The Performance of a Deep Learning-Based Automatic Measurement Model for Measuring the Cardiothoracic Ratio on Chest Radiographs
Abstract
:1. Introduction
2. Materials and Methods
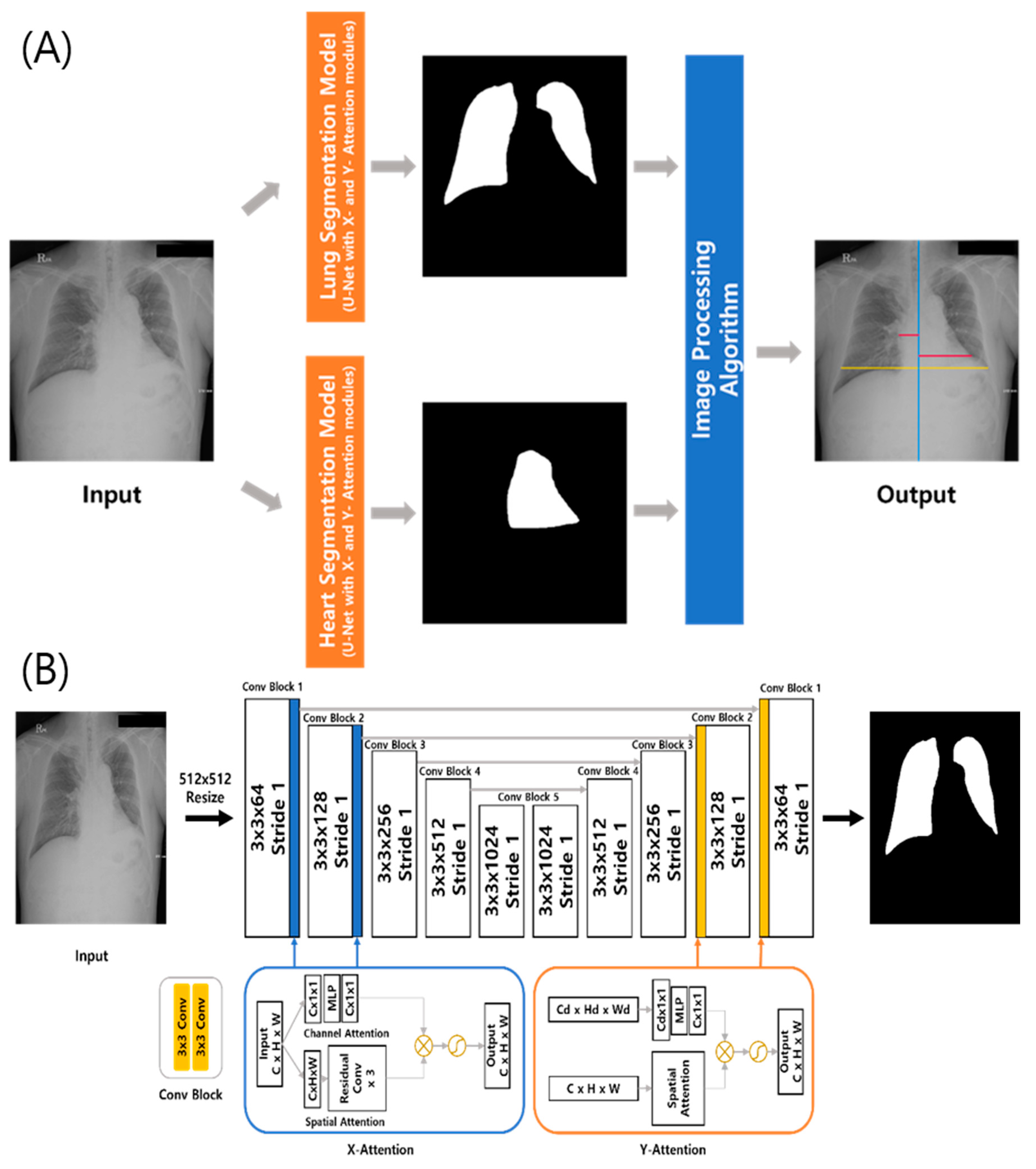
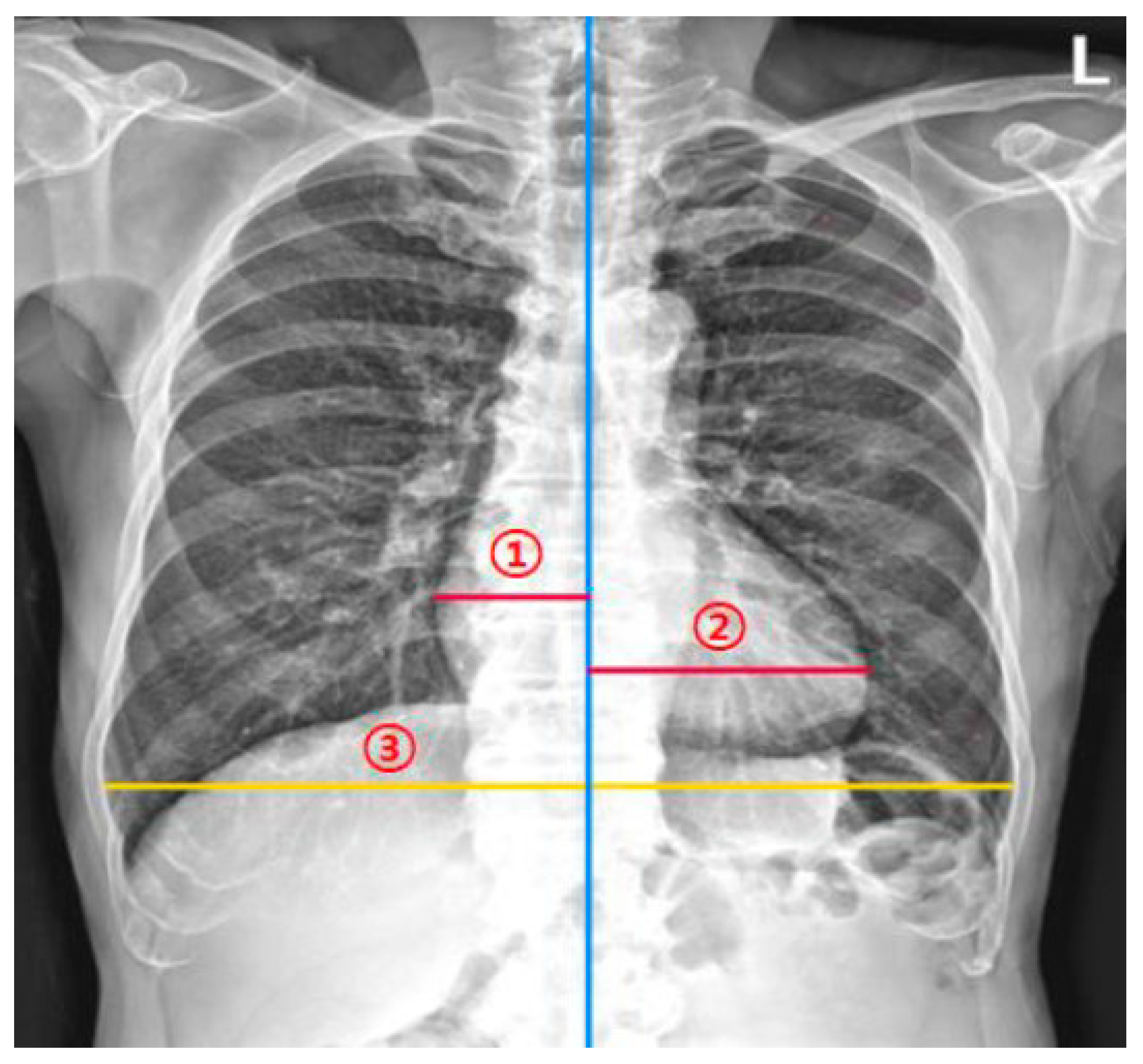
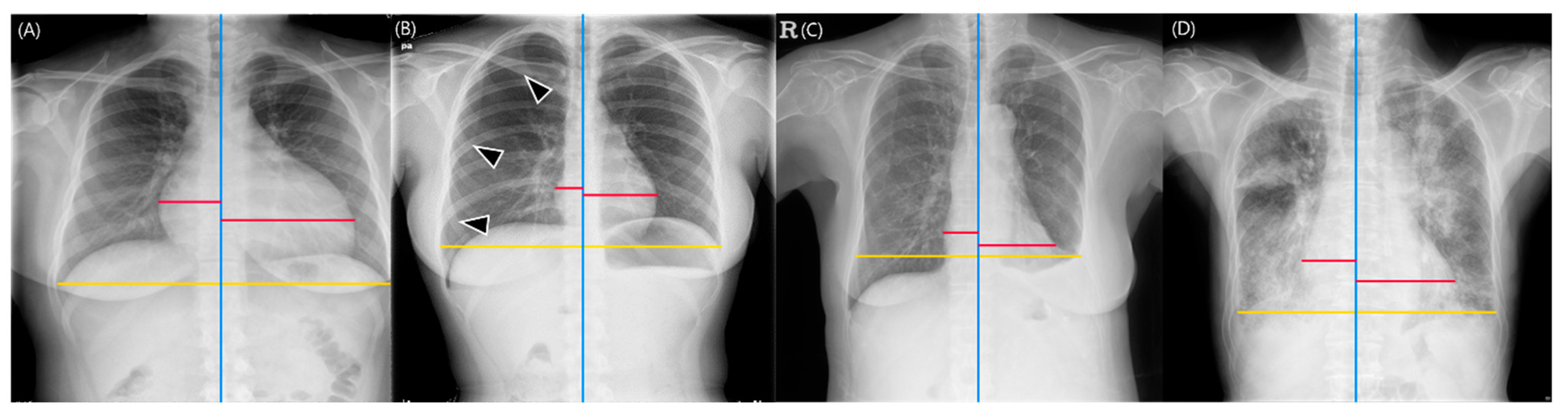
2.1. DL-Based Model Measuring the CTR on Chest Radiographs
2.2. Study Sample
2.3. Measurement of the CTR by Thoracic Radiologists for Agreement Analyses
2.4. Reader Tests
2.5. Statistical Analysis
3. Results
3.1. Study Sample and CTR
3.2. Agreement Analyses
3.3. Reader Tests
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American College of Radiology. ACR Standard for the Performance of Pediatric and Adult Chest Radiography; American College of Radiology: Reston, VA, USA, 2001. [Google Scholar]
- Speets, A.M.; van der Graaf, Y.; Hoes, A.W.; Kalmijn, S.; Sachs, A.P.; Rutten, M.J.; Gratama, J.W.C.; van Swijndregt, A.D.M.; Mali, W.P. Chest radiography in general practice: Indications, diagnostic yield and consequences for patient management. Br. J. Gen. Pract. 2006, 56, 574–578. [Google Scholar] [PubMed]
- Simkus, P.; Gutierrez Gimeno, M.; Banisauskaite, A.; Noreikaite, J.; McCreavy, D.; Penha, D.; Arzanauskaite, M. Limitations of cardiothoracic ratio derived from chest radiographs to predict real heart size: Comparison with magnetic resonance imaging. Insights Imaging 2021, 12, 158. [Google Scholar] [CrossRef] [PubMed]
- Truszkiewicz, K.; Poręba, R.; Gać, P. Radiological cardiothoracic ratio in evidence-based medicine. J. Clin. Med. 2021, 10, 2016. [Google Scholar] [CrossRef]
- Ajmera, P.; Kharat, A.; Gupte, T.; Pant, R.; Kulkarni, V.; Duddalwar, V.; Lamghare, P. Observer performance evaluation of the feasibility of a deep learning model to detect cardiomegaly on chest radiographs. Acta Radiol. Open 2022, 11, 20584601221107345. [Google Scholar] [CrossRef] [PubMed]
