the suggested values from the literature. Li et al. [
108] recently found that L
2-SP is robust across different parameter ranges. However,
α >
β is generally preferable. Therefore, we extracted
α = 0.1 and
β = 0.01 from the original study [
24]. For MARS-PGM, we used the average values of
γj = 9.32 and
γL = 16.89 [
52]. For DELTA, Li et al. [
53] fixed
β = 0.01 and generally used it for all conditions [
53]. We further set
α = 0.04. For Co-Tuning, You et al. [
22] found that
λ = 2.3 is robust across all tested datasets and sampling rates. For BSS, Chen et al. [
55] found that
η = 0.001 and
k = 1 are generally adequate settings under varying conditions. Zhong et al. [
54] universally applied
τ = 0.07 and
m = 0.999 for Bi-Tuning. For a 100% sampling rate, which is comparable to our class size, they found that a high number of sampling keys,
K, is beneficial. In line with this, we therefore used the following defaults:
K = 40 and
L = 128. Wang et al. [
61] used
α = 0.01 and
β = 0.01 for all their experiments.
For our baseline, vanilla fine-tuning, the model weights were copied from the ImageNet domain. The pre-trained classification head was replaced by a new one with randomly initialized weights. The new model was then fine-tuned on the histopathology tasks. For the pre-trained feature extractor, a smaller learning rate (10 times smaller) was chosen.
