Cofactor Metabolic Engineering of Escherichia coli for Aerobic L-Malate Production with Lower CO2 Emissions
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Media and Cultivation Conditions
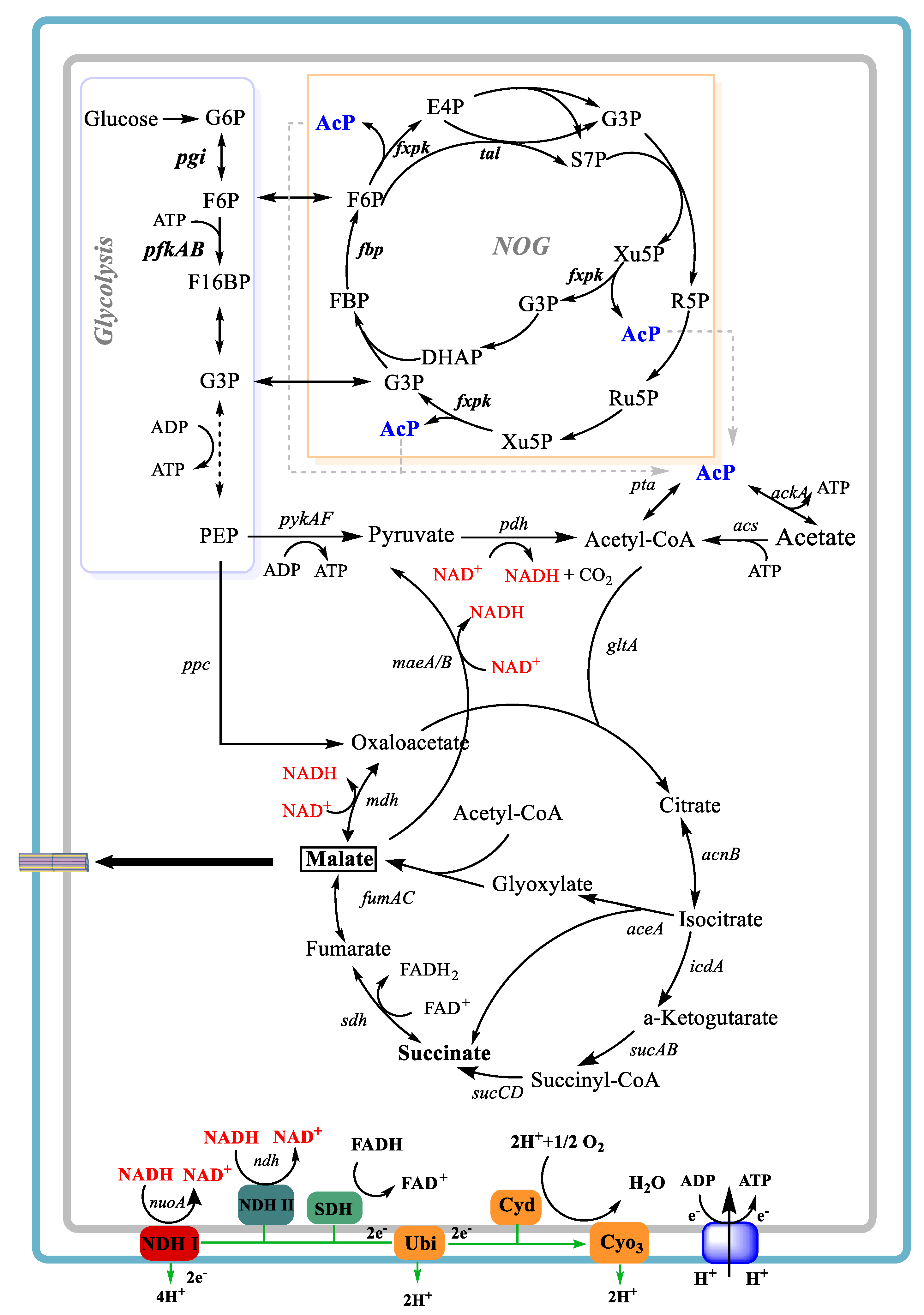
2.3. Construction of a NDH-1 Defective Mutant
2.4. Construction of NOG Expression System
2.5. Cell Biomass and Residual Glucose Concentration
2.6. Metabolites Analysis and Yield Analysis
2.7. Determination of CO2 Emissions
2.8. Phosphoketolase Activity Assay
2.9. Quantification of Intracellular NAD(H)
2.10. Quantification of Intracellular Acetyl-coA and ATP
2.11. Transcriptional Analysis
3. Results
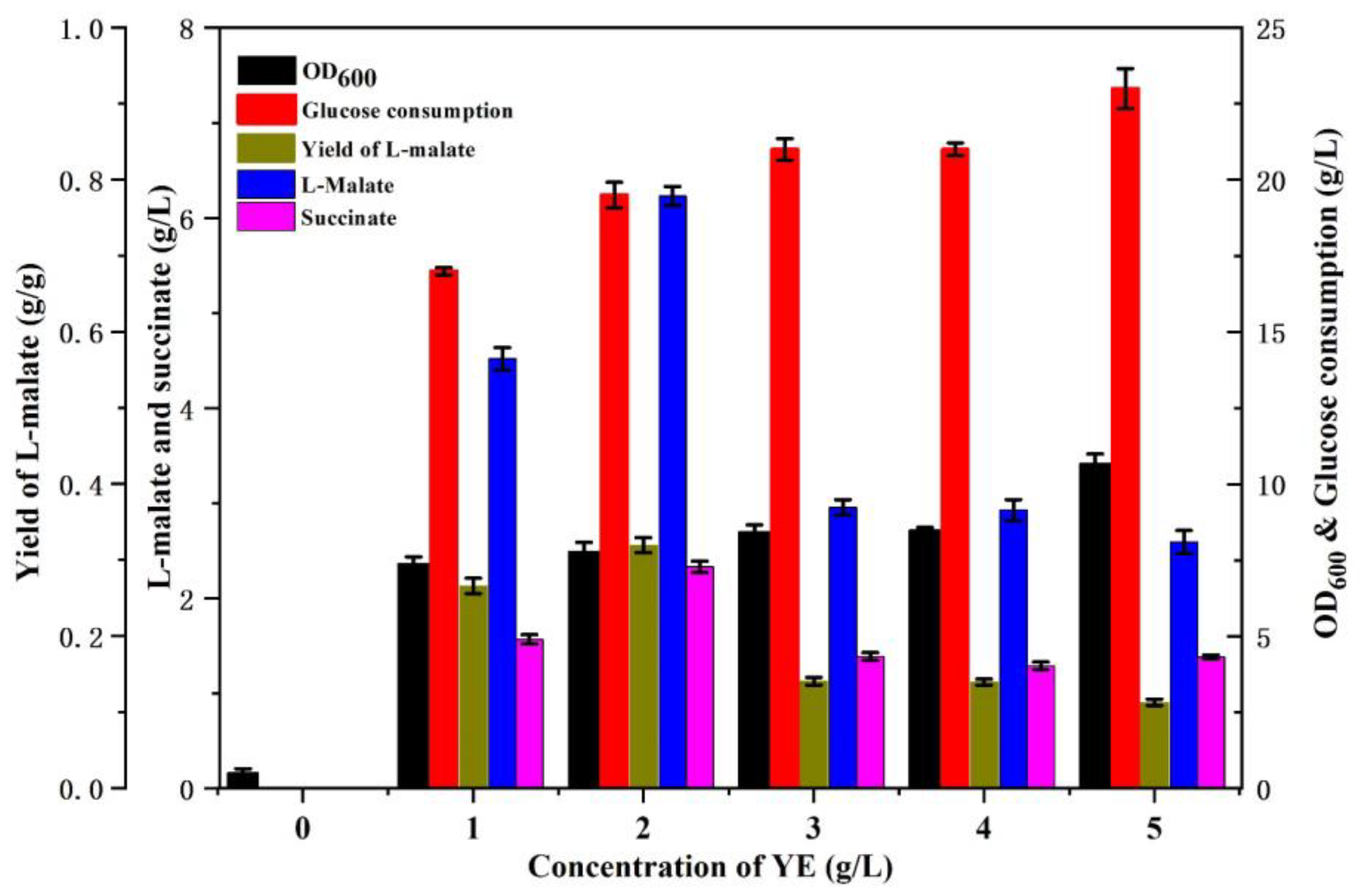
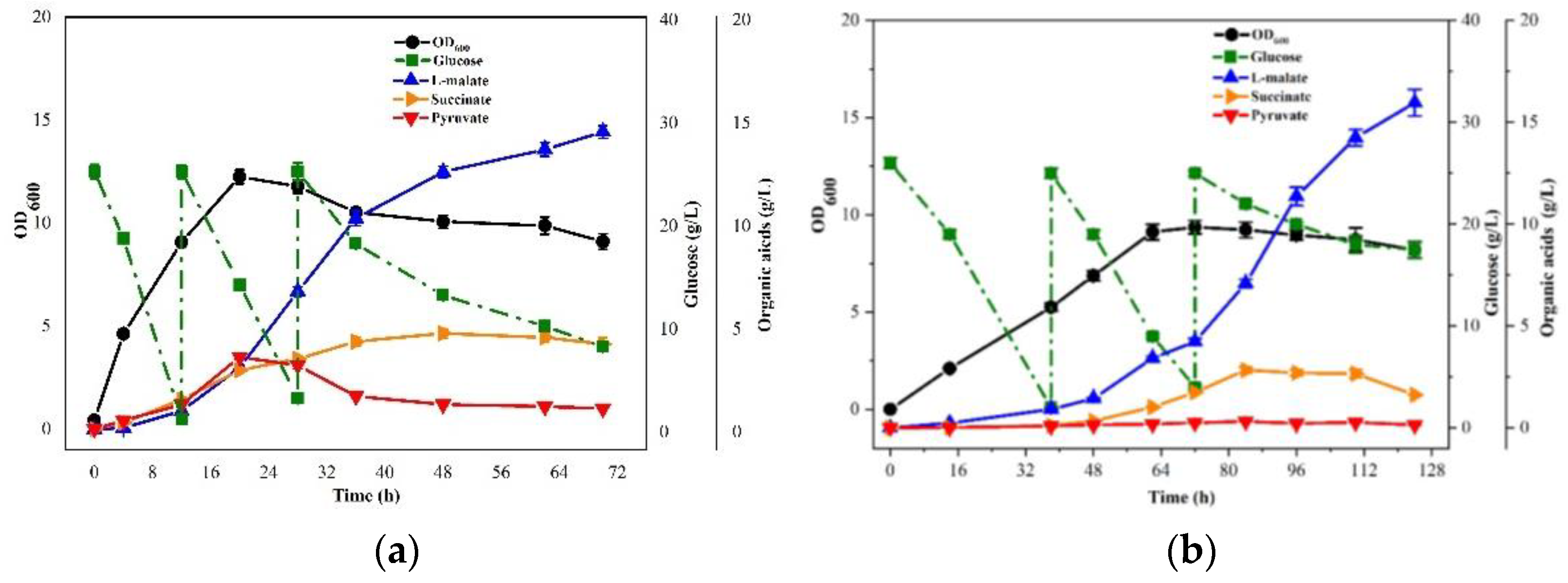
3.1. Metabolic Characteristics of E. coli E23 with Different Concentrations of YE
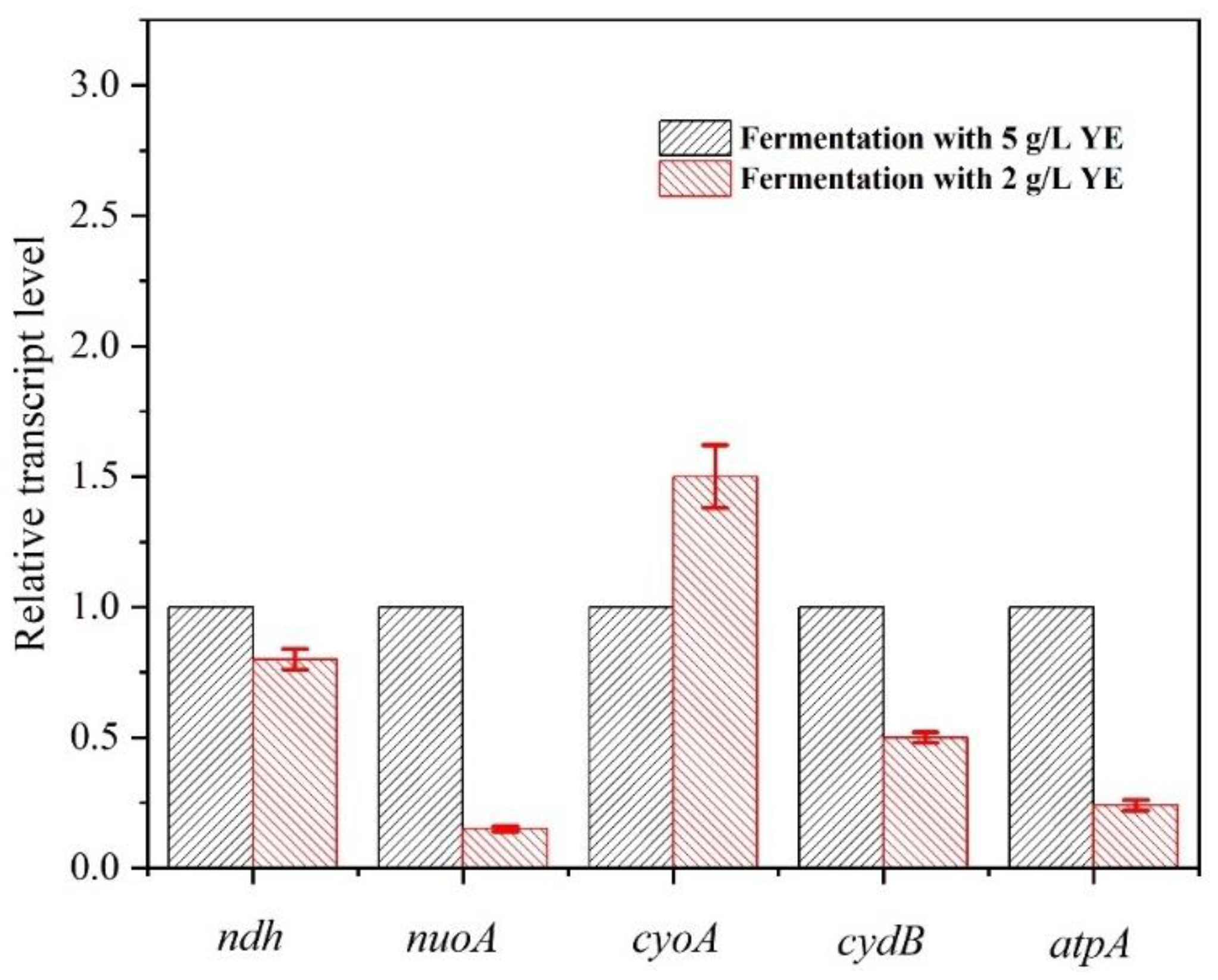
3.2. Transcriptional Analysis of E. coli E23 with Different Concentrations of YE
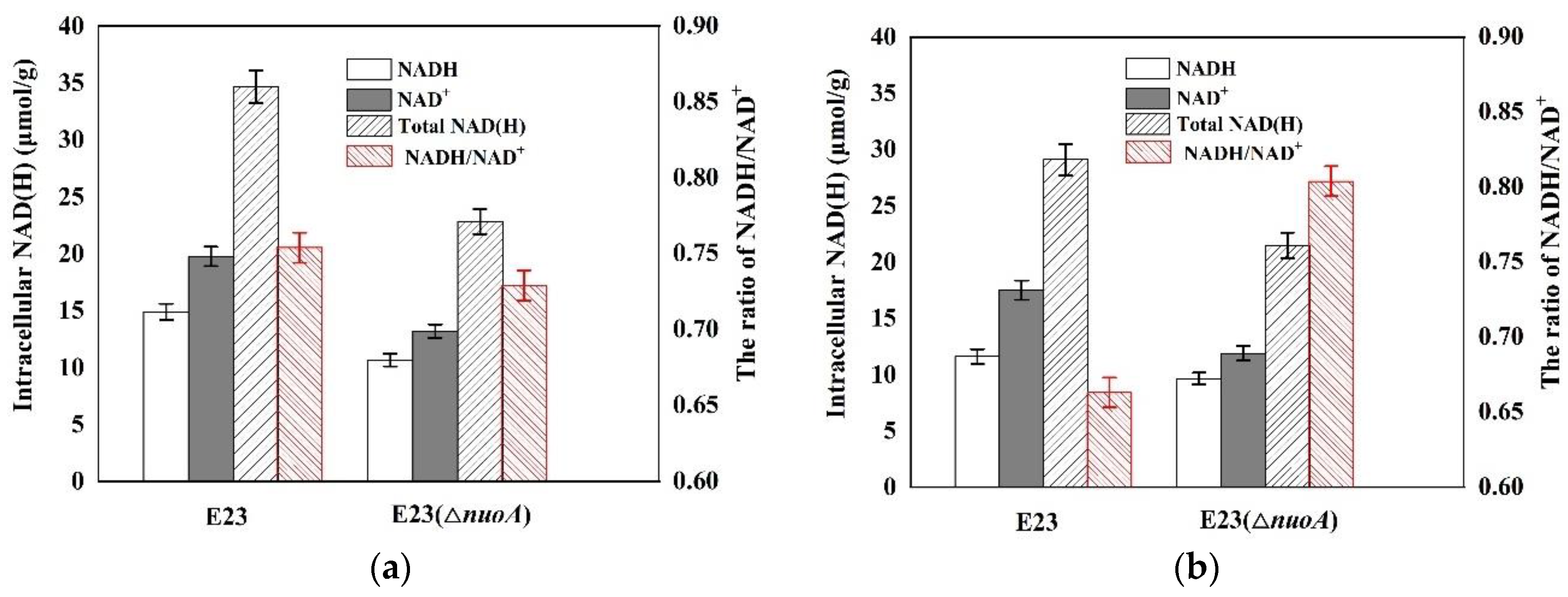
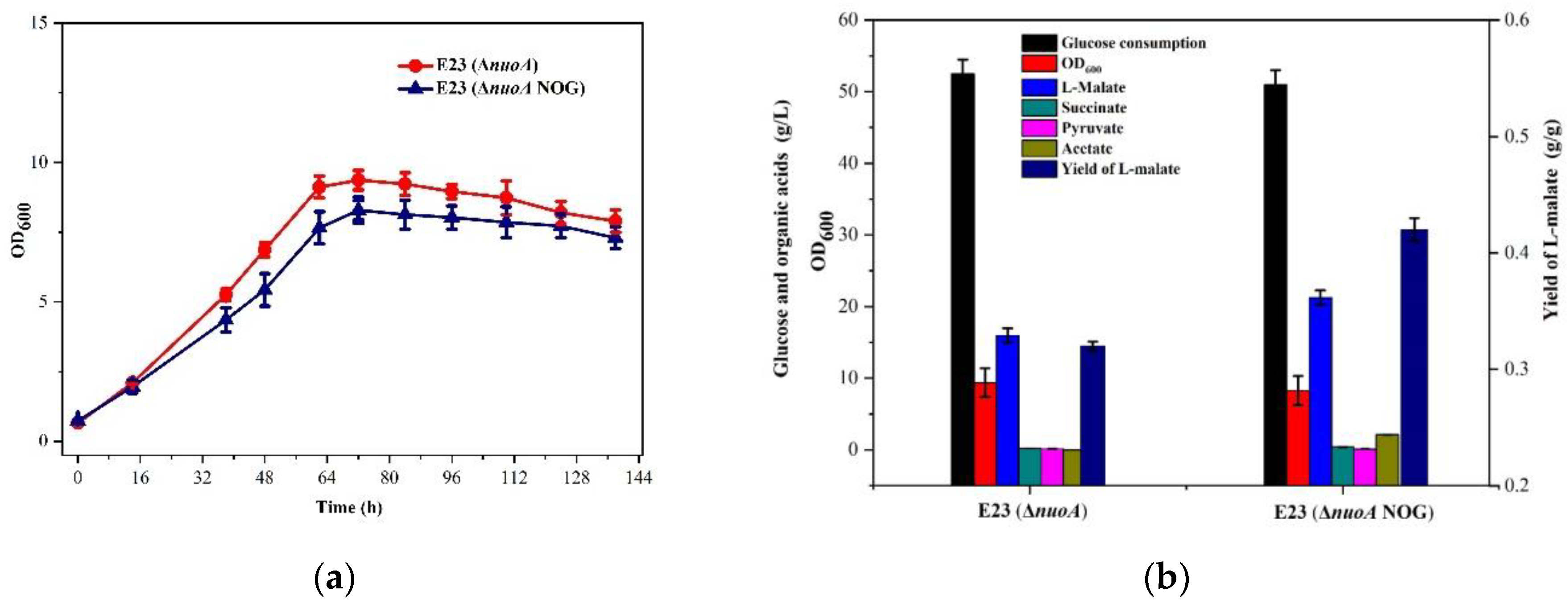
3.3. Engineering E. coli E23 to Decrease NAD+ Supply by the Knockout of nuoA
3.4. Limiting ATP Supply of E. coli E23 by the Introduction of the NOG System
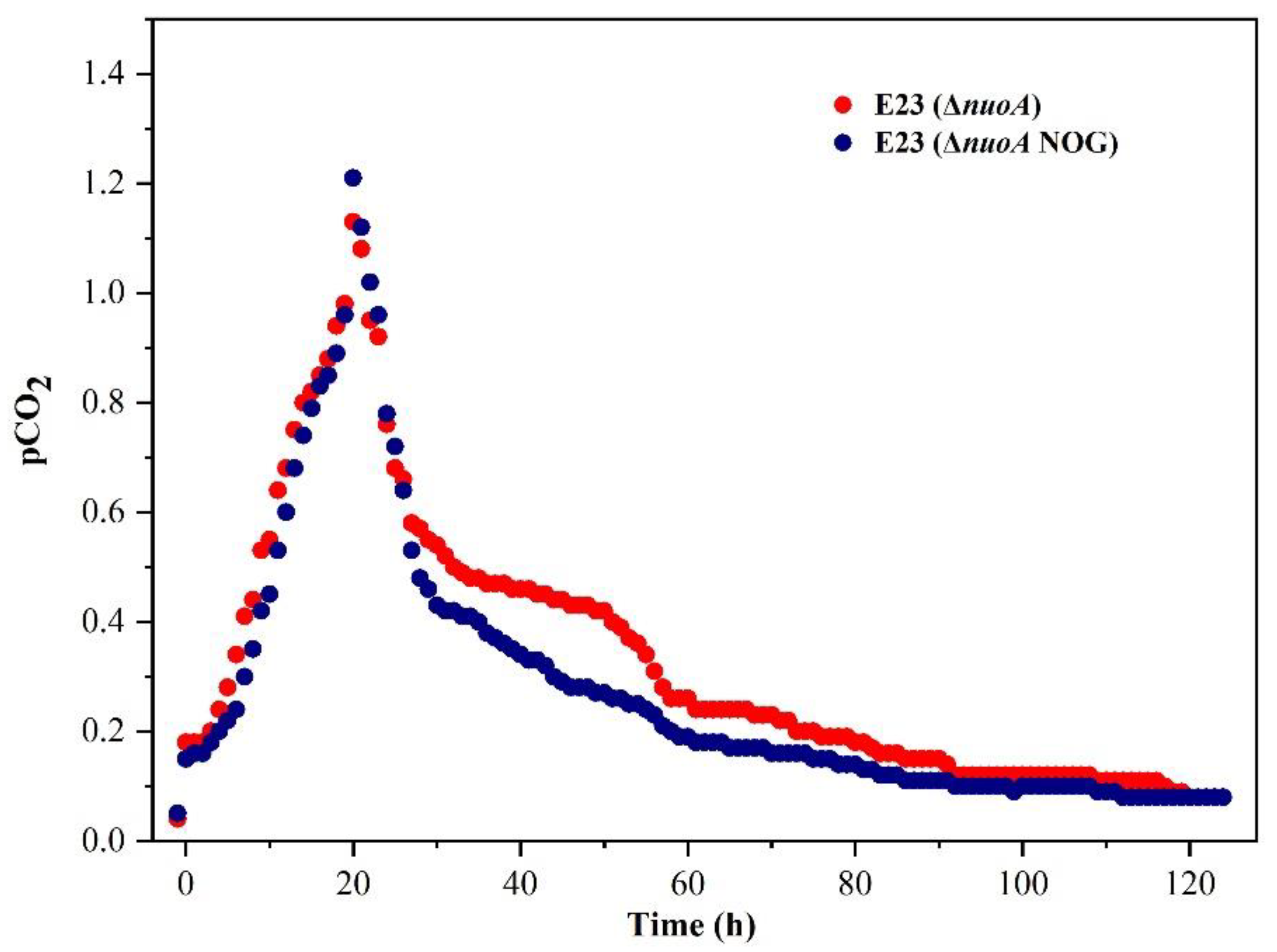
3.5. Metabolic Characteristics of E. coli E23 (ΔnuoA, NOG) in 5 L Fed-Batch Cultivation
