Recent Advancements in Deep Learning Using Whole Slide Imaging for Cancer Prognosis
Abstract
1. Introduction
2. Literature Analysis
2.1. Methodology for Paper Selection
2.2. Analysis of Publications
2.3. Publication Journals
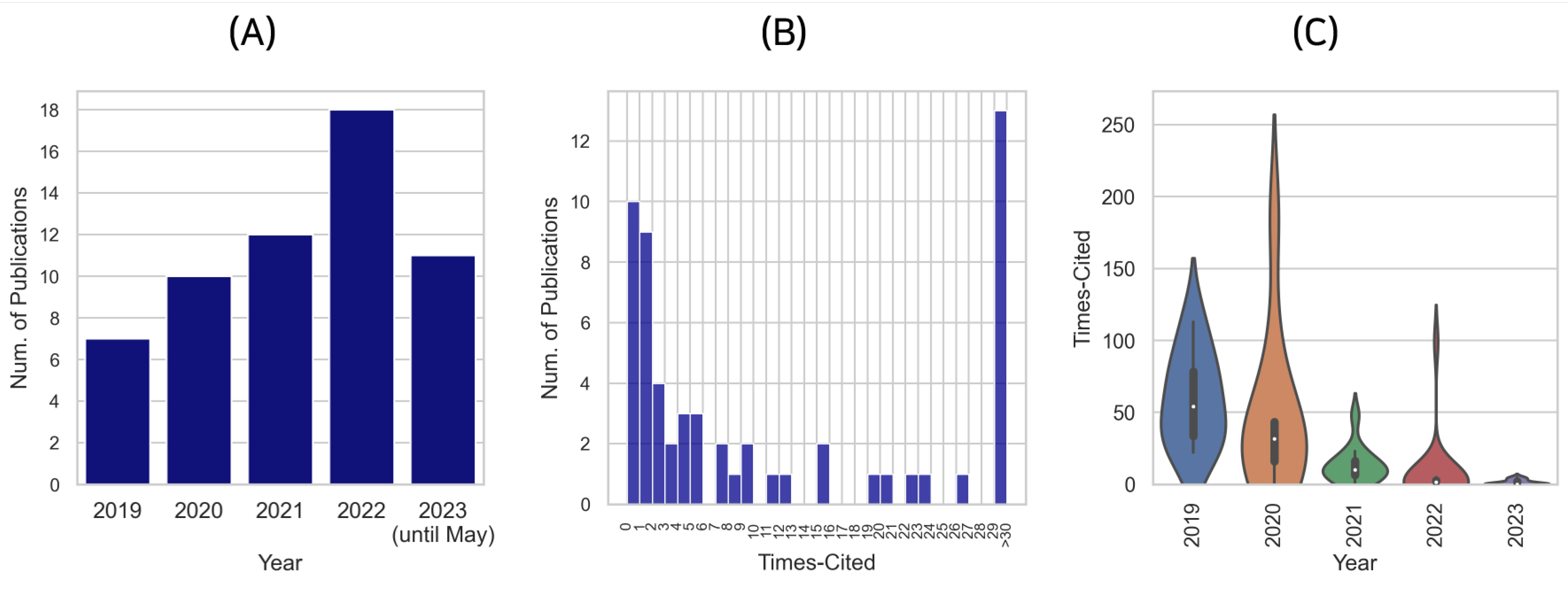
2.4. Publication Years
2.5. Citation Distribution of the Publications
3. Deep Learning with Whole Slide Images in Studies on Cancer Prognosis
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saharia, C.; Chan, W.; Saxena, S.; Li, L.; Whang, J.; Denton, E.L.; Ghasemipour, K.; Gontijo Lopes, R.; Karagol Ayan, B.; Salimans, T.; et al. Photorealistic text-to-image diffusion models with deep language understanding. Adv. Neural Inf. Process. Syst. 2022, 35, 36479–36494. [Google Scholar]
- Dhariwal, P.; Nichol, A. Diffusion models beat gans on image synthesis. Adv. Neural Inf. Process. Syst. 2021, 34, 8780–8794. [Google Scholar]
- Ko, K.; Lee, M. ZIGNeRF: Zero-shot 3D Scene Representation with Invertible Generative Neural Radiance Fields. arXiv 2023, arXiv:2306.02741. [Google Scholar]
- Cai, L.; Gao, J.; Zhao, D. A review of the application of deep learning in medical image classification and segmentation. Ann. Transl. Med. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, M. Deep Learning Approaches for lncRNA-Mediated Mechanisms: A Comprehensive Review of Recent Developments. Int. J. Mol. Sci. 2023, 24, 10299. [Google Scholar] [CrossRef]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A review of deep-learning-based medical image segmentation methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Tran, K.A.; Kondrashova, O.; Bradley, A.; Williams, E.D.; Pearson, J.V.; Waddell, N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021, 13, 152. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Q.; Lam, S.; Cai, J.; Yang, R. A review on application of deep learning algorithms in external beam radiotherapy automated treatment planning. Front. Oncol. 2020, 10, 580919. [Google Scholar] [CrossRef]
- Kim, J.; Lee, M.; Seok, J. Deep learning model with L1 penalty for predicting breast cancer metastasis using gene expression data. Mach. Learn. Sci. Technol. 2023, 4, 025026. [Google Scholar] [CrossRef]
- Dimitriou, N.; Arandjelović, O.; Caie, P.D. Deep learning for whole slide image analysis: An overview. Front. Med. 2019, 6, 264. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Quiroga-Garza, G.M.; Bien, L.; Heled, R.; Laifenfeld, D.; Linhart, C.; Sandbank, J.; Shach, A.A.; Shalev, V.; Vecsler, M.; et al. An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: A blinded clinical validation and deployment study. Lancet Digit. Health 2020, 2, e407–e416. [Google Scholar] [CrossRef] [PubMed]
- Hanna, M.G.; Reuter, V.E.; Hameed, M.R.; Tan, L.K.; Chiang, S.; Sigel, C.; Hollmann, T.; Giri, D.; Samboy, J.; Moradel, C.; et al. Whole slide imaging equivalency and efficiency study: Experience at a large academic center. Mod. Pathol. 2019, 32, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, N.; Elharrouss, O.; Al-Maadeed, S.; Chowdhury, M. A review of deep learning-based detection methods for COVID-19. Comput. Biol. Med. 2022, 143, 105233. [Google Scholar] [CrossRef]
- Lakshmanna, K.; Kaluri, R.; Gundluru, N.; Alzamil, Z.S.; Rajput, D.S.; Khan, A.A.; Haq, M.A.; Alhussen, A. A review on deep learning techniques for IoT data. Electronics 2022, 11, 1604. [Google Scholar] [CrossRef]
- Zheng, W.; Tian, X.; Yang, B.; Liu, S.; Ding, Y.; Tian, J.; Yin, L. A few shot classification methods based on multiscale relational networks. Appl. Sci. 2022, 12, 4059. [Google Scholar] [CrossRef]
- Lee, M. Deep Learning Techniques with Genomic Data in Cancer Prognosis: A Comprehensive Review of the 2021–2023 Literature. Biology 2023, 12, 893. [Google Scholar] [CrossRef]
- Kim, J.; Lee, M. Portfolio optimization using predictive auxiliary classifier generative adversarial networks. Eng. Appl. Artif. Intell. 2023, 125, 106739. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Valenstein, P.N.; Evans, A.J.; Kaplan, K.J.; Pfeifer, J.D.; Wilbur, D.C.; Collins, L.C.; Colgan, T.J. Review of the current state of whole slide imaging in pathology. J. Pathol. Inform. 2011, 2, 36. [Google Scholar] [CrossRef]
- Ghaznavi, F.; Evans, A.; Madabhushi, A.; Feldman, M. Digital imaging in pathology: Whole-slide imaging and beyond. Annu. Rev. Pathol. Mech. Dis. 2013, 8, 331–359. [Google Scholar] [CrossRef]
