1. Introduction
Writing medical image diagnostic reports is time-consuming, laborious, and professionally demanding for radiologists. Thus, automatic medical report generation has become increasingly popular. This method can generate diagnostic text based on natural language for experienced radiologists, assist radiologists in completing their diagnosis, significantly reduce the burden of writing text, and accumulate diagnostic experience for radiologists or medical students who lack clinical experience.
Automatic generation of medical image diagnostic reports is based on traditional image captions [
1,
2,
3,
4,
5], using an encoder–decoder framework [
6].
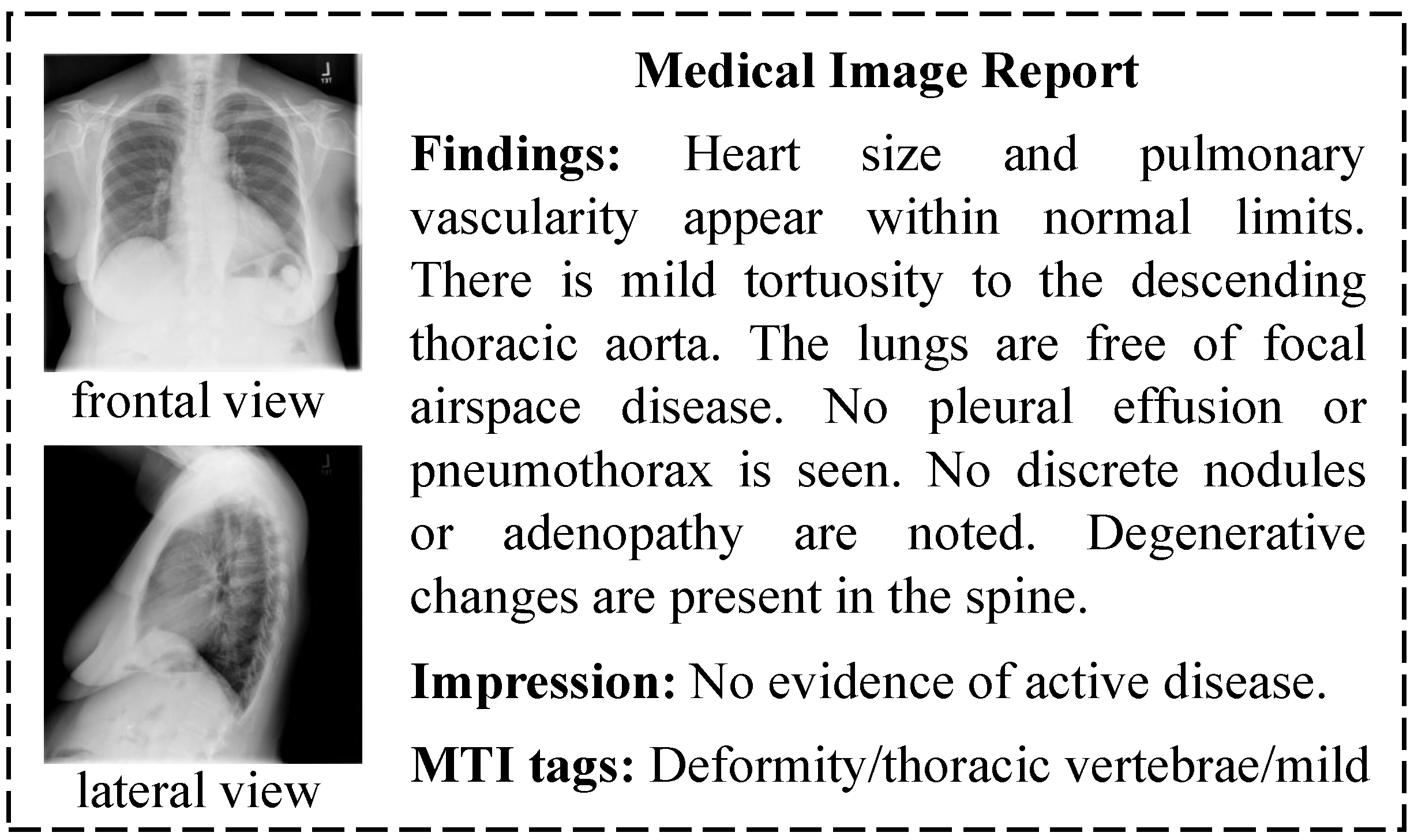
Figure 1 is a report sample in the Indiana University (IU) X-ray dataset, which is widely used in automatic medical report generation. A sample is composed of multi-view images, findings, impressions, and Medical Text Indexer (MTI) tags. Findings and impressions are long diagnostic sentences with fixed sentence patterns. MTI tags are key words generated from the diagnostic sentences. Automatic medical report generation based on the characteristics of medical data has become a hot research topic in recent years. Zhang et al. [
7] used knowledge maps and prior medical knowledge to enhance the features extracted from images. Xue et al. [
8] used hierarchical Long Short Term Memory (LSTM) to generate long diagnostic sentences. Li et al. [
9] used a template method to generate diagnostic sentences with fixed sentence patterns. Jing et al. [
10] embedded the MTI tags predicted by the encoder in order to generate diagnostic reports. Similarly, Yuan et al. [
11] extracted normalized medical concepts like MTI tags from the diagnostic report.