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chi, Z.; Wang, Z.P.; Wang, G.Y.; Khan, I.; Chi, Z.M. Microbial biosynthesis and secretion of L-malic acid and its applications. Crit. Rev. Biotechnol. 2016, 36, 99–107. [Google Scholar] [CrossRef]
- Werpy, T.; Petersen, G. Top Value Added Chemicals from Biomass: Volume I-Results of Screening for Potential Candidates from Sugars and Synthesis Gas; Report; National Renewable Energy Laboratory: Golden, CO, USA, 2004.
- Giorno, L.; Drioli, E.; Carvoli, G.; Cassano, A.; Donato, L. Study of an enzyme membrane reactor with immobilized fumarase for production of L-malic acid. Biotechnol. Bioeng. 2001, 72, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.X.; Zhou, H.Y.; Zhang, S.J.; Gu, H.L.; Yang, Q.; Zhang, W.M.; Dong, W.L.; Ma, J.F.; Fang, Y.; Jiang, M.; et al. Current advance in biological production of malic acid using wild type and metabolic engineered strains. Bioresour. Technol. 2018, 258, 345–353. [Google Scholar] [CrossRef]
- Iyyappan, J.; Baskar, G.; Bharathiraja, B.; Gopinath, M. Enhanced malic acid production using Aspergillus niger coupled with in situ product recovery. Bioresour. Technol. 2020, 308, 123259. [Google Scholar] [CrossRef]
- Khan, I.; Nazir, K.; Wang, Z.P.; Liu, G.L.; Chi, Z.M. Calcium malate overproduction by Penicillium viticola 152 using the medium containing corn steep liquor. Appl. Microbiol. Biotechnol. 2014, 98, 1539–1546. [Google Scholar] [CrossRef]