- Farahani, N.; Parwani, A.V.; Pantanowitz, L. Whole slide imaging in pathology: Advantages, limitations, and emerging perspectives. Pathol. Lab. Med. Int. 2015, 23–33. [Google Scholar]
- The Cancer Genome Atlas. Available online: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (accessed on 28 May 2023).
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.N.; Tomaszewski, J.; González, F.A.; Madabhushi, A. Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent. Sci. Rep. 2017, 7, 46450. [Google Scholar] [CrossRef] [PubMed]
- Kwok, S. Multiclass classification of breast cancer in whole-slide images. In Proceedings of the Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, 27–29 June 2018; Proceedings 15. Springer: Berlin/Heidelberg, Germany, 2018; pp. 931–940. [Google Scholar]
- Li, R.; Yao, J.; Zhu, X.; Li, Y.; Huang, J. Graph CNN for survival analysis on whole slide pathological images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 174–182. [Google Scholar]
- Ronaldson-Bouchard, K.; Baldassarri, I.; Tavakol, D.N.; Graney, P.L.; Samaritano, M.; Cimetta, E.; Vunjak-Novakovic, G. Engineering complexity in human tissue models of cancer. Adv. Drug Deliv. Rev. 2022, 184, 114181. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, Z.; Yao, S.; Wang, Y.; Wu, X.; Xu, Z.; Wu, L.; Huang, Y.; Liang, C.; Liu, Z. Artificial intelligence quantified tumour-stroma ratio is an independent predictor for overall survival in resectable colorectal cancer. Ebiomedicine 2020, 61. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Wu, L.; Huang, Y.; Yao, S.; Xu, Z.; Lin, H.; Wang, H.; Liang, Y.; Xu, Y.; Chen, X.; et al. Deep learning quantified mucus-tumor ratio predicting survival of patients with colorectal cancer using whole-slide images. Precis. Clin. Med. 2021, 4, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, B.; Wei, G.; Qiu, W.; Li, D.; Li, X.; Liu, X.; Wei, W.; Wang, S.; Liu, Z.; et al. Deep learning with whole slide images can improve the prognostic risk stratification with III colorectal cancer. Comput. Methods Programs Biomed. 2022, 221, 106914. [Google Scholar] [CrossRef]
- Yang, J.; Ye, H.; Fan, X.; Li, Y.; Wu, X.; Zhao, M.; Hu, Q.; Ye, Y.; Wu, L.; Li, Z.; et al. Artificial intelligence for quantifying immune infiltrates interacting with stroma in colorectal cancer. J. Transl. Med. 2022, 20, 451. [Google Scholar] [CrossRef]
- Xu, Z.; Li, Y.; Wang, Y.; Zhang, S.; Huang, Y.; Yao, S.; Han, C.; Pan, X.; Shi, Z.; Mao, Y.; et al. A deep learning quantified stroma-immune score to predict survival of patients with stage II-III colorectal cancer. Cancer Cell Int. 2021, 21, 585. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, J.; Qian, C.; Fei, S. Deep learning-based tumor microenvironment analysis in colon adenocarcinoma histopathological whole-slide images. Comput. Methods Programs Biomed. 2021, 204, 106047. [Google Scholar]
- Xu, H.; Cha, Y.J.; Clemenceau, J.R.; Choi, J.; Lee, S.H.; Kang, J.; Hwang, T.H. Spatial analysis of tumor-infiltrating lymphocytes in histological sections using deep learning techniques predicts survival in colorectal carcinoma. J. Pathol. Clin. Res. 2022, 8, 327–339. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, L.; Chen, W.; Huang, S.; Liu, Z.; Zhang, J. Computer-aided detection and prognosis of colorectal cancer on whole slide images using dual resolution deep learning. J. Cancer Res. Clin. Oncol. 2023, 149, 91–101. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, S.; Zhu, Y.; Yao, S.; Li, Y.; Ye, H.; Ye, Y.; Li, Z.; Wu, L.; Zhao, K.; et al. Artificial intelligence for quantifying Crohn’s-like lymphoid reaction and tumor-infiltrating lymphocytes in colorectal cancer. Comput. Struct. Biotechnol. J. 2022, 20, 5586–5594. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, M.; Wang, J.; Xu, M.; Hu, W.; Wee, L.; Dekker, A.; Sheng, W.; Zhang, Z. Automatic Tumor Grading on Colorectal Cancer Whole-Slide Images: Semi-Quantitative Gland Formation Percentage and New Indicator Exploration. Front. Oncol. 2022, 12, 833978. [Google Scholar] [CrossRef] [PubMed]
