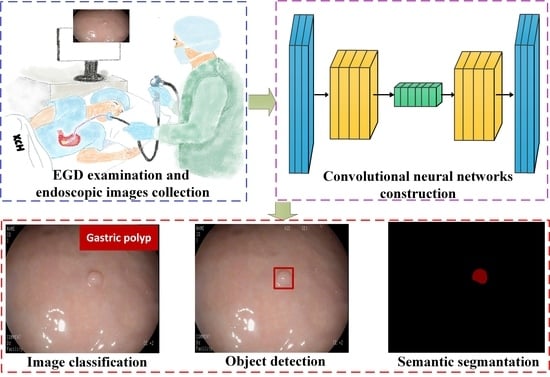
Semantic Segmentation of Gastric Polyps in Endoscopic Images Based on Convolutional Neural Networks and an Integrated Evaluation Approach
Abstract
1. Introduction
- (1)
- A high-quality gastric polyp dataset for training, validation, and testing of the semantic segmentation models is built, and the dataset will be publicly available for further research.
- (2)
- This is pioneering research on gastric polyp segmentation. Additionally, seven semantic segmentation models, including U-Net, UNet++, DeepLabv3, DeepLabv3+, PAN, LinkNet, and MA-Net, with the encoders of ResNet50, MobineNetV2, or EfficientNet-B1, are constructed and compared.
- (3)
- The objective and subjective evaluation methods are combined to propose a novel integrated evaluation approach to evaluate the experimental results, aiming at the determination of the best CNN model for the automated polyp-segmentation system.
2. Related Work
2.1. CNN-Based Gastric Polyp Diagnosis
2.2. MCDM Methods
3. Materials and Methods
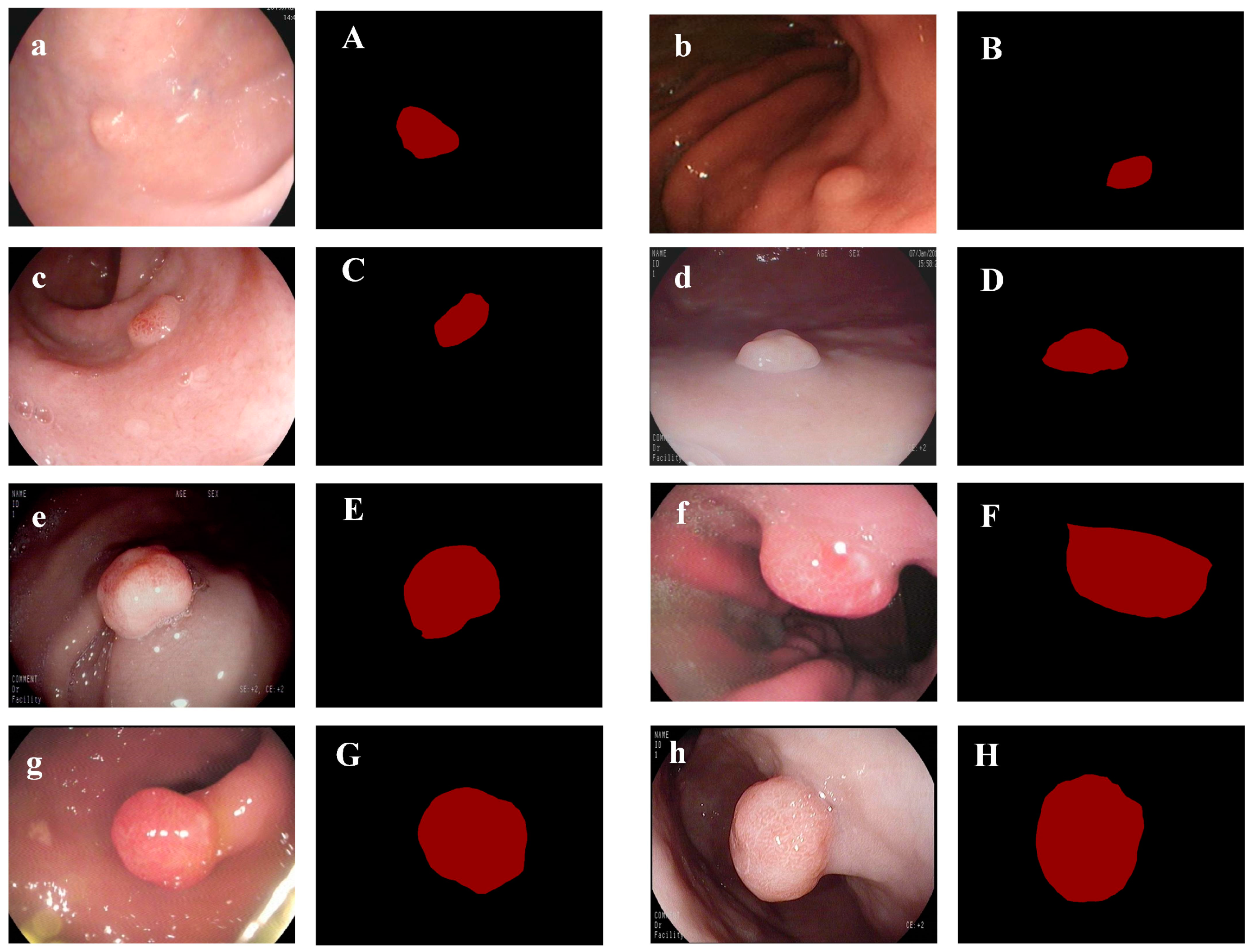
3.1. Dataset
- (1)
- Images that used endoscopic optics other than standard white-light endoscopy.
