1. Introduction
In 2020, prostate cancer (PCa) was the world’s second-most-common tumor among men (accounting for 14.1% of new diagnoses, just behind lung cancer) and the fifth for mortality (6.8%) [
1]. For screening PCa, physicians primarily use the prostate-specific antigen (PSA) test, which measures the amount of PSA in the blood, a marker of potential PCa [
2]. However, PSA levels may also arise due to other conditions, including an enlarged or inflamed prostate [
3]. Therefore, if the PSA test is positive, the patient typically undergoes a multiparametric magnetic resonance imaging (mpMRI) examination [
2]. Here, T2-weighted (T2w) and diffusion-weighted (DWI) images (along with apparent diffusion coefficient (ADC) maps) are acquired to investigate the anatomy and detect the presence of the tumor, respectively. These acquisitions allow radiologists to make a preliminary diagnosis following the Prostate Imaging Reporting and Data System (PI-RADS) guidelines [
4]. According to the PI-RADS standard, the radiologist assigns a numerical value between 1 and 5 to the suspected lesion: the higher the score, the greater the likelihood that the accounted nodule is malignant. If PI-RADS ≥ 3, the lesion is likely to be clinically significant, and the patient undergoes a biopsy [
2]. Based on the two most common patterns in the biopsy specimen, the pathologist assigns a score known as the Gleason Score (GS) to the tumor’s aggressiveness. Along with the GS, it is also recommended to provide the group affiliation of the assigned score defined by the International Society of Urological Pathology (ISUP), as this facilitates predicting patient outcomes and patient communication [
5]. If GS ≥
(ISUP ≥ 2), the tumor is confirmed to be clinically significant [
6]. However, it is often the case that the suspected lesion results are indolent after a biopsy examination. In particular, only about 20% of all PI-RADS 3 patients biopsied show intermediate/severe PCa pathology [
7]. Although mpMRI investigation reduces overdiagnosis [
8], it remains a qualitative diagnostic tool, highly dependent on radiologist experience and acquisition protocols [
9]. For this reason, there is a need for an automated tool to support radiologists in the clinical practice to make diagnosis more robust, reliable, and, above all, non-invasive.
To date, several studies aimed to build machine and deep learning models for the automatic classification of PCa from mpMRI images [
10], exploring various techniques, including utilizing generative methods [
11], which currently represent the forefront of performance enhancement in this field. Most of these distinguish clinically significant from non-significant PCa (GS ≤ 3 + 3, ISUP ≤ 1 vs. GS ≥ 3 + 4, ISUP ≥ 2) [
12,
13,
14,
15,
16,
17,
18]. However, an even more critical task is to distinguish lesions based on their aggressiveness, i.e., low-grade (LG) (GS ≤ 3 + 4, ISUP ≤ 2) vs. high-grade (HG) (GS ≥ 4 + 3, ISUP ≥ 3) lesions, as this is what discriminates the patient’s clinical path. Indeed, all patients with GS ≤ 3 + 4 (ISUP ≤ 2) typically undergo active surveillance [
19], even though a lesion with GS = 3 + 4 is still clinically significant. In [
20], the radiomic approach was exploited by classifying features extracted from mpMRI images employing a k-nearest neighbor (KNN) algorithm. In [
21], the authors employed AlexNet according to a transfer-learning approach by fine-tuning the last classification layer with T2w axial, T2w sagittal images, and ADC maps jointly. In [
22], the authors explored both radiomic and deep learning approaches, training several classical machine learning algorithms and 2D convolutional neural networks (CNNs) (with and without attention gates [
23]) exploiting T2w axial images only, ADC maps only, and the combination of the two modalities.