- Geessink, O.G.F.; Baidoshvili, A.; Klaase, J.M.; Bejnordi, B.E.; Litjens, G.J.S.; van Pelt, G.W.; Mesker, W.E.; Nagtegaal, I.D.; Ciompi, F.; van der Laak, J.A.W.M. Computer aided quantification of intratumoral stroma yields an independent prognosticator in rectal cancer. Cell. Oncol. 2019, 42, 331–341. [Google Scholar] [CrossRef]
- Shapcott, M.; Hewitt, K.J.; Rajpoot, N. Deep Learning With Sampling in Colon Cancer Histology. Front. Bioeng. Biotechnol. 2019, 7, 52. [Google Scholar] [CrossRef]
- Liu, H.; Kurc, T. Deep learning for survival analysis in breast cancer with whole slide image data. Bioinformatics 2022, 38, 3629–3637. [Google Scholar] [CrossRef] [PubMed]
- Jaber, M.I.; Song, B.; Taylor, C.; Vaske, C.J.; Benz, S.C.; Rabizadeh, S.; Soon-Shiong, P.; Szeto, C.W. A deep learning image-based intrinsic molecular subtype classifier of breast tumors reveals tumor heterogeneity that may affect survival. Breast Cancer Res. 2020, 22, 12. [Google Scholar] [CrossRef]
- Balkenhol, M.C.A.; Ciompi, F.; Swiderska-Chadaj, Z.; van de Loo, R.; Intezar, M.; Otte-Holler, I.; Geijs, D.; Lotz, J.; Weiss, N.; de Bel, T.; et al. Optimized tumour infiltrating lymphocyte assessment for triple negative breast cancer prognostics. Breast 2021, 56, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.Y.; Perez-Losada, J.; Abad, M.; Rodriguez-Gonzalez, M.; Rodriguez, C.A.; Mao, J.H.; Chang, H. iCEMIGE: Integration of CEll-morphometrics, MIcrobiome, and GEne biomarker signatures for risk stratification in breast cancers. World J. Clin. Oncol. 2022, 13, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Li, Y.; Wu, F.; Cao, S.; Ye, F. Predicting the prognosis of HER2-positive breast cancer patients by fusing pathological whole slide images and clinical features using multiple instance learning. Math. Biosci. Eng. 2023, 20, 11196–11211. [Google Scholar] [CrossRef]
- Levy-Jurgenson, A.; Tekpli, X.; Kristensen, V.N.; Yakhini, Z. Spatial transcriptomics inferred from pathology whole-slide images links tumor heterogeneity to survival in breast and lung cancer. Sci. Rep. 2020, 10, 18802. [Google Scholar] [CrossRef]
- Fassler, D.J.; Torre-Healy, L.A.; Gupta, R.; Hamilton, A.M.; Kobayashi, S.; Van Alsten, S.C.; Zhang, Y.; Kurc, T.; Moffitt, R.A.; Troester, M.A.; et al. Spatial Characterization of Tumor-Infiltrating Lymphocytes and Breast Cancer Progression. Cancers 2022, 14, 2148. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, S.; Shao, W.; Wu, Y.; Zhang, J.; Han, Z.; Feng, Q.; Huang, K. Deep-Learning-Based Characterization of Tumor-Infiltrating Lymphocytes in Breast Cancers From Histopathology Images and Multiomics Data. JCO Clin. Cancer Inform. 2020, 4, 480–490. [Google Scholar] [CrossRef] [PubMed]
- du Terrail, J.O.; Leopold, A.; Joly, C.; Beguier, C.; Andreux, M.; Maussion, C.; Schmauch, B.; Tramel, E.W.; Bendjebbar, E.; Zaslavskiy, M.; et al. Federated learning for predicting histological response to neoadjuvant chemotherapy in triple-negative breast cancer. Nat. Med. 2023, 29, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Lu, Z.; Shao, W.; Yu, C.Y.; Reiter, J.L.; Feng, Q.; Feng, W.; Huang, K.; Liu, Y. Integrative analysis of histopathological images and chromatin accessibility data for estrogen receptor-positive breast cancer. BMC Med. Genom. 2020, 13, 195. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Huang, S.; Zhu, X.; Cheng, L.; Liu, W.; Chen, Q.; Yang, D. Artificial Intelligence Meets Whole Slide Images: Deep Learning Model Shapes an Immune-Hot Tumor and Guides Precision Therapy in Bladder Cancer. J. Oncol. 2022, 2022. [Google Scholar] [CrossRef]
- Zheng, Q.; Yang, R.; Ni, X.; Yang, S.; Jiao, P.; Wu, J.; Xiong, L.; Wang, J.; Jian, J.; Jiang, Z.; et al. Quantitative Assessment of Tumor-Infiltrating Lymphocytes Using Machine Learning Predicts Survival in Muscle-Invasive Bladder Cancer. J. Clin. Med. 2022, 11, 7081. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Yang, R.; Ni, X.; Yang, S.; Xiong, L.; Yan, D.; Xia, L.; Yuan, J.; Wang, J.; Jiao, P.; et al. Accurate Diagnosis and Survival Prediction of Bladder Cancer Using Deep Learning on Histological Slides. Cancers 2022, 14, 5807. [Google Scholar] [CrossRef]
- Gavriel, C.G.; Dimitriou, N.; Brieu, N.; Nearchou, I.P.; Arandjelovic, O.; Schmidt, G.; Harrison, D.J.; Caie, P.D. Assessment of Immunological Features in Muscle-Invasive Bladder Cancer Prognosis Using Ensemble Learning. Cancers 2021, 13, 1624. [Google Scholar] [CrossRef]