These studies have made progress in producing specific diagnostic reports; however, there are still two challenges in making full use of the characteristics of medical data to improve diagnostic report generation. The first challenge is the reasonable use of multi-perspective medical images. For example, there are two chest X-ray images in the report sample in
Figure 1, so radiologists need to comprehensively evaluate the lesions from both images in the process of medical image diagnosis in order to write an accurate and comprehensive diagnostic report. The second challenge lies in the effective combination of multi-modal data because radiologists need to synthesize the observed image features and the key lesion features when writing the diagnostic report.
To overcome these two challenges, we propose a novel automatic report generation approach, which consists of two modules: Cross-View Attention Module and Medical Visual-Semantic LSTMs (CVAM+MVSL). We first develop CVAM based on the characteristics of multi-view medical images. The encoder receives the input of frontal and lateral chest X-ray images and outputs feature maps. Then, the feature maps are sent into two branches of CVAM. One is the single-view branch, which retains the view features, and the other is the cross-view branch, which integrates the multi-view features. The binary cross-entropy (BCE) loss function is used as the classification loss function, and mean square error (MSE) loss is used to unify the prediction results of the two branches. Next, we propose the module of MVSL to fuse visual features of the images and semantic features of lesions. The input for this structure is the multi-view image feature map given by the encoder and the embedding of predicted medical concepts. MVSL uses three LSTMs to process the multi-view image features and medical concepts, and the hidden layers of the LSTMs are utilized to determine the image area and medical concepts that should be examined at the moment. The medical visual-semantic features calculated by the fully connected layer are used as the input of the Sentence LSTM–Word LSTM to generate the diagnostic sentence.
The main contributions of our work are as follows:
We propose CVAM to process multi-view medical images, which not only maintains the features of images but also makes full use of the complementary information of frontal and lateral chest X-ray images.
We present MVSL to couple the visual features of images and the semantic features of lesions and to employ the hidden layers of LSTMs to determine the important features at the current moment.
We perform extensive experiments and a user study to verify the effect and utility of the proposed methods. Results show that the proposed CVAM can significantly increase the area under curve (AUC) on both Chexpert and IU X-ray datasets. Compared with the previous methods, CVAM+MVSL can generate better medical reports with more information and higher accuracy.
2. Related Work
In this section, we introduce the related work on the topic of automatic medical report generation. The existing report generation methods mainly improve on encoders and decoders, but due to the entanglement of these two components, it is difficult to distinguish the key developments in these two components. Therefore, we introduce the relevant work in encoders and decoders separately.
Medical image analysis and processing based on deep learning play an increasingly important role in medical and health auxiliary diagnosis [
12,
13,
14]. A key application of this technology is the automatic generation of medical images, which has received extensive attention in recent years [
10,
15,
16,
17,
18,
19]. Compared to other medical image analysis and processing tasks, the automatic generation of medical images is more challenging because it requires the modeling of both images and texts. So far, most of the current methods of automatic medical report generation are based on the framework of encoder–decoder [
6,
20,
21,
22,
23,
24,
25].
The encoder is responsible for extracting image features. Jing et al. [
10] used Convolutional Neural Networks (CNNs) to extract features from single-view chest X-ray images, taking out the results of the last layer of convolution as the feature expression of medical images. The majority of studies utilize CNNs as encoders and Recurrent Neural Network (RNNs) as decoders for report generation. However, Alahmadi et al. [
21] employed RNNs as image encoders, adhering to the encoder–decoder machine translation model paradigm for caption generation. Li et al. [
9] worked on multiple graphs to model the data structure and the transformation rules among different graphs. Zhang et al. [
7] used the chest abnormality graph with prior knowledge to characterize image features. Yuan et al. [
11] first proposed using two CNNs to process multi-view images; however, the utilization of multi-view medical images can be further improved. On the basis of these studies, our CVAM uses the characteristics of multi-view medical images to fuse and process multi-view medical image features.