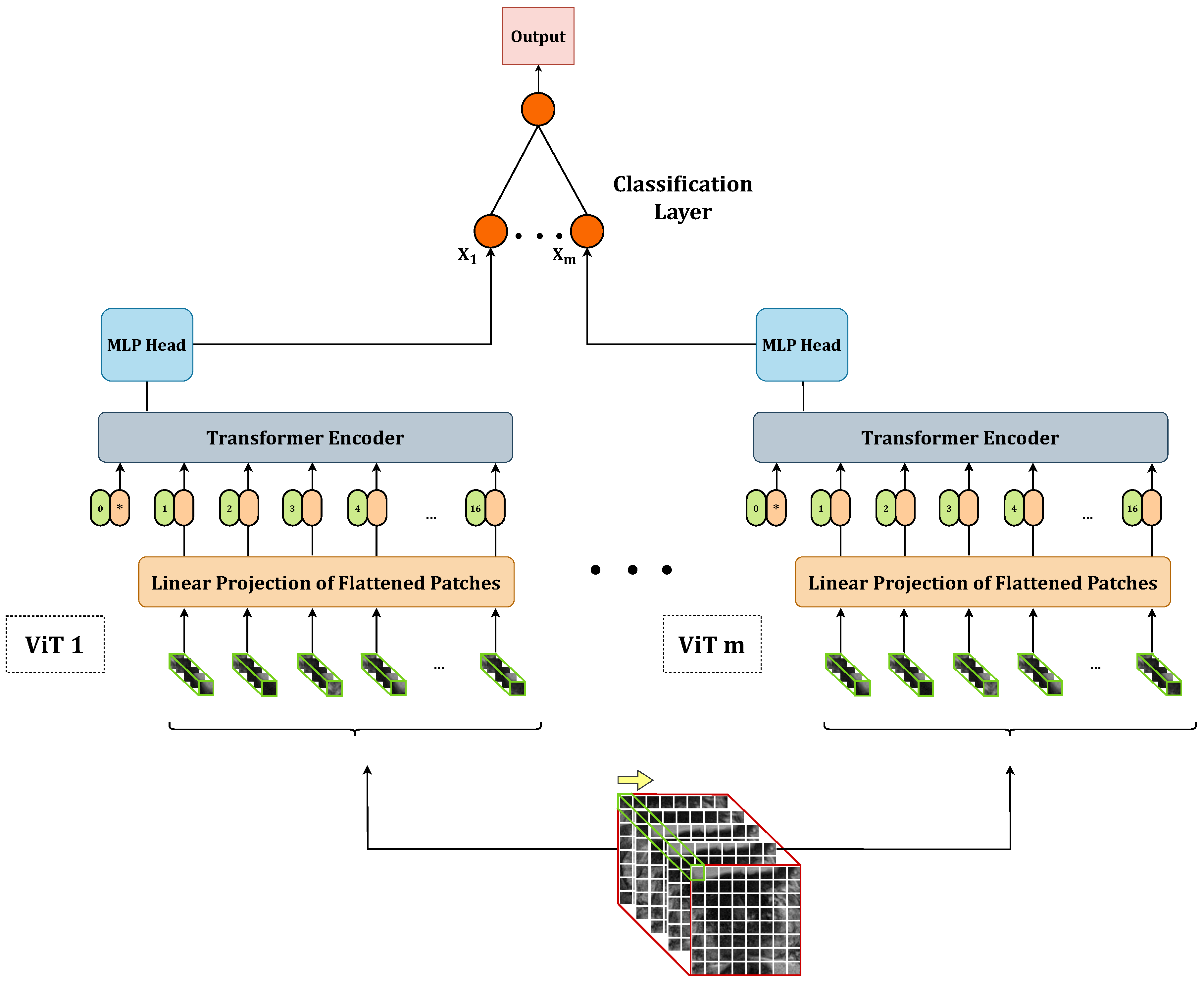
In this work, we propose to perform the PCa aggressiveness classification task from T2w images by exploiting an ensemble of vision transformers(ViTs) [
24]. ViTs are becoming increasingly popular in the medical imaging domain [
25,
26,
27,
28,
29,
30,
31,
32,
33,
34], usually outperforming classical CNNs [
35,
36], which are one of the most significant networks in the deep learning field [
37]. The existing literature typically employs ViTs in transfer learning scenarios by pre-training them on large datasets of natural images and fine-tuning them on specific datasets [
27,
28,
38]. However, due to the limited availability of medical imaging data, we propose training the ViT from scratch by downsizing the architecture. Additionally, since medical data is often acquired in volumetric form, we modify the ViT’s architecture to train it on 3D volumes, leveraging most of the information from the acquisitions. To further enhance the performance of vanilla 3D ViTs, which we will call
base models henceforth, we propose to combine them in stacking ensembles. The aim is to create a meta-model that learns how to best combine the predictions of base 3D ViTs to harness the capabilities of a stack of models. Finally, to assess the models’ confidence in making predictions in addition to the sole accuracy, we propose two confidence metrics based on the models’ output probability.
The key contributions of our work can be summarized as follows:
We introduce a downscaled version of the ViT architecture and train it from scratch using small datasets, addressing the challenge of limited data availability.
We propose modifications to the ViT architecture to handle 3D input, enabling the model to effectively leverage volumetric data in the PCa aggressiveness classification task from T2w images.
We develop stacking ensembles by combining multiple base 3D ViTs, thereby leveraging the strengths of both stronger and weaker base models to improve overall performance.