- Brieu, N.; Gavriel, C.G.; Nearchou, I.P.; Harrison, D.J.; Schmidt, G.; Caie, P.D. Automated tumour budding quantification by machine learning augments TNM staging in muscle-invasive bladder cancer prognosis. Sci. Rep. 2019, 9, 5174. [Google Scholar] [CrossRef]
- Wu, P.; Wu, K.; Li, Z.; Liu, H.; Yang, K.; Zhou, R.; Zhou, Z.; Xing, N.; Wu, S. Multimodal investigation of bladder cancer data based on computed tomography, whole slide imaging, and transcriptomics. Quant. Imaging Med. Surg. 2023, 13, 1023. [Google Scholar] [CrossRef] [PubMed]
- Saillard, C.; Schmauch, B.; Laifa, O.; Moarii, M.; Toldo, S.; Zaslavskiy, M.; Pronier, E.; Laurent, A.; Amaddeo, G.; Regnault, H.; et al. Predicting Survival After Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology 2020, 72, 2000–2013. [Google Scholar] [CrossRef]
- Liang, J.; Zhang, W.; Yang, J.; Wu, M.; Dai, Q.; Yin, H.; Xiao, Y.; Kong, L. Deep learning supported discovery of biomarkers for clinical prognosis of liver cancer. Nat. Mach. Intell. 2023, 5, 408. [Google Scholar] [CrossRef]
- Qu, W.F.; Tian, M.X.; Qiu, J.T.; Guo, Y.C.; Tao, C.Y.; Liu, W.R.; Tang, Z.; Qian, K.; Wang, Z.X.; Li, X.Y.; et al. Exploring pathological signatures for predicting the recurrence of early-stage hepatocellular carcinoma based on deep learning. Front. Oncol. 2022, 12, 968202. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Jia, X.; Xie, Y.; Qin, W. Integrative Histology-Genomic Analysis Predicts Hepatocellular Carcinoma Prognosis Using Deep Learning. Genes 2022, 13, 1770. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.Y.; Wang, X.; Ding, G.Y.; Dong, Z.; Han, J.; Guan, Z.; Ma, L.J.; Zheng, Y.; Zhang, L.; Yu, G.Z.; et al. Exploring prognostic indicators in the pathological images of hepatocellular carcinoma based on deep learning. Gut 2021, 70, 951–961. [Google Scholar] [CrossRef]
- Pham, H.H.N.; Futakuchi, M.; Bychkov, A.; Furukawa, T.; Kuroda, K.; Fukuoka, J. Detection of Lung Cancer Lymph Node Metastases from Whole-Slide Histopathologic Images Using a Two-Step Deep Learning Approach. Am. J. Pathol. 2019, 189, 2428–2439. [Google Scholar] [CrossRef]
- Shim, W.S.; Yim, K.; Kim, T.J.; Sung, Y.E.; Lee, G.; Hong, J.H.; Chun, S.H.; Kim, S.; An, H.J.; Na, S.J.; et al. DeepRePath: Identifying the Prognostic Features of Early-Stage Lung Adenocarcinoma Using Multi-Scale Pathology Images and Deep Convolutional Neural Networks. Cancers 2021, 13, 3308. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Diao, L.; Zhou, X.; Chen, J.N.; Zhou, Y.; Fang, Q.; He, Y.; Dziadziuszko, R.; Zhou, C.; Hirsch, F.R. Artificial intelligence-based analysis for immunohistochemistry staining of immune checkpoints to predict resected non-small cell lung cancer survival and relapse. Transl. Lung Cancer Res. 2021, 10, 2452. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, X.; Lin, H.; Han, C.; An, Y.; Qiu, B.; Feng, Z.; Huang, X.; Xu, Z.; Shi, Z.; et al. Multi-scale pathology image texture signature is a prognostic factor for resectable lung adenocarcinoma: A multi-center, retrospective study. J. Transl. Med. 2022, 20, 595. [Google Scholar] [CrossRef]
- Zadeh Shirazi, A.; Fornaciari, E.; Bagherian, N.S.; Ebert, L.M.; Koszyca, B.; Gomez, G.A. DeepSurvNet: Deep survival convolutional network for brain cancer survival rate classification based on histopathological images. Med. Biol. Eng. Comput. 2020, 58, 1031–1045. [Google Scholar] [CrossRef]
- Chen, R.J.; Lu, M.Y.; Wang, J.; Williamson, D.F.K.; Rodig, S.J.; Lindeman, I.N.; Mahmood, F. Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis. IEEE Trans. Med. Imaging 2022, 41, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.P.; Jin, X.; Ahmadian, S.S.; Yang, X.; Tian, S.F.; Cai, Y.X.; Chawla, K.; Snijders, A.M.; Xia, Y.; van Diest, P.J.; et al. Clinical significance and molecular annotation of cellular morphometric subtypes in lower-grade gliomas discovered by machine learning. Neuro-Oncol. 2023, 25, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zanazzi, G.J.; Hassanpour, S. Predicting prognosis and IDH mutation status for patients with lower-grade gliomas using whole slide images. Sci. Rep. 2021, 11, 16849. [Google Scholar] [CrossRef] [PubMed]
- Tabibu, S.; Vinod, P.K.; Jawahar, C.V. Pan-Renal Cell Carcinoma classification and survival prediction from histopathology images using deep learning. Sci. Rep. 2019, 9, 10509. [Google Scholar] [CrossRef]