3.4. Hardware and Software Implementation
Our server had two AMD EPYC 7402 24-core processors, one terabyte of random-access memory, NVIDIA RTX A6000 graphics cards, and an Ubuntu 22.04 LTS operating system. Various freely available software packages were used. All experiments were implemented in Python (ver. 3.9.16). WSI data were loaded using the Python package tiffslide (ver. 2.1.2) [
109]. Model training was implemented using PyTorch (ver. 2.0.0) [
110], Keras (ver. 2.11.0), and TensorFlow (ver. 2.11.1) [
111] libraries. For PyTorch training, image augmentations used the Transforms module from Torchvision (ver. 0.15.1). For Keras training, the same augmentations were implemented using the Albumentations (ver. 1.3.0) package [
112]. For the calculation of general sample statistics and the Bayesian signed-rank test, we used the implementation of the Python package Autorank (ver. 1.2.0) [
85]. The Bayesian correlated
t-test was calculated using baycomp (ver. 1.0.3) [
113].
5. Discussion
To the best of our knowledge, we are the first to provide extensive empirical evidence on advanced task adaptation techniques in histopathology. We showed that specific techniques are, on average, better suited for histopathology than others. Interestingly, the authors of all methods investigated in this paper observed, in their original studies, an improvement of their method over the baseline fine-tuning technique [
23,
52,
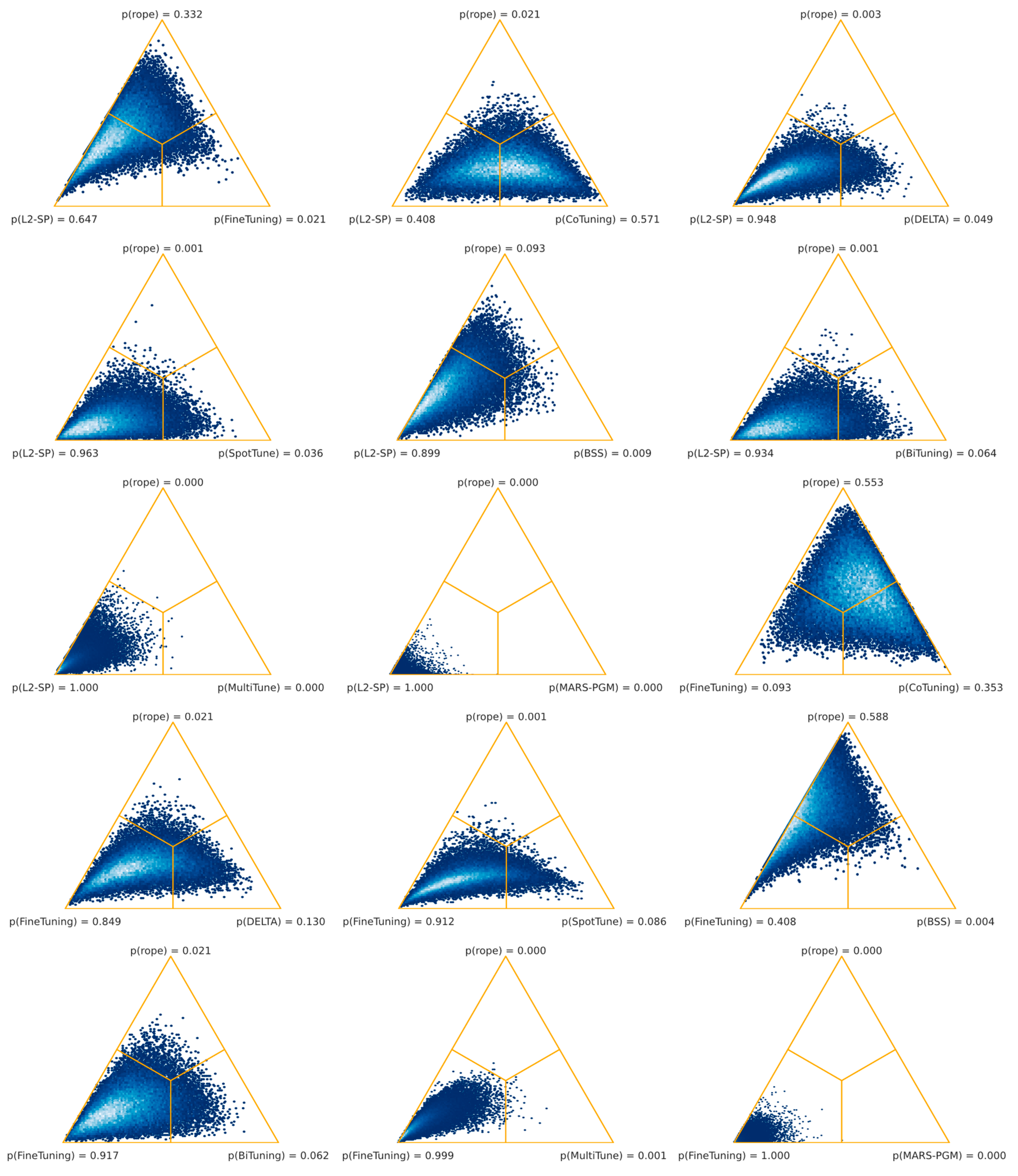
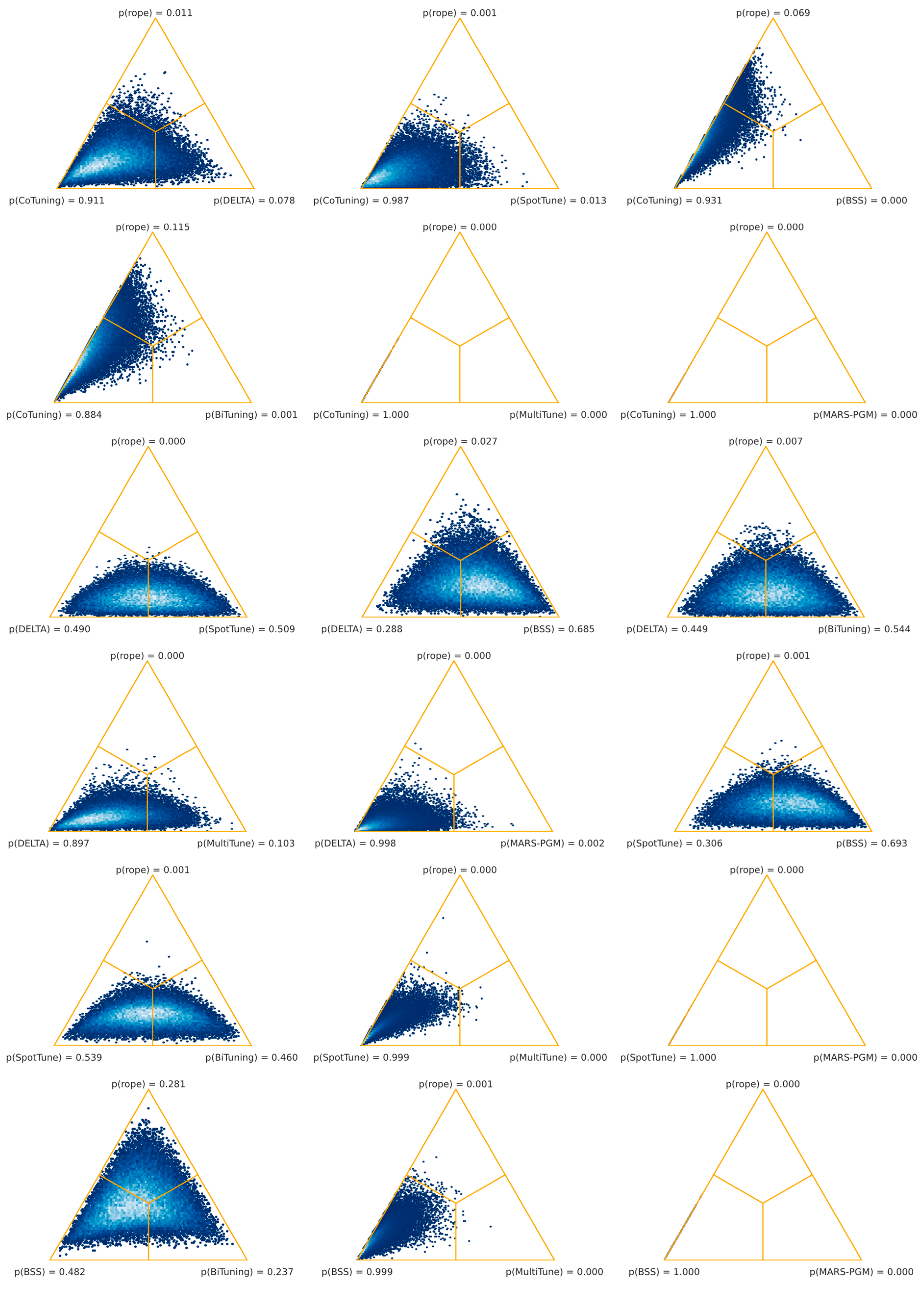
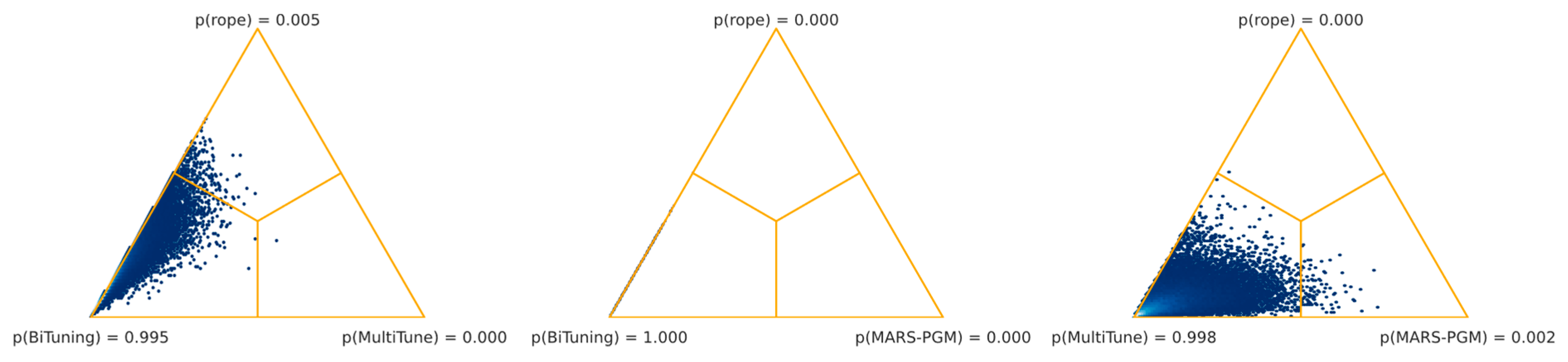
54]. However, as shown in
Table 6, this is not confirmed by our results for the histopathology setting. Vanilla fine-tuning significantly outperformed SpotTune, Bi-Tuning, MultiTune, and MARS-PGM. Three task adaptation techniques combined, namely, L
2-SP, vanilla fine-tuning, and Co-Tuning, out-performed the others regarding classification tasks in digital histopathology. Among the three top-performing algorithms were distance regularization (L
2-SP), shared domains (Co-Tuning), and the baseline technique (fine-tuning). Moreover, the worst-performing algorithms (MultiTune and MARS-PGM) were significantly outperformed by almost all other techniques, whereas MARS-PGM was significantly worse than MultiTune.