We define two novel confidence metrics that provide insights into the models’ confidence in making predictions, offering a more comprehensive evaluation beyond accuracy alone.
We conduct comparative experiments to assess the performance of ensemble 3D ViTs against the base models in the task of PCa aggressiveness assessment from T2w images.
These contributions collectively aim to enhance the classification accuracy and reliability of PCa aggressiveness assessment, utilizing the power of ensemble models and tailored adaptations to the ViT architecture.
4. Experiments
4.1. Dataset Splitting
In our study, we initially divided the dataset into two parts: 90 lesions (80%) for training and validation and 22 lesions (20%) for the test set. To ensure robustness in model evaluation, we employed two different strategies for further splitting the 90-lesion set. The first strategy involved creating two subsets: 90% for training and 10% for validation. The second strategy involved dividing it into five subsets for conducting a five-fold cross-validation (CV). We used the first splitting strategy to train the 18 base 3D ViTs, which we later used in the ensembles. We employed the second splitting strategy for optimizing both the base and ensemble models. To enhance the reliability of our findings, we bootstrapped the training set obtained from the first splitting strategy, generating 100 samples. We utilized these samples for re-training the best-performing base and ensemble models obtained from the five-fold CV. To ensure balanced representation, we performed all splits while stratifying the data based on the lesion class (approximately two-thirds LG and one-third HG) and lesion location (approximately two-fifths PZ, two-fifths AS, and one-fifth TZ). Additionally, we employed a patient-wise splitting approach to prevent any data leakage. A summary of the composition of the training, validation, and test sets with respect to tumour aggressiveness and location can be found in
Table 2.
4.2. Training Setup
Training, validation, and test phases, both for base and ensemble models, were coded in Python by employing the following modules: Pytorch (v. cuda-1.10.0) [
42], Numpy (v.1.20.3) [
43], Scikit-learn (v. 0.24.2) [
44], Pydicom (v. 2.1.2) [
45], Pillow (v. 9.0.1) [
46], and Pandas [
47]. We performed training and test processes employing an Intel Core i7 ASUS Desktop Computer with 32 GB RAM and an NVIDIA GeForce GTX 1650 GPU.
4.2.1. Base Models
We trained each of the 18 base models with the following hyperparameters: learning rate = 1 × 10
, weight decay = 1 × 10
, maximum number of steps = 1000, batch size = 4, warmup steps = 1000, optimization algorithm =
, loss function =
. To make each training reproducible, we exploited the reproducibility flags provided by Pytorch [
42], Numpy [
43], and Random [
48] libraries, choosing a seed equal to 42. We trained each base model according to a five-fold CV.
4.2.2. Ensemble Models
We explored all possible combinations of two- and three-base-model stacking ensembles. Specifically, we evaluated 153 two-model combinations and 816 three-model combinations. Each combination underwent training and evaluation using a five-fold CV approach, maintaining consistency with the dataset splitting and reproducibility seed used for the base models. To train the meta-classifier, we utilized the following set of hyperparameters: learing rate = 1 × 10, weight decay = 1 × 10, number of epochs = 100, batch size = 4, optimization algorithm = , loss function = . These hyperparameters were applied consistently across all training iterations of the meta-classifier to ensure fair and comparable performance evaluation.
4.3. Performance Evaluation
To assess the performance and calibration of the models, we evaluated their accuracy, confidence in predictions, and calibration. The selection of the best-performing base and ensemble model was based on the performance on the five-fold CV process. To compare the performance of the best-performing base and ensemble models, we conducted a 100-sample bootstrap of the entire training set. For each bootstrapped sample, we re-trained both the best-performing base model and the ensemble model. Subsequently, we evaluated each re-trained model on the same hold-out test set generating a performance distribution. From these distributions, we calculated the median performance and the 95% confidence interval (CI). This approach allowed us to obtain a robust estimation of the models’ performance and CI for a reliable performance assessment and comparison.
Statistical Analysis
To evaluate the statistical significance of the difference between the best-performing base and ensemble models, we conducted the Wilcoxon signed-rank test. We considered the difference between the performance distributions of the base and ensemble models statistically significant if the resulting
p-value (
p) was less than 0.050. We performed the statistical analysis exploiting the Scipy library [
49] (v. 1.9.3).