- Yotsueda, R.; Taniguchi, M.; Tanaka, S.; Eriguchi, M.; Fujisaki, K.; Torisu, K.; Masutani, K.; Hirakata, H.; Kitazono, T.; Tsuruya, K. Cardiothoracic ratio and all-cause mortality and cardiovascular disease events in hemodialysis patients: The Q-cohort study. Am. J. Kindey Dis. 2017, 70, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.-J.; Wu, I.-W.; Hsu, K.-H.; Sun, C.-Y.; Chen, C.-Y.; Lee, C.-C. Vitamin D deficiency, cardiothoracic ratio, and long-term mortality in hemodialysis patients. Sci. Rep. 2020, 10, 7533. [Google Scholar] [CrossRef]
- Ito, K.; Ookawara, S.; Ueda, Y.; Miyazawa, H.; Yamada, H.; Goto, S.; Ishii, H.; Shindo, M.; Kitano, T.; Hirai, K.; et al. A higher cardiothoracic ratio is associated with 2-year mortality after hemodialysis initiation. Nephron Extra 2015, 5, 100–110. [Google Scholar] [CrossRef]
- Jiang, L.; Chen, W.-G.; Geng, Q.-S.; Du, G.; He, P.-C.; Feng, D.; Qin, T.-H.; Wei, X.-B. The cardiothoracic ratio: A neglected preoperative risk-stratified method for patients with rheumatic heart disease undergoing valve replacement surgery. Eur. J. Cardiothorac. Surg. 2019, 55, 511–517. [Google Scholar] [CrossRef]
- Winder, M.; Owczarek, A.J.; Chudek, J.; Pilch-Kowalczyk, J.; Baron, J. Are we overdoing it? Changes in diagnostic imaging workload during the years 2010–2020 including the impact of the SARS-CoV-2 pandemic. Healthcare 2021, 9, 1557. [Google Scholar] [CrossRef]
- Huh, J.E.; Lee, J.H.; Hwang, E.J.; Park, C.M. Effects of Expert-Determined Reference Standards in Evaluating the Diagnostic Performance of a Deep Learning Model: A Malignant Lung Nodule Detection Task on Chest Radiographs. Korean J. Radiol. 2023, 24, 155–165. [Google Scholar] [CrossRef]