- Marostica, E.; Barber, R.; Denize, T.; Kohane, I.S.; Signoretti, S.; Golden, J.A.; Yu, K.H. Development of a Histopathology Informatics Pipeline for Classification and Prediction of Clinical Outcomes in Subtypes of Renal Cell Carcinoma. Clin. Cancer Res. 2021, 27, 2868–2878. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, Z.; Yan, Y.; Wang, K.; Wang, A.; Ye, X.; Wang, L.; Wei, W.; Li, B.; Sun, C.; et al. Development of Prognostic Biomarkers by TMB-Guided WSI Analysis: A Two-Step Approach. IEEE J. Biomed. Health Inform. 2023, 27, 1780–1789. [Google Scholar] [CrossRef]
- Yokomizo, R.; Lopes, T.J.S.; Takashima, N.; Hirose, S.; Kawabata, A.; Takenaka, M.; Iida, Y.; Yanaihara, N.; Yura, K.; Sago, H.; et al. O3C Glass-Class: A Machine-Learning Framework for Prognostic Prediction of Ovarian Clear-Cell Carcinoma. Bioinform. Biol. Insights 2022, 16, 11779322221134312. [Google Scholar] [CrossRef]
- Liu, T.; Su, R.; Sun, C.; Li, X.; Wei, L. EOCSA: Predicting prognosis of Epithelial ovarian cancer with whole slide histopathological images. Expert Syst. Appl. 2022, 206, 117643. [Google Scholar] [CrossRef]
- Wu, M.; Zhu, C.; Yang, J.; Cheng, S.; Yang, X.; Gu, S.; Xu, S.; Wu, Y.; Shen, W.; Huang, S.; et al. Exploring prognostic indicators in the pathological images of ovarian cancer based on a deep survival network. Front. Genet. 2023, 13, 1069673. [Google Scholar] [CrossRef]
- Chen, C.; Cao, Y.; Li, W.; Liu, Z.; Liu, P.; Tian, X.; Sun, C.; Wang, W.; Gao, H.; Kang, S.; et al. The pathological risk score: A new deep learning-based signature for predicting survival in cervical cancer. Cancer Med. 2023, 12, 1051–1063. [Google Scholar] [CrossRef]
- Ma, B.; Guo, Y.; Hu, W.; Yuan, F.; Zhu, Z.; Yu, Y.; Zou, H. Artificial Intelligence-Based Multiclass Classification of Benign or Malignant Mucosal Lesions of the Stomach. Front. Pharmacol. 2020, 11, 572372. [Google Scholar] [CrossRef] [PubMed]
- Klimov, S.; Xue, Y.; Gertych, A.; Graham, R.P.; Jiang, Y.; Bhattarai, S.; Pandol, S.J.; Rakha, E.A.; Reid, M.D.; Aneja, R. Predicting Metastasis Risk in Pancreatic Neuroendocrine Tumors Using Deep Learning Image Analysis. Front. Oncol. 2021, 10, 593211. [Google Scholar] [CrossRef] [PubMed]
- Knuutila, J.S.; Riihila, P.; Karlsson, A.; Tukiainen, M.; Talve, L.; Nissinen, L.; Kahari, V.M. Identification of metastatic primary cutaneous squamous cell carcinoma utilizing artificial intelligence analysis of whole slide images. Sci. Rep. 2022, 12, 9876. [Google Scholar] [CrossRef]
- Zhang, F.; Zhong, L.Z.; Zhao, X.; Dong, D.; Yao, J.J.; Wang, S.Y.; Liu, Y.; Zhu, D.; Wang, Y.; Wang, G.J.; et al. A deep-learning-based prognostic nomogram integrating microscopic digital pathology and macroscopic magnetic resonance images in nasopharyngeal carcinoma: A multi-cohort study. Ther. Adv. Med. Oncol. 2020, 12, 1758835920971416. [Google Scholar] [CrossRef]
- Ho, D.J.; Agaram, N.P.; Suser, S.D.; Chu, C.; Vanderbilt, C.M.; Meyers, P.A.; Wexler, L.H.; Healey, J.H.; Fuchs, T.J.; Hameed, M.R. Deep Learning-Based Objective and Reproducible Osteosarcoma Chemotherapy Response Assessment and Outcome Prediction. Am. J. Pathol. 2023, 193, 341–349. [Google Scholar] [CrossRef]
- Shaban, M.; Khurram, S.A.; Fraz, M.M.; Alsubaie, N.; Masood, I.; Mushtaq, S.; Hassan, M.; Loya, A.; Rajpoot, N.M. A Novel Digital Score for Abundance of Tumour Infiltrating Lymphocytes Predicts Disease Free Survival in Oral Squamous Cell Carcinoma. Sci. Rep. 2019, 9, 13341. [Google Scholar] [CrossRef]
- Shao, W.; Wang, T.; Huang, Z.; Han, Z.; Zhang, J.; Huang, K. Weakly Supervised Deep Ordinal Cox Model for Survival Prediction From Whole-Slide Pathological Images. IEEE Trans. Med. Imaging 2021, 40, 3739–3747. [Google Scholar] [CrossRef] [PubMed]
- Cheerla, A.; Gevaert, O. Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics 2019, 35, I446–I454. [Google Scholar] [CrossRef]
- Fu, Y.; Jung, A.W.; Torne, R.V.; Gonzalez, S.; Vohringer, H.; Shmatko, A.; Yates, L.R.; Jimenez-Linan, M.; Moore, L.; Gerstung, M. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 2020, 1, 800. [Google Scholar] [CrossRef]
- Jiang, S.; Suriawinata, A.A.; Hassanpour, S. MHAttnSurv: Multi-head attention for survival prediction using whole-slide pathology images. Comput. Biol. Med. 2023, 158, 106883. [Google Scholar] [CrossRef]