4.4. Results
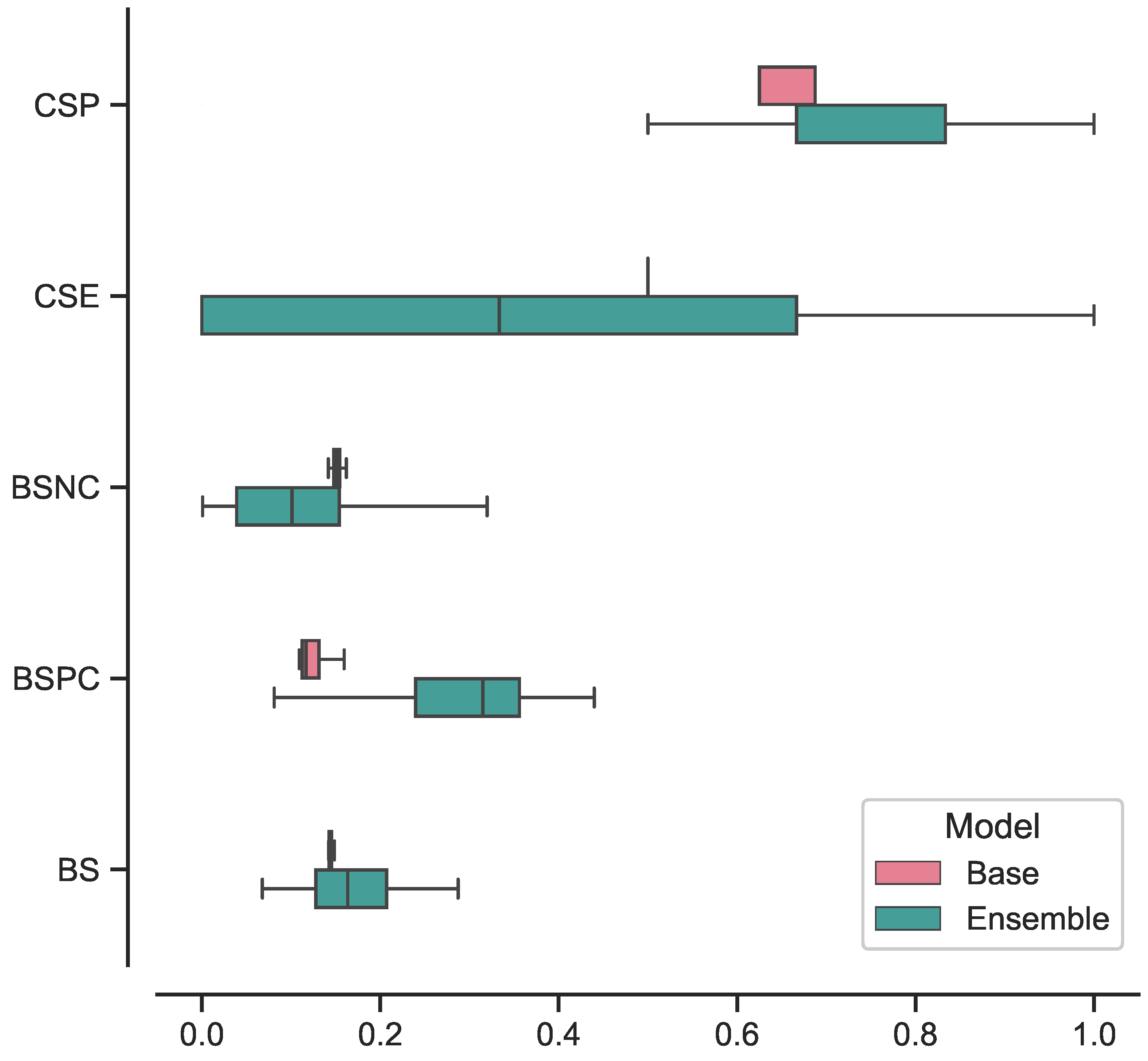
The best-performing ensemble of 3D ViTs, composed of configurations 5, 9, and 11, achieved the following results in terms on median and 95% CI: a specificity of 0.83 [0.67–1], sensitivity of 0.67 [0.67–1], balanced accuracy of 0.75 [0.67–0.96], AUROC of 0.89 [0.64–1], and AUPRC of 0.87 [0.57–1]. In terms of confidence prediction, the ensemble yielded the following results: a CSP of 0.83 [0.41–1], CSE of 0.33 [0–1], a BSNC of 0.10 [0–0.25], a BSPC of 0.31 [0.02–0.41], and an overall BS of 0.16 [0.08–0.24]. For a random classifier, the AUROC and AUPRC values on this test set were 0.500 and 0.273, respectively.