- Gupte, T.; Niljikar, M.; Gawali, M.; Kulkarni, V.; Kharat, A.; Pant, A. Deep learning models for calculation of cardiothoracic ratio from chest radiographs for assisted diagnosis of cardiomegaly. In Proceedings of the 2021 International Conference on Artificial Intelligence, Big Data, Computing and Data Communication Systems (icABCD), Durban, South Africa, 5–6 August 2021; IEEE: Piscataway, NJ, USA, 2021. [Google Scholar] [CrossRef]
- Li, Z.; Hou, Z.; Chen, C.; Hao, Z.; An, Y.; Liang, S.; Lu, B. Automatic cardiothoracic ratio calculation with deep learning. IEEE Access 2019, 7, 37749–37756. [Google Scholar] [CrossRef]
- Lee, M.S.; Kim, Y.S.; Kim, M.; Usman, M.; Byon, S.S.; Kim, S.H.; Lee, B.I.; Lee, B.D. Evaluation of the feasibility of explainable computer-aided detection of cardiomegaly on chest radiographs using deep learning. Sci. Rep. 2021, 11, 16885. [Google Scholar] [CrossRef] [PubMed]
- Saiviroonporn, P.; Rodbangyang, K.; Tongdee, T.; Chaisangmongkon, W.; Yodprom, P.; Siriapisith, T.; Wonglaksanapimon, S.; Thiravit, P. Cardiothoracic ratio measurement using artificial intelligence: Observer and method validation studies. BMC Med. Imaging 2021, 21, 95. [Google Scholar] [CrossRef]
- Bercean, B.; Iarca, S.; Tenescu, A.; Avramescu, C.; Fuicu, S. Assisting radiologists through automatic cardiothoracic ratio calculation. In Proceedings of the 2020 IEEE 14th International Symposium on Applied Computational Intelligence and Informatics (SACI), Timisoara, Romania, 21–23 May 2020; IEEE: Piscataway, NJ, USA, 2020. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Kim, M.; Lee, B.-D. Automatic lung segmentation on chest X-rays using self-attention deep neural network. Sensors 2021, 21, 369. [Google Scholar] [CrossRef] [PubMed]