- Lee, M. Multi-Task Deep Learning Games: Investigating Nash Equilibria and Convergence Properties. Axioms 2023, 12, 569. [Google Scholar] [CrossRef]
| Cancer Type | Studies |
|---|---|
| Colorectal Cancer | Zhao et al. [26], Zhao et al. [27], Sun et al. [28], Yang et al. [29], Xu et al. [30], Jiao et al. [31], Xu et al. [32], Xu et al. [33], Xu et al. [34], Chen et al. [35], Geessink et al. [36], Shapcott, Hewitt, and Rajpoot [37] |
| Breast Cancer | Liu and Kurc [38], Jaber et al. [39], Balkenhol et al. [40], Mao et al. [41], Wang et al. [42], Levy-Jurgenson et al. [43], Fassler et al. [44], Lu et al. [45], du Terrail et al. [46], Xu et al. [47] |
| Bladder Cancer | Jiang et al. [48], Zheng et al. [49], Zheng et al. [50], Gavriel et al. [51], Brieu et al. [52], Wu et al. [53] |
| Liver Cancer | Saillard et al. [54], Liang et al. [55], Qu et al. [56], Hou et al. [57], Shi et al. [58] |
| Lung Cancer | Pham et al. [59], Shim et al. [60], Guo et al. [61], Wang et al. [62] |
| Brain Cancer | Shirazi et al. [63], Chen et al. [64], Liu et al. [65], Jiang et al. [66] |
| Renal Cell Carcinoma | Tabibu et al. [67], Marostica et al. [68], Liu et al. [69] |
| Ovarian Cancer | Yokomizo et al. [70], Liu et al. [71], Wu et al. [72] |
| Cervical Cancer | Chen et al. [73] |
| Gastric Cancer | Ma et al. [74] |
| Pancreatic Cancer | Klimov et al. [75] |
| Skin Cancer | Knuutila et al. [76] |
| Head and Neck Cancer | Zhang et al. [77] |
| Bone Cancer | Ho et al. [78] |
| Oral Cancer | Shaban et al. [79] |
| Multiple Cancers | Shao et al. [80], Cheerla and Gevaert [81], Fu et al. [82], Jiang et al. [83] |
| Ref. | Deep Learning Methods | Expected Strengths | Expected Limitations |
|---|---|---|---|
| [83] | Multihead Attention (Attention Mechanisms) | Comprehensive WSI analysis outperforms existing approaches and contributes to prognosis prediction. | Not specified |
| [55] | General Deep Learning (including MLP) | Potential biomarkers discovered provide enhanced prognostic performance. | Interpretability and generalizability limitations may hinder clinical acceptance. |
| [69] | ResNet | Cost-effective tumor mutation burden measurement and prognostic biomarkers outperform original TMB signature. | Not specified |
| [78] | Deep Multimagnification Network | Highly correlated necrosis ratio estimation and outcome prediction. | Dependence on manual review of necrosis ratio from multiple slides. |
| [46] | Federated Learning | Privacy-preserving multicentric studies with interpretable ML model. | Biases from small-scale study and time-consuming expert annotations. |
| [53] | CNN | Potential for multimodal data use in clinical applications with high diagnostic accuracy. | Not specified |
| [72] | ResNet, Attention Mechanisms | Risk stratification facilitated in ovarian cancer through deep learning framework. | Moderate mean value of C-index; uneven prediction strength across subgroups. |
| [42] | Multiple-Instance Learning (MIL), GAT, Attention Mechanisms | Novel MIL fusion model enables accurate prognostic risk prediction. | Not specified, potential challenges with image segmentation and representation. |
| [62] | ResNet-50 | MPIS integration with clinicopathological variables improves LUAD prognostic stratification. | Transferability of MPIS to all cancer types uncertain. |
| [50] | Weakly Supervised Deep Learning | Accurate bladder cancer diagnosis and personalized treatment decisions. | Not specified |
| [49] | General Deep Learning (including MLP) | The proposed model improves survival prediction in bladder cancer by assessing TILs. | Not specified |
| [71] | CNN, Attention Mechanisms | High-performance prognosis prediction in Epithelial ovarian cancer using AI mechanisms. | Not specified |
| [33] | General Deep Learning (including MLP) | High-accuracy colorectal cancer prognosis using a weakly supervised deep learning network. | Not specified |
| [29] | General Deep Learning (including MLP) | Deep learning-based immune index correlates strongly with colorectal cancer survival rates. | Not specified |
| [57] | General Deep Learning (including MLP) | Multimodality prognostic model provides high-performance survival prediction in hepatocellular carcinoma. | Not specified |