- (2)
- The anatomical position of the image is not in the stomach (like the esophagus).
- (3)
- Images that contain no polyps.
- (4)
- Images that are damaged and low-quality due to halation, mucous, blurring, lack of focus, low insufflation of air, et cetera.
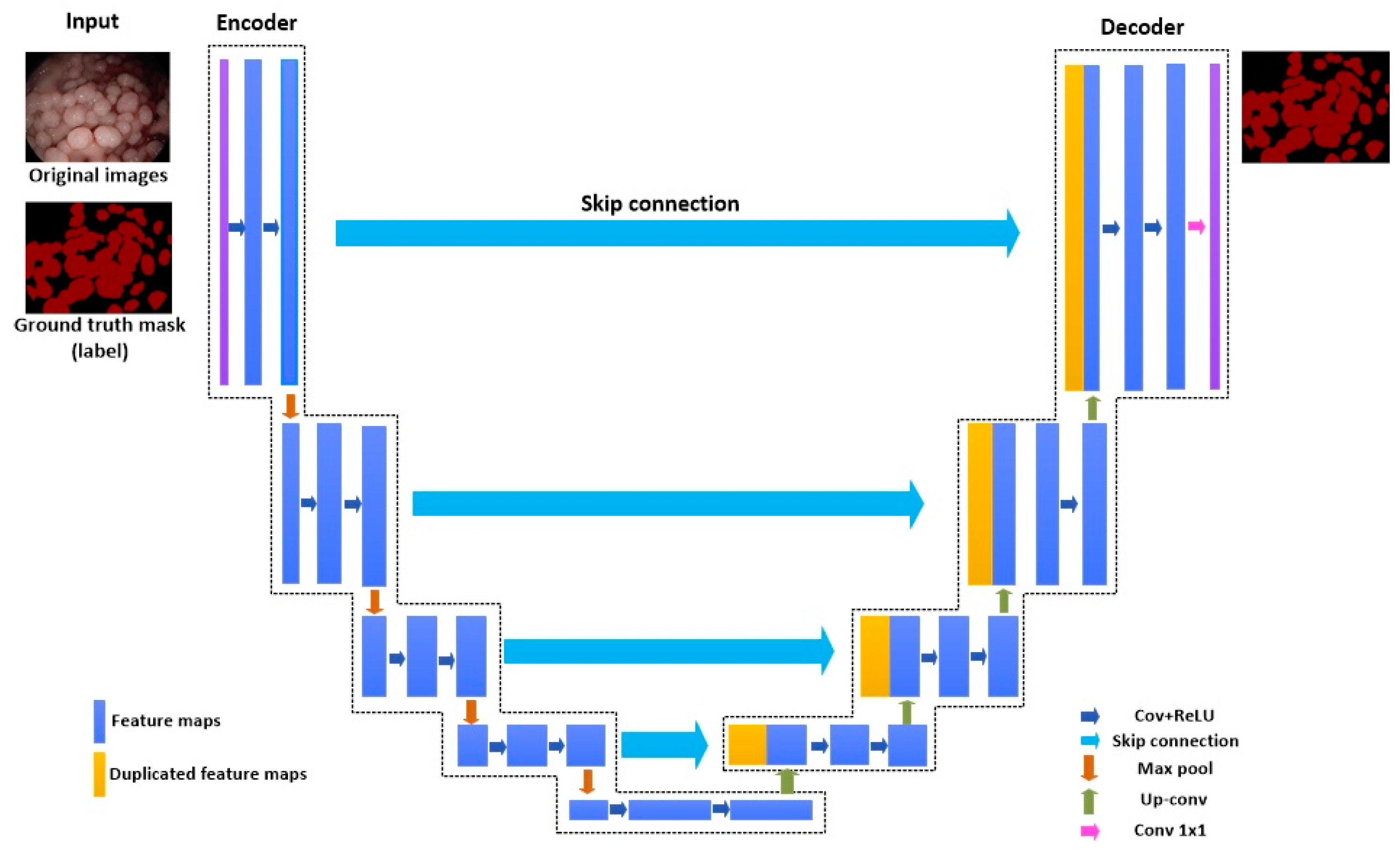
3.2. Existing Semantic Segmentation Methods
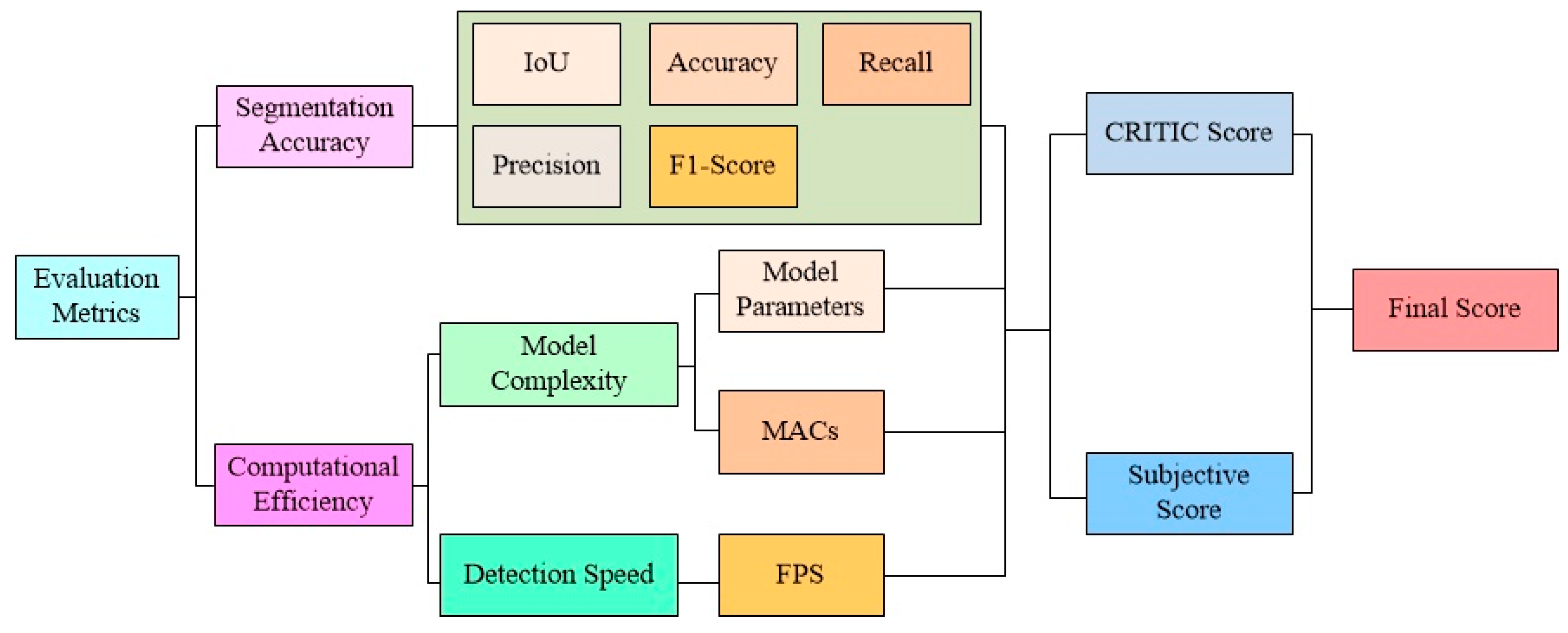
3.3. Integrated Evaluation Approach
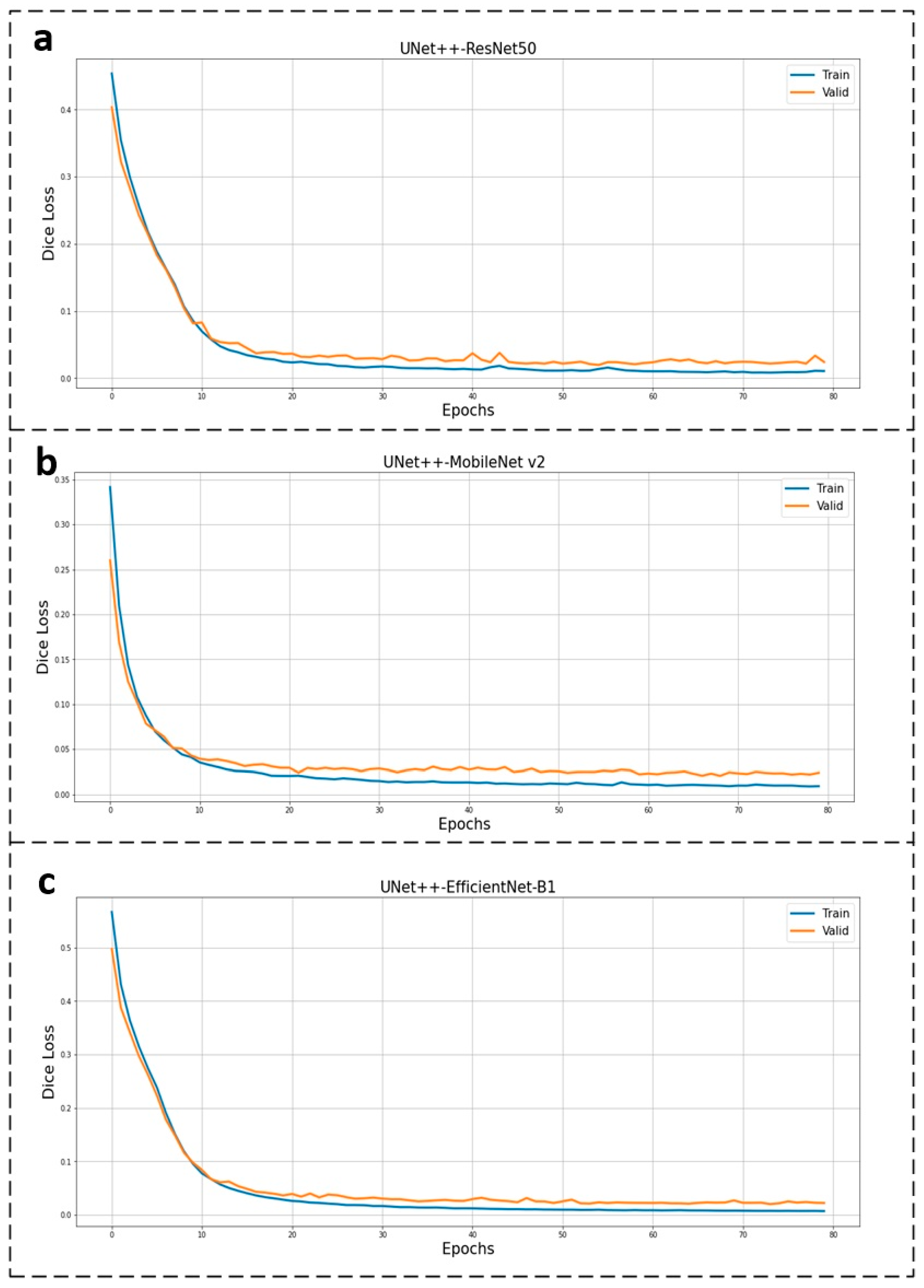