- Dawid, A.P.; Skene, A.M. Maximum likelihood estimation of observer error-rates using the EM algorithm. J. R. Stat. Soc. Ser. C Appl. Stat. 1979, 28, 20–28. [Google Scholar] [CrossRef]
- Seah, J.C.; Tang, C.H.; Buchlak, Q.D.; Holt, X.G.; Wardman, J.B.; Aimoldin, A.; Esmaili, N.; Ahmad, H.; Pham, H.; Lambert, J.F.; et al. Effect of a comprehensive deep-learning model on the accuracy of chest X-ray interpretation by radiologists: A retrospective, multireader multicase study. Lancet Digit. Health 2021, 3, e496–e506. [Google Scholar] [CrossRef]
- Digitalis Investigation Group. The effect of digoxin on mortality and morbidity in patients with heart failure. N. Engl. J. Med. 1997, 336, 525–533. [Google Scholar] [CrossRef]
- Petrie, M.C. It cannot be cardiac failure because the heart is not enlarged on the chest X-ray. Eur. J. Heart Fail. 2003, 5, 117–119. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Agreement between methods of measurement with multiple observations per individual. J. Biopharm. Stat. 2007, 17, 571–582. [Google Scholar] [CrossRef]
- Zou, G. Confidence interval estimation for the Bland–Altman limits of agreement with multiple observations per individual. Stat. Methods Med. Res. 2013, 22, 630–642. [Google Scholar] [CrossRef]
- Wang, L.; Keen, K.J.; Holland, B. Estimation of reliability in a three-factor model. Stat. Med. 2011, 30, 1254–1265. [Google Scholar] [CrossRef] [PubMed]
- Reaching a Consensus in Crowdsourced Data Using the Dawid-Skene Model. Camilleri MPJ. Web Site. Available online: https://michaelpjcamilleri.wordpress.com/2020/06/22/reaching-a-consensus-in-crowdsourced-data-using-the-dawid-skene-model/ (accessed on 23 March 2023).