| [48] | General Deep Learning (including MLP) | Depiction of tumor microenvironment immunophenotypes offers insights into biological pathways in bladder cancer. | Not specified |
| [41] | Sparse Representation Learning | The proposed model improves risk stratification in breast cancer with integrated biomarkers. | Effectiveness tied to biomarker extraction quality; untested outside of breast cancer. |
| [73] | CNN with Autoencoder | Deep learning-based pathological risk score predicts cervical cancer prognosis. | Prediction performance tied to dataset quality; clinical application untested. |
| [65] | Autoencoder with Regularization | CMS discovery allows personalized diagnosis in lower-grade gliomas. | Limitations with validating subtypes for other cancer types and accounting for inter-tumor heterogeneity. |
| [76] | ResNet | The proposed model identifies morphological features associated with metastasis in cSCC. | Performance tied to data quality and diversity; untested outside of cSCC. |
| [38] | Deep Learning with Multiresolution | Deep learning method for breast cancer survival integrates image data, improving model performance. | Needs more validation; performance varies with data quality. |
| [28] | Variational Autoencoder (VAE), Generative Adversarial Network (GAN) | Improved prognostic signature for stratifying outcomes in stage III CRC. | Limited generalizability to other cancer types or stages. |
| [35] | Spatial Pyramid Network | Automated CRC risk stratification approach related to gland formation. | Model may require further refinement despite better discrimination. |
| [44] | ResNet-34 | TIL infiltrates assessment in breast cancer WSIs acts as significant biomarkers. | Dependence on TIL infiltrates; performance in TIL absence unclear. |
| [32] | General Deep Learning (including MLP) | Prognostic utility for CRC PFS prediction based on automatic TIL quantification. | Performance tied to TIL quantification; unclear performance in TIL absence. |
| [64] | General Deep Learning (including MLP) | End-to-end multimodal fusion improves survival outcome prediction. | Performance tied to availability of paired WSI, genotype, and transcriptome data. |
| [34] | CNN | The proposed model for CLR and TIL quantification improves survival prediction in CRC. | Needs further validation on larger cohorts for generalizability and clinical deployment. |
| [70] | ResNet-34 | The proposed model achieves high accuracy for prognosis in OCCC. | Single-institution data may limit model generalizability. |
| [80] | General Deep Learning (including MLP) | The proposed model reduces interoperator variation in survival prediction from WSIs. | Efficiency compromised by WSI size and pattern heterogeneity. |
| [30] | CNN | Stroma-immune score using deep learning improves survival prediction in CRC. | Larger validation cohorts needed for reliable assessment of model’s prognostic value. |
| [66] | ResNet-18 | Improved prognosis and IDH mutation status prediction in lower-grade gliomas. | Small sample size may limit robustness and generalizability. |
| [60] | CNN | The proposed model utilizes multiscale pathology images for prognosis prediction in lung adenocarcinoma. | Not specified |
| [61] | EfficientUnet, ResNet | Efficient analysis of immune checkpoints and prognosis of NSCLC. | Not specified |
| [68] | CNN | Accurate RCC subtype diagnosis and prediction of survival outcomes. | Interrater variability and limitations in capturing all biological signals. |
| [58] | Weakly Supervised Deep Learning | Prognostic indicators from HCC pathological images improve risk stratification. | Efficiency and labor-saving limitations; needs further validation for patient treatment. |
| [51] | Ensemble Learning | Prediction of MIBC prognosis significantly higher than TNM staging system. | Further validation and clinical deployment needed. |
| [40] | CNN | Efficient assessment of TILs in triple negative breast cancer provides valuable prognostic information. | Optimal prognostic information yielding method unclear; lack of objective TIL assessment methods. |
| [75] | CNN | High accuracy in predicting metastasis risk in pancreatic neuroendocrine tumors. | Not specified |
| [27] | General Deep Learning (including MLP) | Accurate mucus proportion quantification in colorectal cancer suitable for clinical application. | Not specified |