Regarding the best-performing base 3D ViT with configuration 5, it achieved the following results on the external test set in terms of median and 95% CI: a specificity of 0.75 [0.75–0.75], sensitivity of 0.83 [0.67–0.83], balanced accuracy of 0.79 [0.71–0.79], AUROC of 0.86 [0.85–0.89], and AUPRC of 0.65 [0.60–0.68]. In terms of confidence prediction, the base model yielded the following results: CSP of 0.68 [0.63–0.69], a CSE of 0.50 [0.50–0.59], a BSNC of 0.15 [0.14–0.16], a BSPC of 0.12 [0.11–0.17], and an overall BS of 0.14 [0.14–0.15].
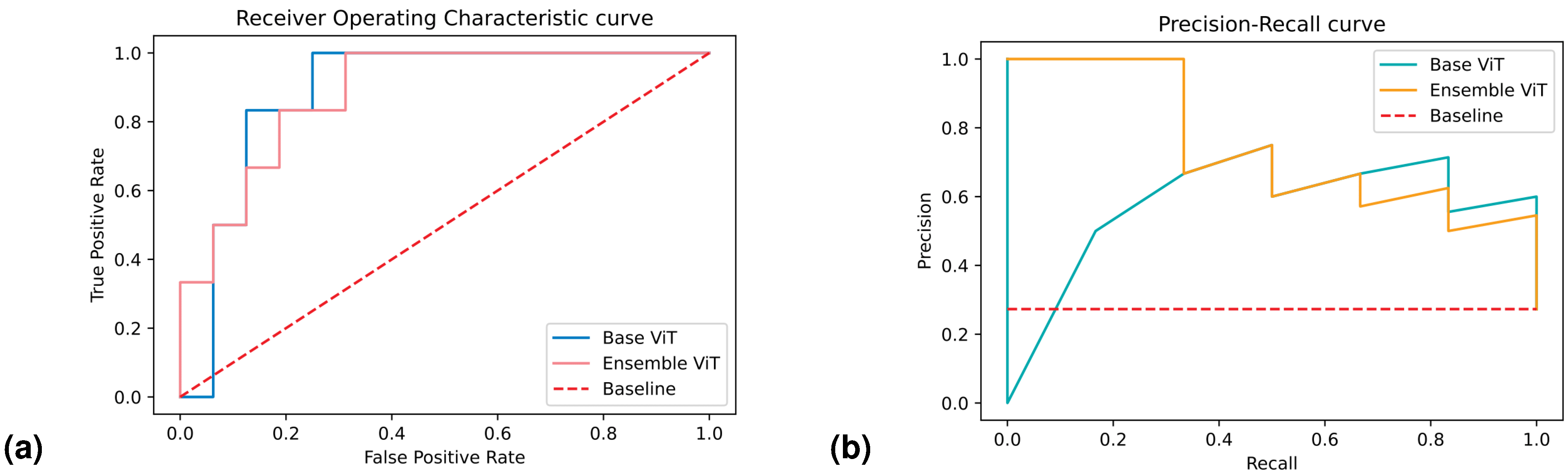
We presented the results for the best-performing base and ensemble models in terms of accuracy and confidence in
Table 3 and
Table 4, respectively. Additionally, in
Figure 4, we included the ROC and PR curves for the two best-performing models when trained on the entire training set (non-bootstrapped). These visualizations provide further insights into the performance and discriminative capabilities of the models.
After conducting the Shapiro–Wilk test, we determined that all the distributions deviated from normality with p < 0.001. Consequently, we employed the Wilcoxon signed-rank test to assess the statistical differences between the base and ensemble models. The test yielded the following statistics for each metric: , for specificity; , for sensitivity; , for balanced accuracy; , for AUROC; , for AUPRC; , for CSP; , for CSE; , for BSNC; , for BSPC; and , for BS.