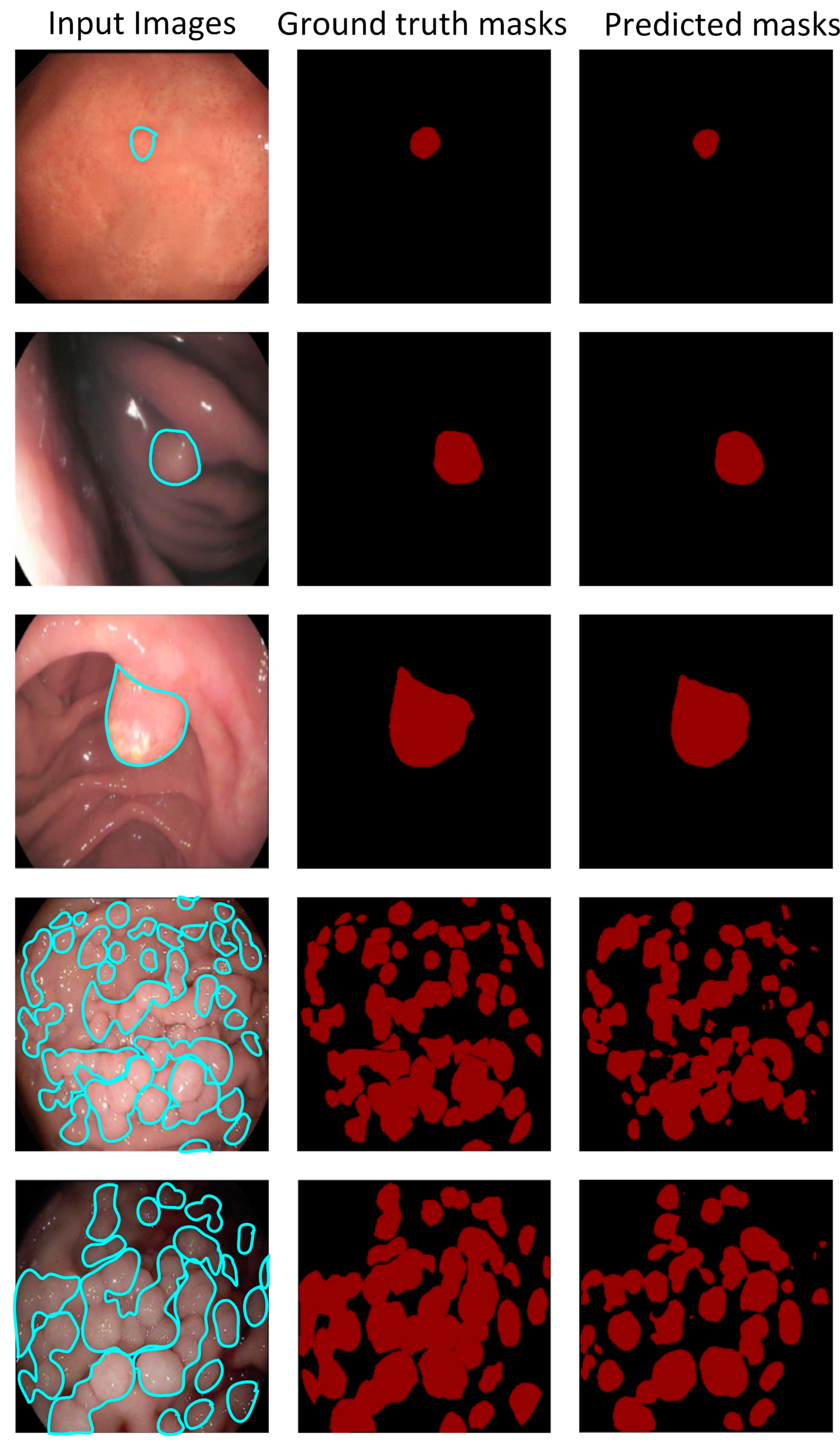
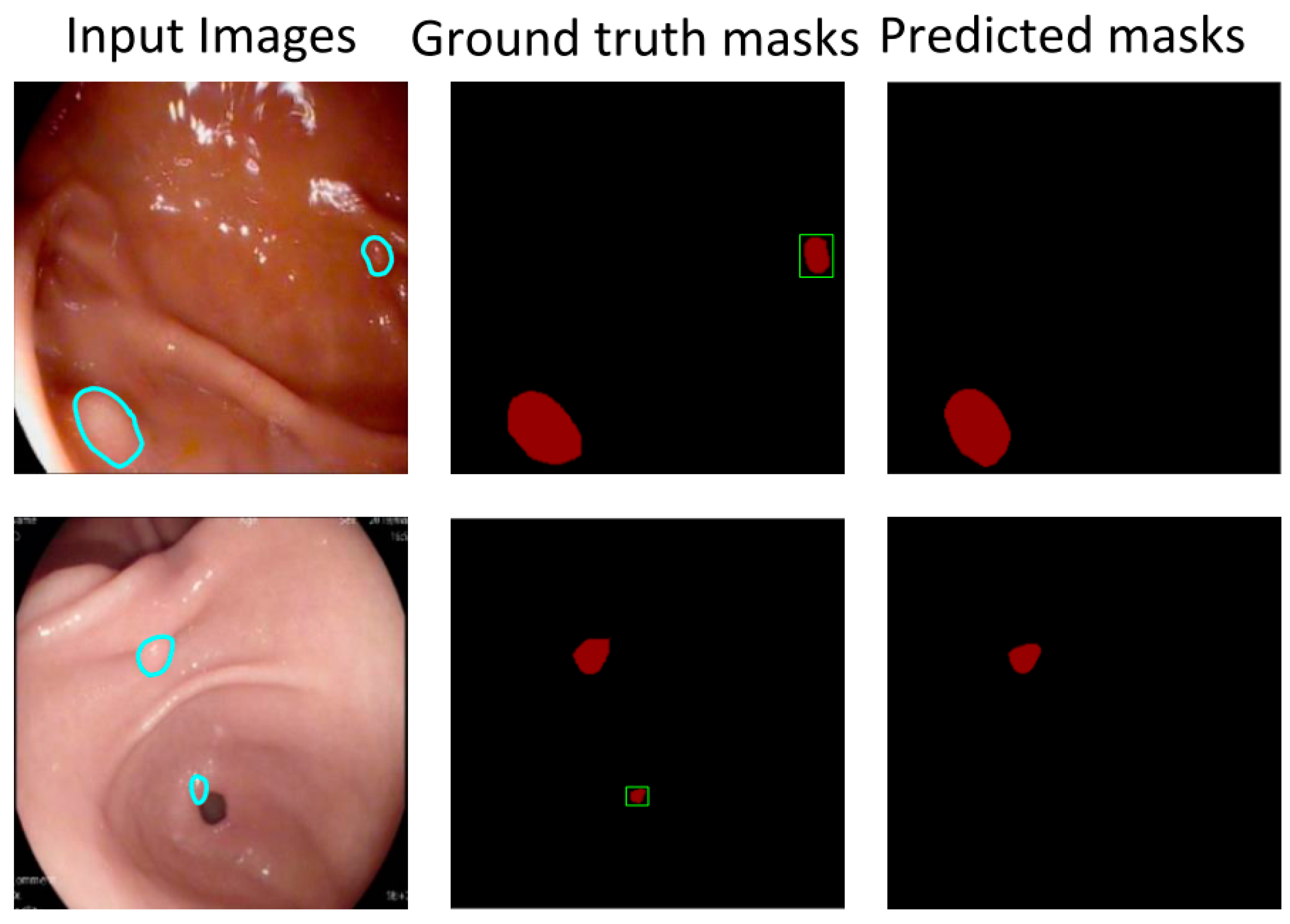
4. Experiments and Results
4.1. Experimental Configuration
4.2. Results
5. Conclusions
- (1)
- This study is pioneering research on gastric polyp segmentation. A high-quality gastric polyp dataset is generated. Seven semantic segmentation models are constructed and evaluated to determine the core model for the automated polyp-segmentation system.