- Lee, J.H.; Park, S.; Hwang, E.J.; Goo, J.M.; Lee, W.Y.; Lee, S.; Kim, H.; Andrews, J.R.; Park, C.M. Deep learning-based automated detection algorithm for active pulmonary tuberculosis on chest radiographs: Diagnostic performance in systematic screening of asymptomatic individuals. Eur. Radiol. 2021, 31, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Sun, H.Y.; Park, S.; Kim, H.; Hwang, E.J.; Goo, J.M.; Park, C.M. Performance of a Deep Learning Algorithm Compared with Radiologic Interpretation for Lung Cancer Detection on Chest Radiographs in a Health Screening Population. Radiology 2020, 297, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Han, K.; Jang, H.Y.; Park, J.E.; Lee, J.; Kim, D.W.; Choi, J. Methods for Clinical Evaluation of Artificial Intelligence Algorithms for Medical Diagnosis. Radiology 2023, 306, 20–31. [Google Scholar] [CrossRef]
- Park, S.H.; Han, K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology 2018, 286, 800–809. [Google Scholar] [CrossRef]
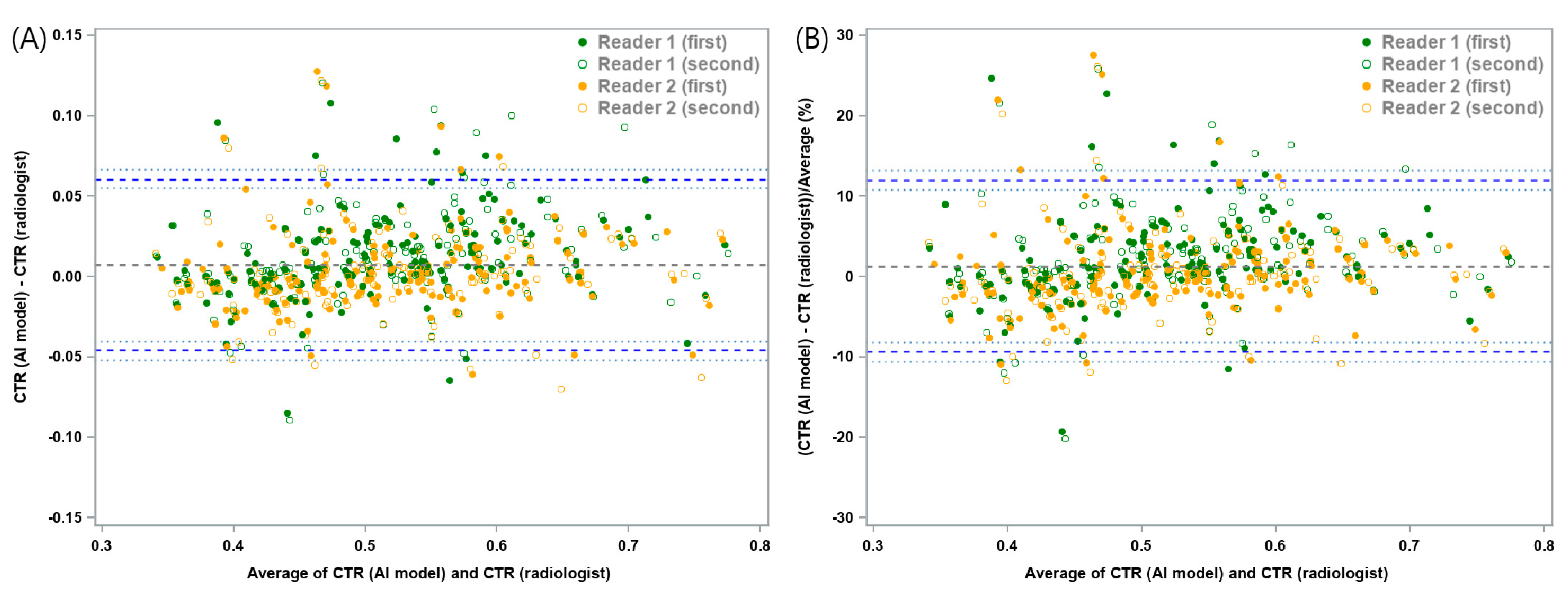
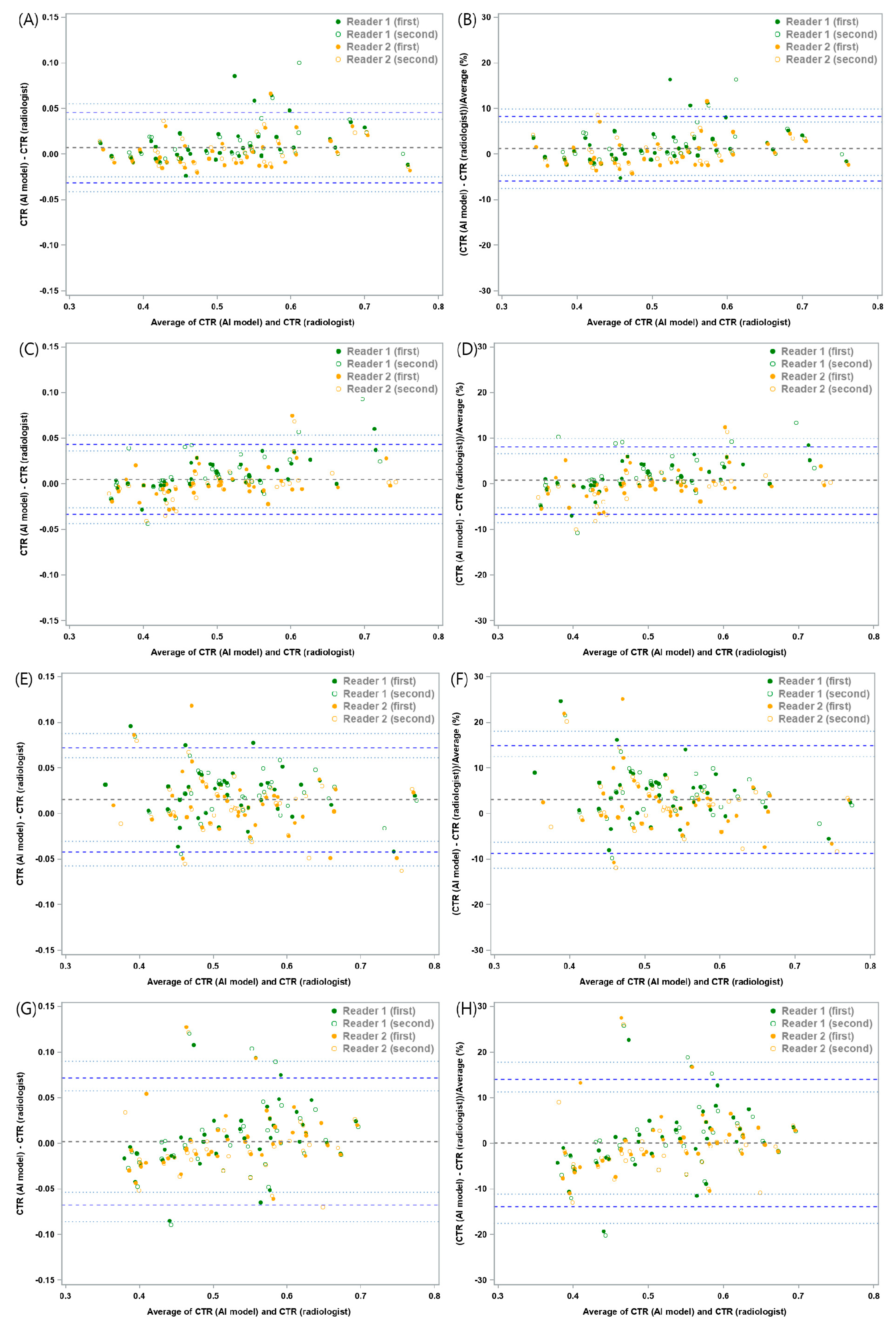
| Mean Absolute Difference | Lower LOA (95% CI) | Upper LOA (95% CI) | |
| Study sample (n = 160) | 0.0074 | −0.0457 (−0.052, −0.0403) | 0.0605 (0.0551, 0.0668) |
| Chest radiographs without any lung or pleural abnormality (n = 40) | 0.0071 | −0.0315 (−0.041, −0.0245) | 0.0458 (0.0387, 0.0553) |
| Chest radiographs with pneumothorax (n = 40) | 0.0051 | −0.0334 (−0.0432, −0.0262) | 0.0436 (0.0363, 0.0533) |
| Chest radiographs with pleural effusion (n = 40) | 0.0153 | −0.0418 (−0.0572, −0.0305) | 0.0724 (0.0611, 0.0877) |
| Chest radiographs with consolidation (n = 40) | 0.0022 | −0.0673 (−0.0857, −0.0537) | 0.0717 (0.0581, 0.0901) |
| Mean Relative Difference (%) | Lower LOA (95% CI) | Upper LOA (95% CI) | |
| Study sample (n = 160) | 1.3 | −9.33 (−10.6, −8.23) | 11.92 (10.82, 13.2) |