- Moon, S.Y.; Hong, S.H.; Kim, T.Y.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of malic acid. Biochem. Eng. J. 2008, 40, 312–320. [Google Scholar] [CrossRef]
- Sun, W.; Jiang, B.; Zhao, D.; Pu, Z.; Bao, Y. Integration of metabolic pathway manipulation and promoter engineering for the fine-tuned biosynthesis of malic acid in Bacillus coagulans. Biotechnol. Bioeng. 2021, 118, 2597–2608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, X.; Shanmugam, K.T.; Ingram, L.O. L-Malate production by metabolically engineered Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 427–434. [Google Scholar] [CrossRef]
- Hu, G.; Zhou, J.; Chen, X.; Qian, Y.; Gao, C.; Guo, L.; Xu, P.; Chen, W.; Chen, J.; Li, Y.; et al. Engineering synergetic CO2-fixing pathways for malate production. Metab. Eng. 2018, 47, 496–504. [Google Scholar] [CrossRef]
- Gao, C.; Wang, S.; Hu, G.; Guo, L.; Chen, X.; Xu, P.; Liu, L. Engineering Escherichia coli for malate production by integrating modular pathway characterization with CRISPRi-guided multiplexed metabolic tuning. Biotechnol. Bioeng. 2018, 115, 661–672. [Google Scholar] [CrossRef]
- Trichez, D.; Auriol, C.; Baylac, A.; Irague, R.; Dressaire, C.; Carnicer-Heras, M.; Heux, S.; Francois, J.M.; Walther, T. Engineering of Escherichia coli for krebs cycle-dependent production of malic acid. Microb. Cell Fact. 2018, 17, 113–124. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, F.; Zhang, C.; Hu, G.; Gao, C.; Chen, X.; Liu, L. Enhancement of malate production through engineering of the periplasmic rTCA pathway in E. coli. Biotechnol. Bioeng. 2018, 115, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hong, P.; Da, Y.; Li, L.; Stephanopoulos, G. Metabolic engineering of Escherichia coli for the production of L-malate from xylose. Metab. Eng. 2018, 48, 25–32. [Google Scholar] [CrossRef]
- Soto-Varela, Z.E.; Cabrera, G.; Romero, A.; Cantero, D.; Valle, A.; Bolivar, J. Identification of enzymatic bottlenecks for the aerobic production of malate from glycerol by the systematic gene overexpression of anaplerotic enzymes in Escherichia coli. Int. J. Mol. Sci. 2021, 22, 2266. [Google Scholar] [CrossRef]
- Amulya, K.; Mohan, S.V. Augmenting succinic acid production by bioelectrochemical synthesis: Influence of applied potential and CO2 availability. Chem. Eng. J. 2021, 411, 128377. [Google Scholar] [CrossRef]
- Bogorad, I.W.; Lin, T.S.; Liao, J.C. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 2013, 502, 693–697. [Google Scholar] [CrossRef]
- Ma, J.; Qian, H.; Zheng, T.; Jiang, Y.; Xin, F.; Dong, W.; Zhang, W.; Fang, Y.; Jiang, M. Novel biosynthesis of L-malate based on overflow metabolism in Escherichia coli. Biochem. Eng. J. 2019, 149, 107255. [Google Scholar] [CrossRef]