- (2)
- To comprehensively evaluate the results, the integrated evaluation approach combined with the CRITIC method and experts’ weight are combined to rank the candidate models, which is the first attempt at a polyp-segmentation task.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Fock, K.M.; Talley, N.; Moayyedi, P.; Hunt, R.; Azuma, T.; Sugano, K.; Xiao, S.D.; Lam, S.K.; Goh, K.L.; Chiba, T.; et al. Asia–Pacific consensus guidelines on gastric cancer prevention. J. Gastroenterol. Hepatol. 2008, 23, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Goddard, A.F.; Badreldin, R.; Pritchard, D.M.; Walker, M.M.; Warren, B. The Management of gastric polyps. Gut 2010, 59, 1270–1276. [Google Scholar] [CrossRef]
- Carmack, S.W.; Genta, R.M.; Schuler, C.M.; Saboorian, M.H. The current spectrum of gastric polyps: A 1-year national study of over 120,000 patients. Am. J. Gastroenterol. 2009, 104, 1524–1532. [Google Scholar] [CrossRef]
- Jung, M.K.; Jeon, S.W.; Park, S.Y.; Cho, C.M.; Tak, W.Y.; Kweon, Y.O.; Kim, S.K.; Choi, Y.H.; Bae, H.I. Endoscopic characteristics of gastric adenomas suggesting carcinomatous transformation. Surg. Endosc. 2008, 22, 2705–2711. [Google Scholar] [CrossRef]
- Carmack, S.W.; Genta, R.M.; Graham, D.Y.; Lauwers, G.Y. Management of gastric polyps: A pathology-based guide for gastroenterologists. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 331–341. [Google Scholar] [CrossRef]
- Shaib, Y.H.; Rugge, M.; Graham, D.Y.; Genta, R.M. Management of gastric polyps: An endoscopy-based approach. Clin. Gastroenterol. Hepatol. 2013, 11, 1374–1384. [Google Scholar] [CrossRef]
- Zheng, B.; Rieder, E.; Cassera, M.A.; Martinec, D.V.; Lee, G.; Panton, O.N.; Park, A.; Swanström, L.L. Quantifying mental workloads of surgeons performing natural orifice transluminal endoscopic surgery (NOTES) procedures. Surg. Endosc. 2012, 26, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Asfeldt, A.M.; Straume, B.; Paulssen, E.J. Impact of observer variability on the usefulness of endoscopic images for the documentation of upper gastrointestinal endoscopy. Scand. J. Gastroenterol. 2007, 42, 1106–1112. [Google Scholar] [CrossRef]
- Wong, P.K.; Chan, I.N.; Yan, H.M.; Gao, S.; Wong, C.H.; Yan, T.; Yao, L.; Hu, Y.; Wang, Z.R.; Yu, H.H. Deep learning based radiomics for gastrointestinal cancer diagnosis and treatment: A minireview. World J. Gastroenterol. 2022, 28, 6363–6379. [Google Scholar] [CrossRef] [PubMed]
- Rees, C.J.; Koo, S. Artificial Intelligence—Upping the game in gastrointestinal endoscopy? Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 584–585. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X. Medical image recognition and segmentation of pathological slices of gastric cancer based on Deeplab v3+neural network. Comput. Methods Programs Biomed. 2021, 207, 106210. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.J.; Zhou, Y.; Guo, Y.R. A brief survey on semantic segmentation with deep learning. Neurocomputing 2020, 406, 302–321. [Google Scholar] [CrossRef]
- Corral, J.E.; Keihanian, T.; Diaz, L.I.; Morgan, D.R.; Sussman, D.A. Management patterns of gastric polyps in the United States. Frontline Gastroenterol. 2019, 10, 16–23. [Google Scholar] [CrossRef]
- Zionts, S. MCDM—If not a roman numeral, then what? Interfaces 1979, 9, 94–101. [Google Scholar] [CrossRef]
- Sitorus, F.; Brito-Parada, P.R. The selection of renewable energy technologies using a hybrid subjective and objective multiple criteria decision making method. Expert Syst. Appl. 2022, 206, 117839. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, F.; Yu, T.; An, J.; Huang, Z.; Liu, J.; Hu, W.; Wang, L.; Duan, H.; Si, J. Real-time gastric polyp detection using convolutional neural networks. PLoS ONE 2019, 14, e0214133. [Google Scholar] [CrossRef]