The decoder receives the image features extracted by the encoder and generates the diagnostic sentence using LSTM. Xue et al. [
8] proposed to use a Sentence LSTM–Word LSTM framework similar to [
26] to generate multiple diagnostic sentences, which is widely used in the literature, such as [
7,
10,
11]. Harzig et al. [
24] contended that distinct patterns and data distributions between normal and abnormal cases can lead to biases in models. They addressed this by employing two separate word LSTMs to differentiate between the generation of abnormal and normal reports. Jing et al. and Yuan et al. [
10,
11] proposed introducing semantic features of lesions in the process of generating reports because these features can provide more abnormal information. Although we use the same basic Sentence LSTM–Word LSTM framework of the previous studies, the proposed MVSL pays more attention to how to effectively combine the visual features of medical images with the semantic features of lesions and provides the Sentence LSTM–Word LSTM with the data of image areas and lesion semantics when generating diagnostic sentences.
3. Materials and Methods
3.1. Datasets
The first dataset we use is IU X-ray [
27], which contains 3959 medical diagnostic reports. Each report is labeled with chest X-ray images, impressions, findings, and MTI tags. We filter out samples of single-view images according to the experimental requirements. Following the conventions of the field of natural language processing, samples with fewer than 3 diagnostic sentences are removed, resulting in a total of 3331 samples. We pre-process the report text by converting it to lowercase text and replacing the words whose frequency is less than 3 with the
token. The filtered 1185 words constitute more than 99% of the word occurrence rate in the corpus. There are 155 independent tags in the MTI annotation of the original dataset, which is thus regarded as multi-label classification annotation. We randomly select 2000 samples for training, 678 samples for validation, and 653 samples for testing. The second dataset is Chexpert [
28], in which a total of 224,316 chest X-ray images were collected and 14 common radiographic observations were labeled. From the entire dataset, 19,811 pairs of data with frontal and lateral images are utilized for training, 6619 pairs for validation, and 6608 pairs for testing. The purpose of using this dataset is to pre-train the encoder so that the model can extract effective medical image features. We then fine-tune the model on the IU X-ray dataset.
3.2. Methods Overview
In
Figure 2, we use CVAM to process the features of multi-view medical images and to predict the tags that represent medical concepts contained in multi-view images as the semantic features of key lesions. The MVSL developed in this study receives image features and semantic features of lesions and generates a medical visual-semantic feature representing the sentence through the joint action of the two features. Using the characteristics of LSTMs, the module also records historical information to ensure information independence between diagnostic sentences. We then use the Sentence LSTM–Word LSTM to generate diagnostic reports.
3.3. Cross-View Attention Module (CVAM)
As shown in
Figure 2, we use two CNNs to extract the features of the frontal-view image and lateral-view image, respectively. The last convolutional layer yields the feature map
and
({
,
} ∈
, where
N is the
of the feature map and
D is the depth). Each feature map then enters two branches. One is a single-view branch, where the multi-classification predictions
and
of
M medical concepts are obtained by the fully connected layer. The other is a cross-view branch, as shown in
Figure 3;
and
use SE-Attention [
29] to enhance different lesion features channel-wise and then employ the following formula to complete the cross-view attention:
where
f is the SE-Attention and
is a hyperparameter of [0.5, 1], which represents how many visual features of images are retained. By introducing (
) visual features from complementary perspectives,
and
can be calculated by a fully connected layer to obtain the multi-classification predictions
and
of
M medical concepts.