- Li, N.; Zhang, B.; Chen, T.; Wang, Z.; Tang, Y.J.; Zhao, X. Directed pathway evolution of the glyoxylate shunt in Escherichia coli for improved aerobic succinate production from glycerol. J. Ind. Microbiol. Biotechnol. 2013, 40, 1461–1475. [Google Scholar] [CrossRef]
- Virzintiene, E.; Trane, M.; Hägerhäll, C. Revised transmembrane orientation of the NADH:quinone oxidoreductase subunit NuoA. FEBS Lett. 2011, 585, 3277–3283. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Chu, Z.; Peng, J.; Jin, W. Screen-printed biosensor chips with prussian blue nanocubes for the detection of physiological analytes. Sens. Actuators B Chem. 2016, 228, 679–687. [Google Scholar] [CrossRef]
- Yang, P.; Peng, J.; Chu, Z.; Jiang, D.; Jin, W. Facile synthesis of prussian blue nanocubes/silver nanowires network as a water-based ink for the direct screen-printed flexible biosensor chips. Biosens. Bioelectron. 2017, 92, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xu, J.; Sun, Z.; Luan, Q.; Li, Y.; Wang, J.; Liang, Q.; Qi, Q. Engineering an in vivo EP-bifido pathway in Escherichia coli for high-yield acetyl-CoA generation with low CO2 emission. Metab. Eng. 2019, 51, 79–87. [Google Scholar] [CrossRef]