- Laddha, M.; Jindal, S.; Wojciechowski, J. Gastric polyp detection using deep convolutional neural network. In Proceedings of the 2019 4th International Conference on Biomedical Imaging, Nagoya, Japan, 17–19 October 2019; pp. 55–59. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, W.; Nie, W.; Yu, Y. Gastric polyps detection by improved faster R-CNN. In Proceedings of the 2019 8th International Conference on Computing and Pattern Recognition, Beijing, China, 23–25 October 2019; pp. 128–133. [Google Scholar] [CrossRef]
- Cao, C.; Wang, R.; Yu, Y.; Zhang, H.; Yu, Y.; Sun, C. Gastric polyp detection in gastroscopic images using deep neural network. PLoS ONE 2021, 16, e0250632. [Google Scholar] [CrossRef]
- Durak, S.; Bayram, B.; Bakırman, T.; Erkut, M.; Doğan, M.; Gürtürk, M.; Akpınar, B. Deep neural network approaches for detecting gastric polyps in endoscopic images. Med. Biol. Eng. Comput. 2021, 59, 1563–1574. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Q.; Chen, S.; Liu, X.; Cao, Y.; Liu, B.; Zhang, H. Automatic disease detection in endoscopy with light weight transformer. Smart Health 2023, 28, 100393. [Google Scholar] [CrossRef]
- Triantaphyllou, E. Multi-criteria decision making methods. In Multi-Criteria Decision Making Methods: A Comparative Study: Applied Optimization; Springer: Boston, MA, USA, 2000; pp. 5–21. [Google Scholar]
- Saaty, R.W. The analytic hierarchy process—What it is and how it is used. Math. Model. 1987, 9, 161–176. [Google Scholar] [CrossRef]
- Shafaghat, A.; Keyvanfar, A.; Ket, C.W. A decision support tool for evaluating the wildlife corridor design and conservation performance using analytic network process (ANP). J. Nat. Conserv. 2022, 70, 126280. [Google Scholar] [CrossRef]
- Akter, S.; Debnath, B.; Bari, A.B.M.M. A grey decision-making trial and evaluation laboratory approach for evaluating the disruption risk factors in the emergency life-saving drugs supply chains. Healthc. Anal. 2022, 2, 100120. [Google Scholar] [CrossRef]
- Behzadian, M.; Otaghsara, S.K.; Yazdani, M.; Ignatiusc, J. A state-of-the-art survey of TOPSIS applications. Expert Syst. Appl. 2012, 39, 13051–13069. [Google Scholar] [CrossRef]
- Goodyear, M.D.; Krleza-Jeric, K.; Lemmens, T. The declaration of Helsinki. BMJ 2007, 335, 624–625. [Google Scholar] [CrossRef]
- Yeung, M.; Sala, E.; Schönlieb, C.B.; Rundo, L. Focus U-Net: A novel dual attention-gated CNN for polyp segmentation during colonoscopy. Comput. Biol. Med. 2021, 137, 104815. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2020, 39, 1856–1867. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.K.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with Atrous separable convolution for semantic image segmentation. arXiv 2018, arXiv:1802.02611. [Google Scholar] [CrossRef]
- Li, H.; Xiong, P.; An, J.; Wang, L. Pyramid attention network for semantic segmentation. arXiv 2018, arXiv:1805.10180. [Google Scholar] [CrossRef]
- Chaurasia, A.; Culurciello, E. LinkNet: Exploiting encoder representations for efficient semantic segmentation. arXiv 2017, arXiv:1707.03718. [Google Scholar] [CrossRef]
- Fan, T.L.; Wang, G.L.; Li, Y.; Wang, H.R. MA-Net: A multi-scale attention network for liver and tumor segmentation. IEEE Access 2020, 8, 179656–179665. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. arXiv 2014, arXiv:1911.07069. [Google Scholar] [CrossRef]
- Livni, R.; Shalev-Shwartz, S.; Shamir, O. On the computational efficiency of training neural networks. arXiv 2014, arXiv:1410.1141. [Google Scholar] [CrossRef]
- Lyakhov, P.; Valueva, M.; Valuev, G.; Nagornov, N. A method of increasing digital filter performance based on truncated multiply-accumulate units. Appl. Sci. 2020, 10, 9052. [Google Scholar] [CrossRef]
- Molchanov, P.; Tyree, S.; Karras, T.; Aila, T.; Kautz, J. Pruning convolutional neural networks for resource efficient inference. arXiv 2016, arXiv:1611.06440. [Google Scholar] [CrossRef]
- Diakoulaki, D.; Mavrotas, G.; Papayannakis, L. Determining objective weights in multiple criteria problems—The CRITIC method. Comput. Oper. Res. 1995, 22, 763–770. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.F. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Keys, R.G. Cubic convolution interpolation for digital image processing. IEEE Trans. Acoust. Speech Signal Process. 1981, 29, 1153–1160. [Google Scholar] [CrossRef]