| Chest radiographs without any lung or pleural abnormality (n = 40) | 1.22 | −5.85 (−7.54, −4.59) | 8.29 (7.03, 9.98) |
| Chest radiographs with pneumothorax (n = 40) | 0.73 | −6.63 (−8.5, −5.24) | 8.08 (6.7, 9.96) |
| Chest radiographs with pleural effusion (n = 40) | 3.12 | −8.68 (−11.93, −6.29) | 14.91 (12.52, 18.16) |
| Chest radiographs with consolidation (n = 40) | 0.12 | −13.84 (−17.58, −11.09) | 14.09 (11.34, 17.83) |
| Agreement | ICC (95% Confidence Interval) | p-Value * | |
|---|---|---|---|
| Study sample | Intra-observer agreement (thoracic radiologist 1) | 0.988 (0.983–0.991) | |
| Intra-observer agreement (thoracic radiologist 2) | 0.977 (0.968–0.983) | ||
| Inter-observer agreement (thoracic radiologist 1 vs. thoracic radiologist 2) | 0.972 (0.953–0.982) | 0.192 | |
| Inter-observer agreement (a deep learning-based model versus two thoracic radiologists) | 0.959 (0.945–0.971) | ||
| Chest radiographs without any lung or pleural abnormality | Intra-observer agreement (thoracic radiologist 1) | 0.980 (0.962–0.989) | |
| Intra-observer agreement (thoracic radiologist 2) | 0.998 (0.997–0.999) | ||
| Inter-observer agreement (thoracic radiologist 1 vs. thoracic radiologist 2) | 0.983 (0.968–0.997) | 0.868 | |
| Inter-observer agreement (a deep learning-based model versus two thoracic radiologists) | 0.982 (0.970–0.991) | ||
| Chest radiographs with pneumothorax | Intra-observer agreement (thoracic radiologist 1) | 0.990 (0.981–0.995) | |
| Intra-observer agreement (thoracic radiologist 2) | 0.992 (0.984–0.996) | ||
| Inter-observer agreement (thoracic radiologist 1 vs. thoracic radiologist 2) | 0.985 (0.976–0.992) | 0.534 | |
| Inter-observer agreement (a deep learning-based model versus two thoracic radiologists) | 0.982 (0.971–0.990) | ||
| Chest radiographs with pleural effusion | Intra-observer agreement (thoracic radiologist 1) | 0.993 (0.987–0.996) | |
| Intra-observer agreement (thoracic radiologist 2) | 0.969 (0.943–0.984) | ||