We then use MSE loss to unify the learning results of the single-view and cross-view branches. The loss function is shown below:
3.4. Medical Visual-Semantic LSTMs (MVSL)
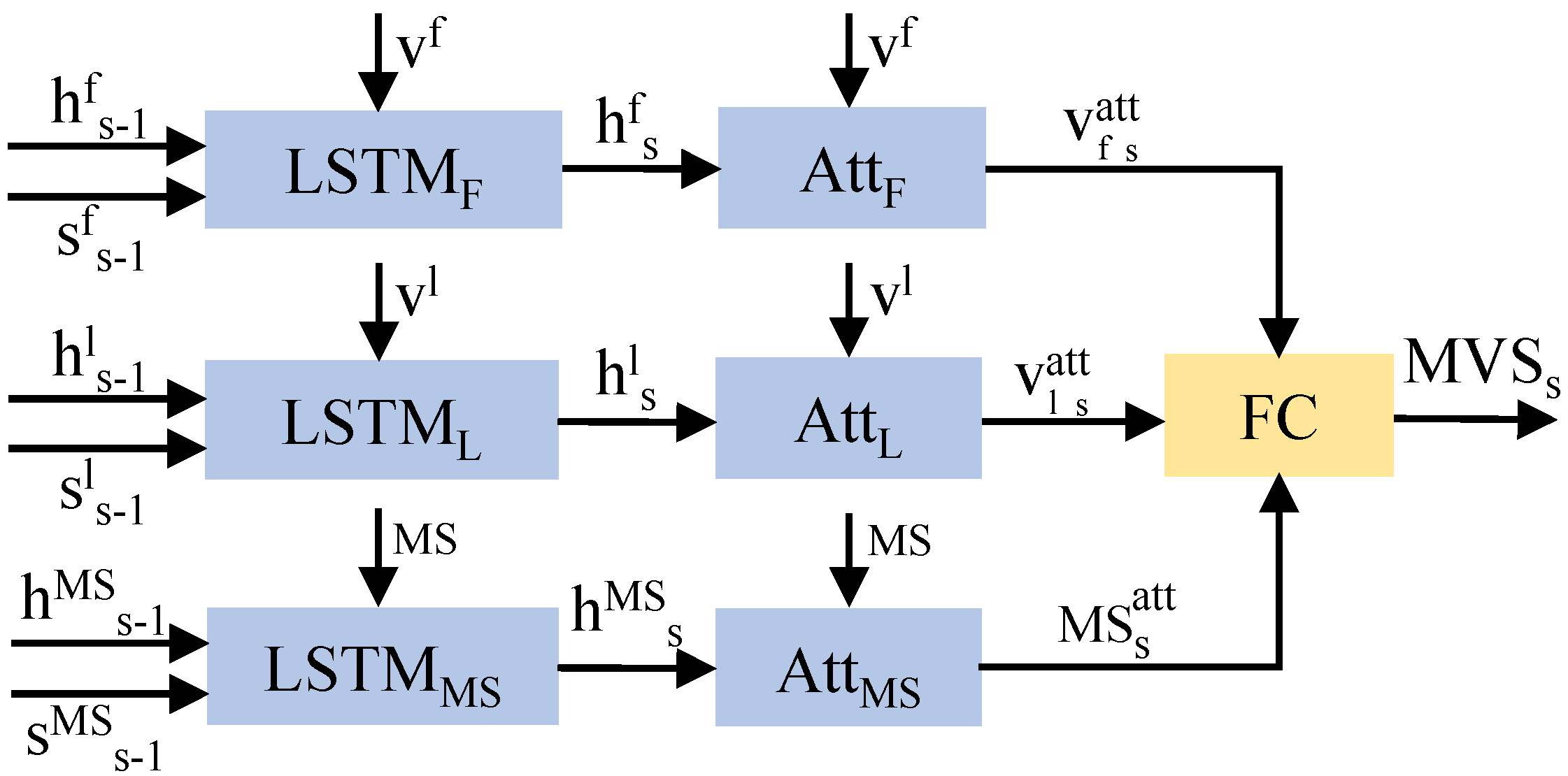
Visual features of images include information about objects and locations, and medical concepts can be directly used as the semantic information for key lesions. The fusion of these features can produce a diagnostic sentence that includes the location and type of the disease. In
Figure 2, the MVSL receives the visual features of medical images and the semantic features of lesions from CVAM. The visual features of different perspectives and branches are integrated into a visual feature vector through the fully connected layer, and the semantic features of the lesions are embedded into a semantic feature vector. As shown in
Figure 4, three LSTMs handle the visual feature vectors and semantic feature vectors, which can be defined as:
where
s refers to the diagnostic sentence that is currently being generated.
F ∈ {
,
,
} represents variables related to the frontal view, lateral view, or medical semantic feature. Then, three hidden layers are used to calculate the visual vector attention (
,
) and semantic vector attention (
):
where
is the function of softmax layer and
,
,
are parameter matrices. Then, the visual and semantic attention vectors are obtained by the following formulas:
With a fully connected layer
W, the three vectors are then integrated to obtain the medical visual-semantic feature (
):
3.5. Sentence LSTM–Word LSTM
The Sentence LSTM generates the topic of the current diagnostic sentence and the control vector of whether to continue to generate the diagnostic sentence. The topic of this sentence is generated by the hidden layer in the Sentence LSTM and medical visual-semantic feature:
where
s is the sequence number of the sentence being generated and
and
are weight parameters. The control vector is generated with the current
h and the previous
h of the LSTM:
where
,
,
are weight parameters.