- Liang, L.; Liu, R.; Ma, J.; Chen, K.; Jiang, M.; Wei, P. Increased production of succinic acid in Escherichia coli by overexpression of malate dehydrogenase. Biotechnol. Lett. 2011, 33, 2439–2444. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yuan, Q.; Zheng, Y.; Ma, H.; Chen, T.; Zhao, X. An engineered non-oxidative glycolysis pathway for acetone production in Escherichia coli. Biotechnol. Lett. 2016, 38, 1359–1365. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Shimizu, K. Transcriptional regulation of main metabolic pathways of cyoA, cydB, fnr, and fur gene knockout Escherichia coli in C-limited and N-limited aerobic continuous cultures. Microb. Cell Fact. 2011, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Larsson, C.; Von, S.U.; Marison, I.; Gustafsson, L. Growth and metabolism of Saccharomyces cerevisiae in chemostat cultures under carbon-, nitrogen-, or carbon- and nitrogen-limiting conditions. J. Bacteriol. 1993, 175, 4809–4816. [Google Scholar] [CrossRef]
- Sauer, U.; Lasko, D.R.; Fiaux, J.; Hochuli, M.; Glaser, R.; Szyperski, T.; Wuthrich, K.; Bailey, J.E. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 1999, 181, 6679–6688. [Google Scholar] [CrossRef]
- Ji, L.; Wang, J.; Luo, Q.; Ding, Q.; Tang, W.; Chen, X.; Liu, L. Enhancing L-malate production of Aspergillus oryzae by nitrogen regulation strategy. Appl. Microbiol. Biotechnol. 2021, 105, 3101–3113. [Google Scholar] [CrossRef]