| Inter-observer agreement (thoracic radiologist 1 vs. thoracic radiologist 2) | 0.975 (0.959–0.999) | 0.066 | |
| Inter-observer agreement (a deep learning-based model versus two thoracic radiologists) | 0.949 (0.916–0.982) | ||
| Chest radiographs with consolidation | Intra-observer agreement (thoracic radiologist 1) | 0.989 (0.980–0.994) | |
| Intra-observer agreement (thoracic radiologist 2) | 0.945 (0.899–0.971) | ||
| Inter-observer agreement (thoracic radiologist 1 vs. thoracic radiologist 2) | 0.939 (0.890–0.978) | 0.812 | |
| Inter-observer agreement (a deep learning-based model versus two thoracic radiologists) | 0.925 (0.878–0.971) |
| Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value | Accuracy | ||
|---|---|---|---|---|---|---|
| Study sample | Deep learning-based model | 96.3% (89.7%–99.2%) | 83.3% (73.2%–90.8%) | 85.9% (77.0%–92.3%) | 95.6% (87.6%–99.1%) | 90.0% (84.3%–94.2%) |
| Radiologists | 97.8% (95.1%–99%) | 85.1% (78.8%–89.8%) | 87.4% (81.1%–91.8%) | 97.4% (93.9%–98.9%) | 91.6% (88.1%–94.2%) | |
| p-value * | <0.001 | 0.005 | <0.001 | <0.001 | <0.001 | |
| Chest radiographs without any lung or pleural abnormality | Deep learning-based model | 95.5% (77.2%–99.9%) | 94.4% (72.7%–99.9%) | 95.5% (77.2%–99.9%) | 94.4% (72.7%–99.9%) | 95% (83.1%–99.4%) |
| Radiologists | 97.3% (83.3%–99.6%) | 97.8% (92%–99.4%) | 98.2% (92.6%–99.6%) | 96.7% (79.7%–99.5%) | 97.5% (91.4%–99.3%) | |
| p-value * | <0.001 | 0.07 | 0.021 | <0.001 | <0.001 | |
| Chest radiographs with pneumothorax | Deep learning-based model | 100% (80.5%–100%) | 82.6% (61.2%–95%) | 81% (58.1%–94.6%) | 100% (82.4%–100%) | 90% (76.3%–97.2%) |
| Radiologists | 98.8% (92.5%–99.8%) | 87% (75.8%–93.4%) | 84.8% (69.3%–93.3%) | 99% (93.1%–99.9%) | 92% (84.8%–96%) | |
| p-value * | <0.001 | 0.132 | 0.083 | <0.001 | 0.004 | |