The Word LSTM uses the topic and embedding vectors to continuously calculate and output words:
where
t is the
t-th word being generated in one sentence and
is the embedding vector of the input. Then, the
h of each time is used to complete the word generation:
where
,
are weight parameters.
3.6. Training Loss
The input of our model includes: (1) frontal and lateral images and ; (2) tag T for medical concepts; (3) a diagnostic report with s sentences; (4) stop signal P.
The model extracts image features from
and
by CNN. Then, the features enter the two branches, and the multi-label classification is conducted. BCE loss is used to calculate the predicted value and the loss of
T:
With the
, the loss of the encoder is formulated as:
Next, the Sentence LSTM generates the medical topic vector and control vector
s times and utilizes cross-entropy loss to calculate the loss of the control vector:
Finally, in the Word LSTM, we use cross-entropy to calculate the loss of words and ground truth of
S sentences:
Combining all the loss described above yields the total training loss:
4. Results
4.1. Evaluation Metrics
We use AUC as the evaluation metric for encoder training. AUC is a numerical value ranging from 0 to 1, representing the area under the receiver operating characteristic curve, which illustrates the relationship between the model’s true positive rate and false positive rate.
To evaluate medical report generation, we use standard image caption evaluation metrics BLEU [
30], ROUGR [
31], and CIDER [
32]. BLEU is an evaluation metric based on n-grams, where n can be 1, 2, 3, and so on. It assesses the degree of n-gram overlap between the generated text and multiple reference texts, serving to evaluate the quality of generated text that corresponds to precision in classification tasks. We use the ROUGE-L metric from the ROUGE metric family to assess the similarity between the texts in this paper. ROUGE-L is an evaluation metric based on the longest common subsequence. It calculates the length of the longest common subsequence between the generated text and the reference text to measure their similarity. CIDER is a fusion of BLEU and the vector space model. It treats each sentence as a document and calculates the cosine angle of TF-IDF vectors (with terms being n-grams instead of words), obtaining the similarity between candidate and reference sentences. Additionally, CIDER leverages inverse document frequency, enhancing the significance of pivotal vocabulary terms within the corpus.
4.2. Baselines
4.2.1. Multi-Label Classification
The first baseline is a multi-task learning model for processing the input of frontal and lateral chest radiographs. The second comparative model adds MSE loss to the multi-task learning model used in [
11]. Because of the design of single-view and cross-view in CVAM, we perform comparative experiments to verify the effectiveness of using two branches. We also investigate the effect of only using ImageNet during the fine-tuning on the IU X-ray dataset and compare this effect with those of the other models.
4.2.2. Medical Report Generation
We compare our method with several basic models of image caption and the state-of-the-art methods for automatic medical report generation. For the method TieNet [
16], CARG [
33], and SentSAT+KG [
7], we compare them with the results reported in [
7]. In addition, we reproduce HLSTM [
26] and CoAtt [
10] (the original method is changed to frontal and lateral input) for comparison. Our baseline can be regarded as a type of encoder (pre-trained on Chexpert) + Sentence LSTM + Word LSTM. Our CVAM adds CVAM to integrate multi-perspective features based on the baseline. Our CVAM+MVSL is a method for verifying the effectiveness of the proposed method.
4.3. Implementation Details
We employ ResNet50 [
34] and use the size of 256 × 256 to train the encoder. The parameter
in CVAM is 0.6 according to the experimental results shown in
Table 1. Then, the 8 × 8 × 2048 feature map is output by the last convolutional layer as medical image features. The other output of the encoder is the prediction of 155 medical concept tags. After the softmax layer, 10 prediction tags with the highest probability are selected as the semantic features of the key lesions, which are expressed by 512-dimensional word embedding. In the decoder, the dimensions of the hidden state and word embedding are set to 512. We set the Sentence LSTM to generate 6 sentences for each sample during training. Each sentence retains the first 30 words processed and uses the
token to fill in when a sentence contains fewer than 30 words.