| [47] | General Deep Learning (including MLP) | Integrative analysis of histopathological images and genomic data improves understanding of disease progression. | Might not identify all potential regulatory regions in the human genome. |
| [54] | General Deep Learning (including MLP) | Two deep learning algorithms aid risk stratification for hepatocellular carcinoma patients. | Not specified |
| [77] | Convolutional Neural Networks (CNN) | Prognostic model predicts treatment failure in nasopharyngeal carcinoma better than existing clinical models. | Not specified |
| [43] | General Deep Learning (including MLP) | The models developed can spatially characterize tumor heterogeneity. Showed a significant statistical link between heterogeneity and survival. | Lack of automated methods for characterizing tumor heterogeneity. |
| [26] | CNN, Transfer Learning | Automated deep learning method for TSR quantification in colorectal cancer reduces pathologist workload. | Not specified |
| [74] | CNN | CNN-based system distinguishes tissue types with high accuracy in gastric diseases. | Not specified |
| [82] | Transfer Learning | Deep transfer learning quantifies histopathological patterns across a range of cancer types. | Not specified |
| [45] | CNN | High-resolution TIL map generation on WSIs strongly associates with immune response pathways and genes. | Not specified |
| [63] | CNN | Exceptional accuracy in brain cancer survival rate classification based on histopathological images. | Challenges in generalizability on unseen samples and practical clinical application. |
| [39] | General Deep Learning (including MLP) | Deep learning classifier identifies breast cancer molecular subtypes and heterogeneity. | Potential inaccuracies due to intratumoral heterogeneity. |
| [59] | General Deep Learning (including MLP) | Two-step deep learning approach accurately detects lung cancer metastases. | Presence of false positives in model predictions. |
| [79] | General Deep Learning (including MLP) | TILAb score predicts disease-free survival in OSCC patients better than manual TIL score. | Accuracy tied to quality and clarity of WSIs. |
| [67] | Convolutional Neural Networks (CNN) | High accuracy distinguishing renal cell carcinoma subtypes and predicting patient survival. | Class imbalance issues in medical datasets. |
| [81] | Multimodal Neural Network | Model combining clinical, mRNA, microRNA data, and WSIs predicts survival for 20 cancer types. | Not specified; potential complexity in interpreting multiple data modalities. |
| [36] | General Deep Learning (including MLP) | Automated approach determines TSR as an independent prognosticator in rectal cancer. | Applicable only in user-provided stroma hot-spots; performance tied to input image quality. |
| [37] | General Deep Learning (including MLP) | Deep learning algorithm for cell identification in colon cancer images improves performance. | Patch selection for analysis may impact results. |
| [52] | CNN | Quantification of tumor buds in bladder cancer adds prognostic value to traditional TNM staging. | Not specified |
| [56] | CNN | Recurrence-related histological score allows for clinical decision making in HCC recurrence prediction. | Prediction accuracy varies; potential bias towards trained data and diseases. |
| [31] | CNN | Automatic evaluation of the tumor microenvironment in WSIs aids in predicting disease progression. | Varied strength of predictors; potential bias towards specific cancer types. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M. Recent Advancements in Deep Learning Using Whole Slide Imaging for Cancer Prognosis. Bioengineering 2023, 10, 897. https://doi.org/10.3390/bioengineering10080897
Lee M. Recent Advancements in Deep Learning Using Whole Slide Imaging for Cancer Prognosis. Bioengineering. 2023; 10(8):897. https://doi.org/10.3390/bioengineering10080897
Chicago/Turabian StyleLee, Minhyeok. 2023. "Recent Advancements in Deep Learning Using Whole Slide Imaging for Cancer Prognosis" Bioengineering 10, no. 8: 897. https://doi.org/10.3390/bioengineering10080897
APA StyleLee, M. (2023). Recent Advancements in Deep Learning Using Whole Slide Imaging for Cancer Prognosis. Bioengineering, 10(8), 897. https://doi.org/10.3390/bioengineering10080897