| Chest radiographs with pleural effusion | Deep learning-based model | 95.2% (76.2%–99.9%) | 68.4% (43.4%–87.4%) | 76.9% (56.4%–91%) | 92.9% (66.1%–99.8%) | 82.5% (67.2%–92.7%) |
| Radiologists | 96.2% (91%–98.4%) | 73.7% (57.6%–85.2%) | 80.2% (64.1%–90.2%) | 94.6% (85.4%–98.1%) | 85.5% (76%–91.7%) | |
| p-value * | 0.015 | 0.277 | 0.071 | 0.079 | 0.055 | |
| Chest radiographs with consolidation | Deep learning-based model | 95.5% (77.2%–99.9%) | 88.9% (65.3%–98.6%) | 91.3% (72%–98.9%) | 94.1% (71.3%–99.9%) | 92.5% (79.6%–98.4%) |
| Radiologists | 99.1% (94%–99.9%) | 82.2% (67.2%–91.2%) | 87.2% (73.2%–94.4%) | 98.7% (90.8%–99.8%) | 91.5% (83.2%–95.9%) | |
| p-value * | 0.082 | 0.01 | 0.003 | 0.173 | 0.004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Lee, J.H.; Jang, M.-j.; Park, J.; Hong, W.; Lee, C.S.; Yang, S.Y.; Park, C.M. The Performance of a Deep Learning-Based Automatic Measurement Model for Measuring the Cardiothoracic Ratio on Chest Radiographs. Bioengineering 2023, 10, 1077. https://doi.org/10.3390/bioengineering10091077
Kim D, Lee JH, Jang M-j, Park J, Hong W, Lee CS, Yang SY, Park CM. The Performance of a Deep Learning-Based Automatic Measurement Model for Measuring the Cardiothoracic Ratio on Chest Radiographs. Bioengineering. 2023; 10(9):1077. https://doi.org/10.3390/bioengineering10091077
Chicago/Turabian StyleKim, Donguk, Jong Hyuk Lee, Myoung-jin Jang, Jongsoo Park, Wonju Hong, Chan Su Lee, Si Yeong Yang, and Chang Min Park. 2023. "The Performance of a Deep Learning-Based Automatic Measurement Model for Measuring the Cardiothoracic Ratio on Chest Radiographs" Bioengineering 10, no. 9: 1077. https://doi.org/10.3390/bioengineering10091077
APA StyleKim, D., Lee, J. H., Jang, M.-j., Park, J., Hong, W., Lee, C. S., Yang, S. Y., & Park, C. M. (2023). The Performance of a Deep Learning-Based Automatic Measurement Model for Measuring the Cardiothoracic Ratio on Chest Radiographs. Bioengineering, 10(9), 1077. https://doi.org/10.3390/bioengineering10091077