We use the Adam optimizer for parameter learning. The learning rates of the encoder, MVSL, and Sentence LSTM–Word LSTM are set to , , and , respectively. After training the encoder on Chexpert, we fine-tune it on the IU X-ray dataset. The loss of the encoder includes BCE loss of multi-label classification and MSE loss of CVAM. The loss of the decoder includes the cross-entropy loss of the stop control variable in the Sentence LSTM and words in the Word LSTM. In the experiment, we set and in to 1 and 0.05, respectively. In , the values of , , and are 1.
4.4. Quantitative Analysis
4.4.1. Multi-Label Classification
The AUC of multi-label classification is shown in
Table 1. As can be seen in the table, the CVAM structure can achieve excellent results on both datasets. On the Chexpert dataset, our model performs 3.7% better than the baseline Multitask. A gap exists between the classification performance of the baseline for frontal and lateral medical images, but our method can effectively make up for this gap. Our naive model only uses the cross-view branch, and the performance of the model in processing frontal images is slightly reduced. When the two branches are utilized, our model can achieve the best performance. When fine-tuned on the IU X-ray dataset, the model using CVAM performs 3.1% better than Multitask–ImageNet. These results show that adding CVAM in the feature extraction stage can effectively process multi-view medical images, improve classification performance while making up for the performance gap of multi-view images, and obtain more useful lesion features for diagnosis.
4.4.2. Medical Report Generation
Table 2 shows the results of the generated report. Note that because some methods do not release the source code or lack important details, we compare the results reported in [
7]. The methods in the middle part of
Table 1 are reproduced with the same data partition as our method. Compared with the previous work, our CVAM+MVSL method can achieve better performance on BLEU-n and CIDER, although the comparison with TieNet, CARG, and SentSAT+KG may be unfair due to different experimental settings. In the reproduced HLSTM and CoAtt, we change the input to multi-view medical images in order to make a more reasonable comparison. Our CVAM+MVSL shows better performance on BLEU-n and CIDER, indicating that the diagnostic report generated by our method has a higher degree of lexical overlap with the real diagnostic report. CIDER utilizes reverse document frequency, which increases the significance of important words in the corpus. The results show that our method can effectively predict important words in the medical corpus. In ROUGE, the effect of our method is slightly reduced compared with that in CoAtt. This might be because ROUGE is a metric that considers the recall rate, which can provide more information for the report generated by our method.
4.5. Qualitative Analysis
As shown in
Figure 5, we randomly selected the visual results of automatically generated medical diagnostic reports, along with the visualization of text and visual-semantic attention. As can be seen from the figure, our CVAM+MVSL method possesses the following characteristics. Firstly, CVAM enables the model to extract more effective lesion features and predict lesions more accurately. In our model, the report generated for the first sample accurately described the symptom of “cardiomegaly”, while the report for the second sample correctly depicted “scarring” and “emphysema”, which CoAtt failed to recognize. We believe that this improvement is due to the utilization of three-dimensional information provided by both frontal and lateral views. Secondly, MVSL iteratively selects different visual and semantic features to generate more diverse diagnostic reports. For the third sample without diseases, our method generates valuable descriptions beyond conventional descriptions, such as the diagnosis of “cardiac mediastinal contour and pulmonary vascular system”. Finally, our method can make full use of the joint and complementary information of multi-view medical images to complete the diagnosis. The features of “cardiac hypertrophy” are captured in the frontal and lateral view of the second sample. When the third sample is generated to describe the “visible bone structure of the chest”, the lateral image, instead of the frontal image, captures the bone structure characteristics.
4.6. Clinical Validation
We invited six radiologists to evaluate the diagnostic reports generated by our method and designed a questionnaire with five questions concerning “fluency of generated reports”, “comprehensiveness of generated reports”, “accuracy of generated reports in describing diseased sites”, “accuracy of attention map in capturing diseased sites”, and “utility of automatic generation of diagnostic reports”. The response to each question was set with a score ranging from 0 to 10, with 0 being the worst and 10 the most satisfactory. As
Table 3 shows, radiologists gave a high evaluation to the fluency of the generated report and the effect of the algorithm. The lowest score is for “comprehensiveness of report generation”. This can be attributed to the limitation of the IU X-ray dataset, as the model cannot effectively generate the words outside the dataset well. However, this problem can be alleviated by using more diagnostic texts to pre-train the decoder. The scores of the third and fourth questions are not bad; we think that the encoder focuses on the discriminant features when extracting features and thus cannot effectively capture all the key features. This can be solved by using methods proposed by [
35]. The average ICC [
36] value of the user study is 0.966, which confirms the consistency and reliability of our results (greater than 0.75).