- Liu, L.M.; Li, Y.; Du, G.C.; Chen, J. Increasing glycolytic flux in Torulopsis glabrata by redirecting ATP production from oxidative phosphorylation to substrate-level phosphorylation. J. Appl. Microbiol. 2006, 100, 1043–1053. [Google Scholar] [CrossRef]
- Aoki, R.; Wada, M.; Takesue, N.; Tanaka, K.; Yokota, A. Enhanced glutamic acid production by a H+-ATPase-defective mutant of Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 2005, 69, 1466–1472. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Benisch, F.; Boles, E. The bacterial Entner-Doudoroff pathway does not replace glycolysis in Saccharomyces cerevisiae due to the lack of activity of iron-sulfur cluster enzyme 6-phosphogluconate dehydratase. J. Biotechnol. 2014, 171, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.P.; Jaeger, A.J.; Wu, T.Y.; Xu, S.; Lee, A.S.; Gao, F.; Chen, P.W.; Liao, J.C. Construction and evolution of an Escherichia coli strain relying on nonoxidative glycolysis for sugar catabolism. Proc. Natl. Acad. Sci. USA 2018, 115, 3538–3546. [Google Scholar] [CrossRef] [PubMed]
| Strains | Feature | Source or Reference |
|---|---|---|
| E. coli DH5α | Cloning host | Lab collection |
| E. coli E2 | Evolved BL21(DE3), Δppc, aceBAK:trc | Li et al. 2013 [19] |
| E. coli E23 | E2, ppc:trc, ΔmaeAB | Chen’s Lab collection |
| E. coli E23 (ΔnuoA) | E23, ΔnuoA | This study |