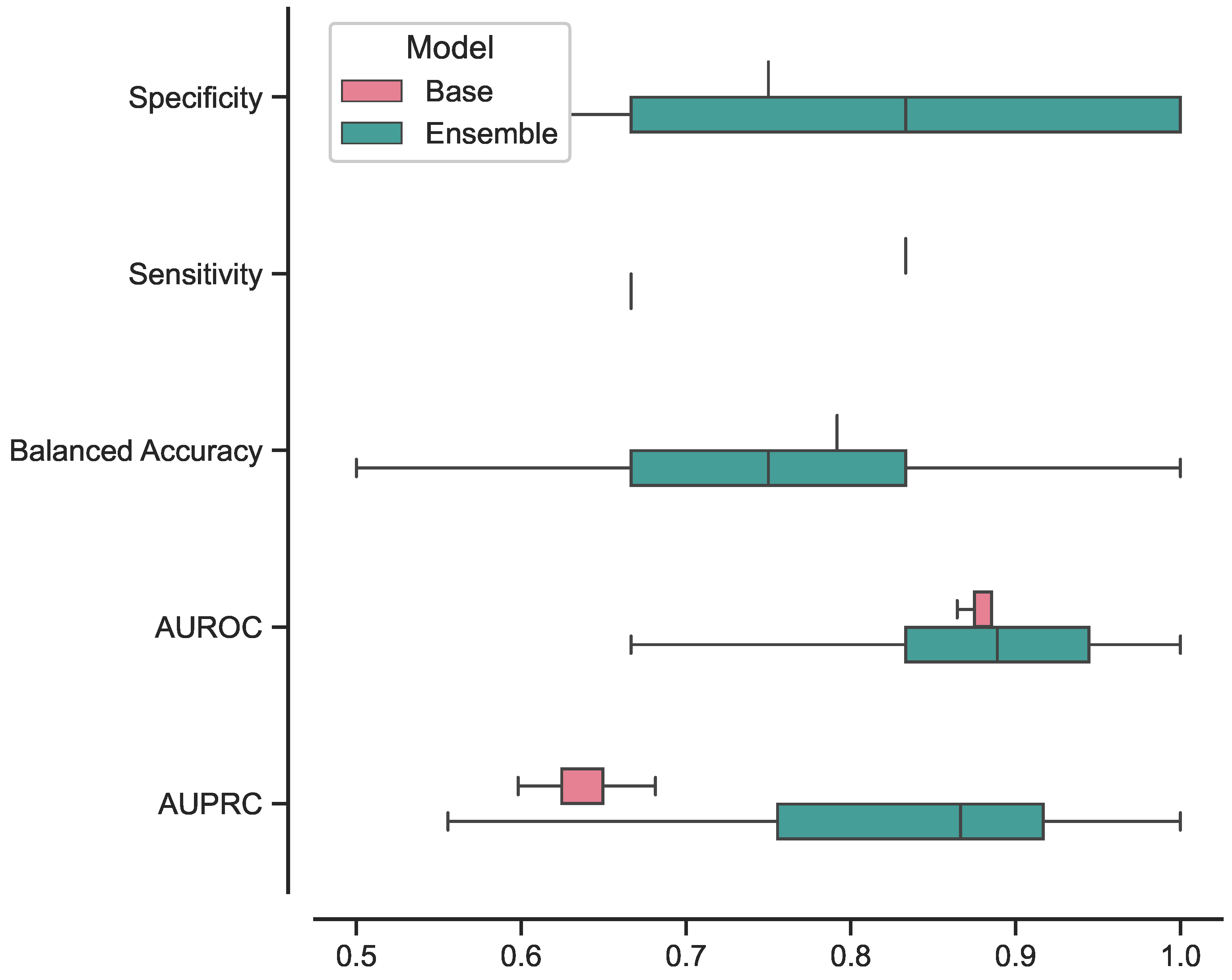
In
Figure 5, we present the box plots illustrating the distributions of results for the best-performing base and ensemble models based on accuracy metrics. Similarly, in
Figure 6, we provide the box plots representing the distributions of results for the base and ensemble models with respect to confidence metrics.
For clarity and comparison purposes, we present the five-fold CV accuracy and confidence results for all the base models in
Table 5 and
Table 6, respectively. Similarly, we provide the five-fold CV results of the top 10 best-performing ensembles in terms of accuracy and prediction confidence in
Table 7 and
Table 8, respectively. All reported values are presented as the mean and SD across the five folds.
5. Discussion
In this study, we proposed a trained-from-scratch 3D ViT in a stacking ensemble configuration and assessed its effectiveness in assessing PCa aggressiveness from T2w MRI acquisitions. The approach employed in this study involved training vanilla 3D ViT in various configurations and subsequently combining them into 2- and 3-model ensembles. We concatenated the features extracted from each of these base models and provided it to a single fully-connected layer, which yielded the final prediction. The evaluation of the trained models centred around assessing their accuracy performance, specifically focusing on measures such as specificity, sensitivity, balanced accuracy, AUROC, and AUPRC. Additionally, we measured the calibration of these models using BS. We further computed BS with respect to the negative class only BSNC and the positive class only (BSPC), allowing us to gain insights into their calibration for each class separately. To enhance the model evaluation process, we proposed the introduction of two novel metrics, CSP and CSE. The primary purpose of these metrics was to provide a comprehensive measure that combined the model’s prediction capabilities with its confidence level. By doing so, we aimed to highlight only those predictions made with a confidence level above a predefined threshold. The implementation of CSP and CSE aimed to offer more detailed and reliable information about the model’s performance, with the ultimate goal of bringing its practical application in clinical settings closer. These metrics were designed to provide valuable insights into the accuracy and confidence of the model’s predictions, enabling a more informed and cautious approach when utilizing the model’s outputs in real-world medical scenarios.