5. Conclusions
We propose a novel automatic report generation approach composed of Cross-View Attention Module (CVAM) and Medical Visual-Semantic LSTMs (MVSL). The CVAM integrates multi-view medical image features to provide more effective visual features of images and semantic features of key lesions. The MVSL can integrate visual and semantic data to provide discriminative information for diagnostic report generation. Applied to the open-source IU X-ray dataset, our model achieved an average improvement of over the state of the art (SOTA) on six evaluation metrics. This demonstrates that our model is capable of generating more detailed and accurate diagnostic reports. We attribute this improvement to the introduction of multi-perspective information, which enables the model to focus on pathological changes that cannot be captured by a single perspective. In the future, we plan to use the weakly supervised localization method to extract more discriminative features from the encoder and use more medical texts to pre-train the decoder so that the model can generate more varied sentences.
Author Contributions
Conceptualization, Y.G.; methodology, Y.G. and X.W.; software, R.L.; validation, X.W.; formal analysis, R.L. and X.W.; investigation, R.L.; resources, Z.Z.; data curation, R.L. and X.W.; writing—original draft preparation, R.L.; writing—review and editing, Y.G.; visualization, X.W.; supervision, Y.G.; project administration, Z.Z.; funding acquisition, Y.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC were funded by Technological Innovation 2030—“New Generation Artificial Intelligence” Major Project, grant number 2022ZD0161902 and CAMS Innovation Fund for Medical Sciences (CIFMS), grant number 2019-I2M-5-016.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
In this paper, the authors wish to gratefully acknowledge CAMS Innovation Fund for Medical Sciences (CIFMS) under Grant 2019-I2M-5-016.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lu, J.; Xiong, C.; Parikh, D.; Socher, R. Knowing when to look: Adaptive attention via a visual sentinel for image captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 375–383. [Google Scholar]
- Chen, L.; Zhang, H.; Xiao, J.; Nie, L.; Shao, J.; Liu, W.; Chua, T.S. Sca-cnn: Spatial and channel-wise attention in convolutional networks for image captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5659–5667. [Google Scholar]
- Feng, Y.; Ma, L.; Liu, W.; Luo, J. Unsupervised image captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 4125–4134. [Google Scholar]
- Wang, W.; Chen, Z.; Hu, H. Hierarchical attention network for image captioning. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 8957–8964. [Google Scholar]
- Wang, Z.; Huang, Z.; Luo, Y. Human Consensus-Oriented Image Captioning. In Proceedings of the Twenty-Ninth International Conference on International Joint Conferences on Artificial Intelligence, Yokohama, Japan, 7–15 January 2021; pp. 659–665. [Google Scholar]
- Vinyals, O.; Toshev, A.; Bengio, S.; Erhan, D. Show and tell: A neural image caption generator. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3156–3164. [Google Scholar]
- Zhang, Y.; Wang, X.; Xu, Z.; Yu, Q.; Yuille, A.; Xu, D. When Radiology Report Generation Meets Knowledge Graph. arXiv 2020, arXiv:2002.08277. [Google Scholar] [CrossRef]
- Xue, Y.; Xu, T.; Long, L.R.; Xue, Z.; Antani, S.; Thoma, G.R.; Huang, X. Multimodal recurrent model with attention for automated radiology report generation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 457–466. [Google Scholar]
- Li, C.Y.; Liang, X.; Hu, Z.; Xing, E.P. Knowledge-driven encode, retrieve, paraphrase for medical image report generation. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 6666–6673. [Google Scholar]
- Jing, B.; Xie, P.; Xing, E. On the automatic generation of medical imaging reports. arXiv 2017, arXiv:1711.08195. [Google Scholar]
- Yuan, J.; Liao, H.; Luo, R.; Luo, J. Automatic radiology report generation based on multi-view image fusion and medical concept enrichment. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 721–729. [Google Scholar]
- Xie, X.; Niu, J.; Liu, X.; Chen, Z.; Tang, S. A Survey on Domain Knowledge Powered Deep Learning for Medical Image Analysis. arXiv 2020, arXiv:2004.12150. [Google Scholar]
- Meng, Q.; Shin’ichi, S. ADINet: Attribute Driven Incremental Network for Retinal Image Classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 14–19 June 2020; pp. 4033–4042. [Google Scholar]