However, our results reveal that the superiority of an algorithm in histopathology depends on the task. Our systematic comparison, summarized in
Table 6, did not show a general superiority of one task adaptation technique over the others. Also, none of the evaluated algorithms showed performances that were significantly superior to our baseline. However, the comparison between the three highest-ranking algorithms (L
2-SP, fine-tuning, and Co-Tuning) was inconclusive as well. At first glance, this might be either due to our experimental setup, or performances might further depend on other influencing factors. Zhang et al. [
33] already mentioned that, in addition to domain divergence and source/target data quality, the fit between the TL algorithm and the task is essential. Our task-level analysis confirmed this for histopathology (see
Table 7). At least one technique obtained significantly better results than the baseline in about half of the cases.
Advanced task adaptation algorithms can also be helpful when a large amount of data are available. Our comparison of the dataset’s size showed that, by increasing the dataset size by a factor of ten, Co-Tuning was able to outperform the baseline technique significantly. This is in line with the results of You et al. [
22]. They found that additional supervision by Co-Tuning helps to further increase performances in large-scale classification settings where regularization techniques, like L
2-SP, do not help. Two other methods, which we did not examine in this large-scale setting, reported related results: the study on Bi-Tuning also reported a superior performance [
54], while the study on StochNorm [
56] obtained mixed findings.
The architecture of a neural network might be an influencing factor for the effectiveness of task adaptation. By choosing ConvNeXt, our investigation was based on an up-to-date CNN architecture from the field of CV. In general, ImageNet performance is a good indicator of the accuracy of CNN architectures relative to a downstream task [
8]. However, Ding et al. [
97] recently showed that CNNs with larger kernel sizes (like ConvNeXt) have higher shape biases compared to texture biases. In general research on CV, this is generally seen as beneficial due to its similarity with human perception [
97,
100,
115]. While this might be true for photograph-like image data, histopathological analysis at a high magnification relies on relatively small cell structures and the visual texture of the tissue for decisions.
The algorithm-specific influence of histopathology pre-training needs to be further clarified. Image features from ImageNet and histopathology are likely to be different, and domain divergence is assumed to affect the result negatively [
33]. For vanilla fine-tuning, three studies found that histopathology benefits from domain-specific pre-training [
34,
35,
116], while two did not [
117,
118]. The question arises with respect to how advanced task adaptation techniques benefit from domain-specific pretraining and whether this effect diverges between algorithms.