We trained the base 3D ViT models according to a grid search, varying architecture parameters such as d, D, L, and k, for a total of 18 base 3D ViTs. By training all possible combinations of two and three models, we created 966 ensembles from these base models. We trained each ensemble following a five-fold CV and re-trained the one with the highest performance on a 100-sample bootstrapped training set. For comparison, we optimized the 18 base ViTs as well using a five-fold CV and re-trained the best-performing base ViT on the same bootstrapped training set. We evaluated each of the 100 models from both the ensemble and base ViT on a separate hold-out test set. To determine whether there was a significant difference in performance between the base and ensemble models, we conducted a statistical analysis on the distributions of results obtained from the test set evaluations.
We evaluated our approach using the ProstateX-2 challenge dataset [
39] appropriately divided into training, validation and test set, ensuring strict separation between patients.
According to the results, the ensemble model demonstrated strong performance in classifying LG and HG lesions, as evidenced by its high median AUROC (0.89). A high AUROC indicates a robust ability to accurately identify positive instances while effectively minimizing false positive predictions. This performance metric holds great importance, particularly in tasks characterized by imbalanced class distributions or situations where the costs associated with false positives and false negatives are substantial, as exemplified in this case. The model also demonstrated good results in classifying the positive class specifically, with a median AUPRC of 0.87. Apart from its general performance, the model displayed notable proficiency in accurately classifying the positive class. The median AUPRC was recorded at a median value of 0.87. The AUPRC is a crucial evaluation metric, especially when dealing with imbalanced datasets. To elaborate further, the AUPRC measures the area under the precision–recall curve, which plots the precision against the recall (sensitivity) at various classification thresholds. In cases where the class distribution is imbalanced, a high AUPRC becomes crucial as it signifies that the model effectively achieves a high precision rate while maintaining a reasonable recall rate. This means that when the model makes a positive prediction, it is highly likely to be correct (high precision), and it successfully captures a significant portion of the actual positive instances (high sensitivity).
Regarding the calibration aspect, the model exhibited strong calibration performance, as evidenced by a BS of 0.16. The BS is a widely used metric that assesses the calibration of probabilistic predictions made by a classification model by measuring the mean squared difference between the predicted probabilities and the actual binary outcomes. A lower Brier score indicates better calibration, implying that the model’s predicted probabilities align closely with the actual outcomes.
Concerning the model’s confidence in its predictions, CSP revealed a remarkable value equal to the classical specificity metric, reaching 0.83. This outcome signifies that all correct predictions related to the negative class (specificity) were accompanied by high confidence levels, i.e., the model returned one with an output probability less than or equal to 0.3.
Upon comparing the ensemble model to the best-performing base 3D ViT, we conducted a Wilcoxon signed-rank test to assess the statistical significance of their performance differences. In terms of AUROC, the results of this analysis revealed no statistically significant difference between the two models (p = 0.73). This indicates that both the ensemble model and the base ViT performed similarly in terms of overall discriminative ability. However, a notable contrast emerged when evaluating the models’ proficiency in classifying HG lesions. Indeed, the ensemble model outperformed the base ViT by a 22% improvement in AUPRC, which resulted in statistically significant (p < 0.001). The substantial improvement in AUPRC for HG lesions highlights the ensemble model’s particular strength in accurately identifying and distinguishing severe lesions from the base ViT. This is crucial in medical applications where detecting HG lesions can significantly impact patient outcomes and treatment decisions.
Indeed, while the ensemble model exhibited a significant performance improvement in classifying HG lesions, it is essential to consider its impact on calibration and confidence in predictions. A closer examination of the calibration metrics reveals that the ensemble model’s performance comes at the expense of poorer calibration towards the HG class. This is evident from the higher BSPC and the lower CSE values compared to the base ViT. The higher BSPC suggests that the ensemble model’s probability predictions for positive instances in the HG class may be less well-calibrated. Consequently, this may lead to less reliable probability estimates for high-grade lesions. Furthermore, the lower CSE value indicates reduced confidence in the ensemble model’s predictions for the HG class, i.e., the model might not be as certain when making predictions for positive instances in this category. On the contrary, the ensemble model demonstrated improved confidence in its predictions for the negative class when compared to the base ViT. This was evident from the lower BSNC and CSP values relative to the base model. The lower BSNC suggests that the ensemble model’s probability predictions for the negative class are better calibrated and align closely with the actual outcomes. Moreover, the lower CSP metric signifies that the ensemble model confidently assigns lower probabilities (less than or equal to 0.3) to the correct negative predictions. All differences in cited metrics values resulted in statistically significant according to the Wilcoxon signed-rank test (p < 0.001).