| E. coli E23 (ΔnuoA NOG) | E23, ΔnuoA, harboring pTrc-fxpk-fbp | This study |
| Strains | Acetyl-coA (nmol/g DCW) | ATP (nmol/g DCW) |
|---|---|---|
| E. coli E23 (ΔnuoA) | 12.20 ± 0.76 | 1685 ± 416 |
| E. coli E23 (ΔnuoA NOG) | 14.30 ± 0.94 | 1370 ± 320 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Z.; Jiang, Y.; Wu, H.; Zhang, W.; Xin, F.; Ma, J.; Jiang, M. Cofactor Metabolic Engineering of Escherichia coli for Aerobic L-Malate Production with Lower CO2 Emissions. Bioengineering 2023, 10, 881. https://doi.org/10.3390/bioengineering10080881
Jiang Z, Jiang Y, Wu H, Zhang W, Xin F, Ma J, Jiang M. Cofactor Metabolic Engineering of Escherichia coli for Aerobic L-Malate Production with Lower CO2 Emissions. Bioengineering. 2023; 10(8):881. https://doi.org/10.3390/bioengineering10080881
Chicago/Turabian StyleJiang, Zhiming, Youming Jiang, Hao Wu, Wenming Zhang, Fengxue Xin, Jiangfeng Ma, and Min Jiang. 2023. "Cofactor Metabolic Engineering of Escherichia coli for Aerobic L-Malate Production with Lower CO2 Emissions" Bioengineering 10, no. 8: 881. https://doi.org/10.3390/bioengineering10080881
APA StyleJiang, Z., Jiang, Y., Wu, H., Zhang, W., Xin, F., Ma, J., & Jiang, M. (2023). Cofactor Metabolic Engineering of Escherichia coli for Aerobic L-Malate Production with Lower CO2 Emissions. Bioengineering, 10(8), 881. https://doi.org/10.3390/bioengineering10080881