- Zhou, H.Y.; Yu, S.; Bian, C.; Hu, Y.; Ma, K.; Zheng, Y. Comparing to learn: Surpassing imagenet pretraining on radiographs by comparing image representations. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 398–407. [Google Scholar]
- Pavlopoulos, J.; Kougia, V.; Androutsopoulos, I. A Survey on Biomedical Image Captioning. In Proceedings of the Second Workshop on Shortcomings in Vision and Language, Minneapolis, MN, USA, 6 June 2019; pp. 26–36. [Google Scholar]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Summers, R.M. Tienet: Text-image embedding network for common thorax disease classification and reporting in chest X-rays. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018; pp. 9049–9058. [Google Scholar]
- Li, Y.; Liang, X.; Hu, Z.; Xing, E.P. Hybrid retrieval-generation reinforced agent for medical image report generation. In Proceedings of the Neural Information Processing Systems (NIPS), Montréal, QC, Canada, 3–8 December 2018; pp. 1530–1540. [Google Scholar]
- Portet, F.; Reiter, E.; Gatt, A.; Hunter, J.; Sripada, S.; Freer, Y.; Sykes, C. Automatic generation of textual summaries from neonatal intensive care data. Artif. Intell. 2009, 173, 789–816. [Google Scholar] [CrossRef]
- van Doorn, S.C.; van Vliet, J.; Fockens, P.; Dekker, E. A novel colonoscopy reporting system enabling quality assurance. Endoscopy 2014, 46, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Ba, J.; Kiros, R.; Cho, K.; Courville, A.; Salakhudinov, R.; Zemel, R.; Bengio, Y. Show, attend and tell: Neural image caption generation with visual attention. In Proceedings of the International Conference on Machine Learnin (ICML), Lille, France, 6–11 July 2015; pp. 2048–2057. [Google Scholar]
- Alahmadi, R.; Park, C.H.; Hahn, J.K. Sequence-to-sequence image caption generator. In Proceedings of the International Conference on Machine Vision, Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Shin, H.C.; Roberts, K.; Lu, L.; Demner-Fushman, D.; Yao, J.; Summers, R.M. Learning to Read Chest X-rays: Recurrent Neural Cascade Model for Automated Image Annotation. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2497–2506. [Google Scholar]
- Singh, S.; Karimi, S.; Ho-Shon, K.; Hamey, L. From Chest X-rays to Radiology Reports: A Multimodal Machine Learning Approach. In Proceedings of the 2019 Digital Image Computing: Techniques and Applications (DICTA), Perth, Australia, 2–4 December 2019; pp. 1–8. [Google Scholar]
- Harzig, P.; Chen, Y.Y.; Chen, F.; Lienhart, R. Addressing Data Bias Problems for Chest X-ray Image Report Generation. arXiv 2019, arXiv:1908.02123. [Google Scholar]
- Nooralahzadeh, F.; Gonzalez, N.A.P.; Frauenfelder, T.; Fujimoto, K.; Krauthammer, M. Progressive Transformer-Based Generation of Radiology Reports. arXiv 2021, arXiv:2102.09777. [Google Scholar]
- Krause, J.; Johnson, J.; Krishna, R.; Fei-Fei, L. A hierarchical approach for generating descriptive image paragraphs. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 317–325. [Google Scholar]
- Demner-Fushman, D.; Kohli, M.D.; Rosenman, M.B.; Shooshan, S.E.; Rodriguez, L.; Antani, S.; Thoma, G.R.; McDonald, C.J. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 2016, 23, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 590–597. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Papineni, K.; Roukos, S.; Ward, T.; Zhu, W.J. BLEU: A method for automatic evaluation of machine translation. In Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics (ACL), Philadelphia, PA, USA, 6–12 July 2002; pp. 311–318. [Google Scholar]
- Lin, C.Y. Rouge: A package for automatic evaluation of summaries. In Proceedings of the Text Summarization Branches Out, Barcelona, Spain, 25–26 July 2004; pp. 74–81. [Google Scholar]
- Vedantam, R.; Lawrence Zitnick, C.; Parikh, D. Cider: Consensus-based image description evaluation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 4566–4575. [Google Scholar]
- Liu, G.; Hsu, T.M.H.; McDermott, M.; Boag, W.; Weng, W.H.; Szolovits, P.; Ghassemi, M. Clinically accurate chest X-ray report generation. arXiv 2019, arXiv:1904.02633. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Choe, J.; Shim, H. Attention-based dropout layer for weakly supervised object localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 2219–2228. [Google Scholar]
- Weir, J.P. Quantifying test-retest reliability using the intraclass correlation coefficient and the SEM. J. Strength Cond. Res. 2005, 19, 231–240. [Google Scholar] [PubMed]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).