Furthermore, we investigated the performance of the ensemble model compared to the base model focusing on the consistency of its predictions. The results, illustrated in
Figure 5 and
Figure 6, showcased the ensemble model displayed larger CIs with respect to to the base model. This suggests that the ensemble model’s performance is characterized by higher variability, making its predictions less stable and more susceptible to fluctuations. While the ensemble model demonstrated superior performance in certain aspects, the broader range of its CIs indicates that its predictions may be less consistent, leading to varying results across different iterations or datasets. This variability in performance could be attributed to the complexity introduced by the ensemble configuration, as it involves combining multiple models, each with its unique characteristics.
Performance of best-performing base and ensemble models. The best-performing ensemble model contains the best-performing base model, indicating that this specific configuration of architecture parameters achieves high performance. This configuration stands out as it is present in three out of the ten best ensemble combinations, as shown in
Table 7.
Impact of the number attention heads and embedding size. Configurations 5 and 8, both having eight attention heads and an embedding size of 32, are the most frequently recurring setups in the top-performing ensembles (three out of ten combinations each). Additionally, configurations 4, 7, and 16, all having an embedding size of 64 and four attention heads, also occur frequently (three out of ten ensembles). These architecture parameter combinations seem to contribute to improving the ensembles’ performance, even though they might not perform well when used in isolation.
Influence of MLP size. The size of the MLP (represented by the parameter d) does not significantly impact performance. The results indicate that there is no noticeable difference in performance between models trained with d = 2048 or d = 3072, whether in the base or ensemble models. This suggests that increasing the MLP size beyond a certain point does not lead to significant improvements in accuracy.
Stacking ensemble effect. Comparing
Table 5 and
Table 7, it is evident that combining weaker models in stacking ensembles significantly improves accuracy performance. This finding highlights the benefits of leveraging ensemble methods to improve model performance, even when individual base models might not be as strong.
We compared the performance of our best-performing ensemble 3D ViT model with state-of-the-art (SoTA) studies that addressed the same clinical task of assessing PCa aggressiveness from MRI images. To ensure a fair comparison, we considered all the studies that employ T2w images alone or combined with other images modalities. In addition, we compared our results with studies employing both radiomics and deep learning for a comprehensive evaluation. In radiomic studies, Jensen et al. [
20] achieved an AUROC of 0.830 using a K-nearest neighbors (KNN) classifier with only T2w images. Bertelli et al. [
22] reported the best machine learning model with an AUROC of 0.750 (90% CI: [0.500–1.000]) for T2w images alone and 0.63 (90% CI: [0.17–1]) when combining T2w and ADC images. As for deep learning models, Yuan et al. [
21] used an AlexNet in a transfer-learning approach, achieving an AUROC of 0.809 with T2w images alone and 0.90 when combining T2w and ADC acquisitions. Bertelli et al. [
22] utilized CNNs with attention gates, resulting in an AUROC of 0.875 (90% CI: [0.639–1.000]) for T2w images only and 0.67 (90% CI: [0.30–1]) when combining T2w and ADC. In comparison, our best-performing ensemble 3D ViT model displayed SoTA performance with an AUROC that outperformed all the other models when using only T2w images. Unfortunately, a direct comparison for the positive class only was not possible due to the lack of available data in the literature. The comparison results are summarized in
Table 9, providing an overview of our model’s performance concerning the SoTA studies.
This study is subject to several limitations that should be acknowledged. Firstly, we did not conduct a hyperparameter optimization, which may have limited the overall performance of our models. Instead, we kept the hyperparameter values fixed throughout all the training phases and focused solely on optimizing the architecture parameters. Moreover, in our investigation of stacking ensembles, we only considered two- and three-model combinations. Expanding the ensemble to include more base models might lead to further improvements in the results. Indeed, utilizing a larger ensemble could enhance model diversity and potentially increase predictive performance. Furthermore, one of the limitations lies in the choice of using only axial T2w images for training. In contrast, many of the cited works combined multiple modalities, such as T2w and ADC images, to improve model performance. Expanding the dataset to incorporate additional modalities could potentially enhance the model’s ability to capture diverse and complementary information, thereby leading to more robust predictions. Overall, recognizing these limitations is essential to interpreting the findings accurately and understanding the scope of the study. Future research could address these limitations and explore new avenues for improving the performance of the 3D ViT model in assessing PCa aggressiveness.