Our results are in line with other medical studies. Several authors already provided evidence for the usefulness of advanced task adaptation techniques in a broader medical context. Most examples can be found in radiology. Liao et al. [
25] used L
2-SP to avoid catastrophic forgetting in a multitask setting called “MUSCLE”. They found that L
2-SP was beneficial in all tested use cases. Sagie et al. [
26] evaluated L
2-SP in a supervised task adaptation setting for segmentation. L
2-SP was not beneficial for ImageNet pre-training. However, it improved performances with respect to medical pre-training. Su et al. [
27] successfully used L
2-SP to carry out the supervised adaptation of an MRI-pre-trained GAN relative to CT image generation. Unfortunately, they did not compare their results to vanilla fine-tuning. An et al. [
28] evaluated SpotTune in a supervised task adaptation setting for segmentation, which was pre-trained on a related medical dataset. SpotTune performed better than vanilla fine-tuning. Another example can be found in lung sound classification. Nguyen and Pernkopf [
29] evaluated Co-Tuning, Stochastic Normalization, and a combination of both for supervised task adaptation with ImageNet pre-training. While Co-Tuning and StochNorm were superior to vanilla fine-tuning, their performance varied between classification tasks.
Our results add empirical evidence to CV research for the general relationship between domain divergence and negative transfer. The findings are based on eight techniques applied to the medical domain. We showed that the relative effectiveness of task adaptation techniques in histopathology diverges from general CV research. This observation confirms the assumption that performance depends on the domain [
33]. Our findings might have implications for evaluating new transfer learning techniques. Increasing the variety in benchmark tasks has been proposed before [
14,
19]. In this paper, we confirmed that the evaluation of commonly used datasets containing animals, cars, aircraft, plants, etc., is insufficient.
However, the findings on some techniques also deviate from earlier findings in CV research in some ways. First, Li et al. [
17] reported that distance regularization does not perform well for dissimilar source and target domains. Plested et al. [
16] thus suggested that distance regularization should be limited to earlier layers, and the number of layers depends on the downstream dataset. Surprisingly, regular L
2-SP distance regularization performed comparatively well in histopathology. Second, our results also contradict the conclusion of Gouk et al. [
52], who reported that hard constraints perform better than distance constraints using the penalty term. MARS-PGM performed the worst of the algorithms compared (see
Table 6).
6. Conclusions
This study is the first to provide empirical evidence on the suitability of up-to-date task adaptation techniques for histopathological classification tasks using CNNs. Analysis at the task level showed that, in half of the cases, one or multiple techniques, including Bi-Tuning, Co-Tuning, DELTA, L2-SP, and SpotTune, could outperform vanilla fine-tuning, which is the standard procedure for task adaptation. Co-Tuning can further increase performance when a large amount of data in the target domain are available. Our results align with earlier studies mainly from the radiological field, showing that medical AI can benefit from advanced task adaptation techniques. We further provided evidence on the general relationship between domain divergence and negative transfer. In this paper, we found some deviations from the literature on distance regularization.
6.1. Limitations
Our study was limited to supervised task adaptation techniques and excluded related research areas, like domain adaptation and zero-, one-, and few-shot learning. As we narrowed down the evaluated task adaptation techniques to the most promising ones in
Section 3.1 for theoretical and practical reasons, our study did not cover 100% of all available methods from the literature. Furthermore, we limited our study to techniques that were initially designed for image classification. Our experiments did not combine BSS with distance regularization using L
2-SP or DELTA. Although the classification tasks in
Section 3.2 were based on 12 different datasets covering various cancer types and classification tasks, not all possible scenarios from histopathology were fully covered.
While we demonstrated relative improvements over the baseline approach (fine-tuning) for several tasks, our results do not show the best possible performance improvement due to the substantial computational costs required in our experimental setup for extensive hyperparameter optimization. We did not further investigate performances under varying influencing factors beyond dataset size—like different source domains, unsupervised pre-training, and the network’s architecture and size—or other downstream ML tasks—like instance segmentation. In our experiments, the number of layers using distance regularization was not adjusted relative to downstream tasks.
6.2. Outlook
This relationship between the histopathological task and the suitability of task adaptation techniques needs to be better understood in the future for task adaptation to be used in a targeted manner. More generally, the differentiated evaluation of transfer learning algorithms should continue so that more domains beyond ImageNet-like photographs can benefit more from task adaptation. Future studies should examine the impact of pre-training in pathology on task adaptation outcomes. More techniques, including Bi-Tuning [
54] and StochNorm [
56], should be investigated concerning their behavior in a large-scale setting.