- Jadon, S. A survey of Loss functions for semantic segmentation. In Proceedings of the 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology, Via del Mar, Chile, 27–29 October 2020; pp. 115–121. [Google Scholar] [CrossRef]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.L.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted residuals and linear bottlenecks. In Proceedings of the 2018 IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Tan, M.X.; Le, Q.V. EfficientNet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946. [Google Scholar] [CrossRef]
- Chowdhary, C.L.; Acharjya, D.P. Segmentation and feature extraction in medical imaging: A systematic review. Procedia Comput. Sci. 2020, 167, 26–36. [Google Scholar] [CrossRef]
- Sarwinda, D.; Paradisa, R.H.; Bustamam, A.; Anggia, P. Deep Learning in Image Classification using Residual Network (ResNet) Variants for Detection of Colorectal Cancer. Procedia Comput. Sci. 2021, 179, 423–431. [Google Scholar] [CrossRef]
- Mihara, M.; Yasuo, T.; Kitaya, K. Precision Medicine for Chronic Endometritis: Computer-Aided Diagnosis Using Deep Learning Model. Diagnostics 2023, 13, 936. [Google Scholar] [CrossRef] [PubMed]

| Reference | Dataset | Objective | Baseline | Constraint |
|---|---|---|---|---|
| Zhang et al. [18] | 404 images | Real-time detection of gastric polyps | SSD | 1. Most of the research focused on object detection, which is unable to portray the boundaries of polyps precisely. 2. The evaluation metrics are solitary without adequate consideration of clinical requirements. |
| Laddha et al. [19] | 654 images | Detection of gastric polyps | YOLOv3 and YOLOv3-tiny | |
| Wang et al. [20] | 1941 images | Detection of gastric polyps | Faster R-CNN | |
| Cao et al. [21] | 2270 images | Stomach classification and detection of gastric polyps | YOLOv3 | |
| Durak et al. [22] | 2195 image | Detection of gastric polyps | YOLOv4, CenterNet, EfficientNet, Cross Stage ResNext50-SPP, YOLOv3, YOLOv3-SPP, Single Shot Detection, and Faster Regional CNN |
| Model | Strength | Weakness |
|---|---|---|
| U-Net |
|
|
| UNet++ |
|
|
| DeepLabv3 |
|
|
| DeepLabv3+ |
|
|
| PAN |
|
|
| LinkNet |
|
|
| MA-Net |
|
|
| Metrics | Equations |
|---|---|
| IoU | |
| ACC | |
| RE | |
| PR | |
| F1 |
| First-Level Metrics | Second-Level Metrics | Weights |
|---|---|---|
| Segmentation accuracy | IoU | 0.3 |
| Accuracy | 0.05 | |
| Recall | 0.05 | |
| Precision | 0.05 | |
| F1-Score | 0.05 | |
| Computational efficiency | Number of parameters | 0.1 |
| Number of MACs | 0.1 | |
| FPS | 0.3 | |
| Sum of weight | 1 | |
| Configuration | Version |
|---|---|
| CPU | 11th Gen Intel(R) Core (TM) i9-11900 @ 2.50 GHz |
| GPU | NVIDIA GeForce RTX 3080 |
| RAM | 64.0 GB |
| Operating System | Windows 10 |
| Programing Language | Python 3.9 |
| Frame | Pytorch-1.10.0 |
| CUDA | 11.4.1 |
| cuDNN | 11.4 |
| Model | Encoder | IoU (%) | ACC (%) | RE (%) | PR (%) | F1 (%) | No. of Parameters (M) | GMACs | FPS | CRITIC Score | Subjective Score | Final Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U-Net | ResNet50 | 94.96 | 97.29 | 97.31 | 97.27 | 97.29 | 32.52 | 10.7 | 24 | 0.59 | 0.63 | 0.61 |
| MobileNet v2 | 95.56 | 97.56 | 97.57 | 97.55 | 97.56 | 6.63 | 3.39 | 26 | 0.75 | 0.81 | 0.78 | |
| EfficientNet-B1 | 96.53 | 98.14 | 98.16 | 98.12 | 98.14 | 8.76 | 2.53 | 22 | 0.77 | 0.74 | 0.75 | |
| UNet++ | ResNet50 | 96.57 | 98.18 | 98.20 | 98.16 | 98.18 | 48.99 | 57.54 | 20 | 0.53 | 0.53 | 0.53 |
| MobileNet v2 | 96.27 | 98.00 | 98.03 | 97.98 | 98.00 | 6.82 | 4.5 | 26 | 0.84 | 0.88 | 0.86 | |
| EfficientNet-B1 | 96.79 | 98.11 | 98.14 | 98.09 | 98.11 | 9.08 | 5.1 | 21 | 0.73 | 0.70 | 0.72 | |
| DeepLabv3 | ResNet50 | 96.23 | 97.95 | 97.98 | 97.93 | 97.96 | 39.63 | 40.99 | 24 | 0.65 | 0.70 | 0.67 |
| MobileNet v2 | 95.79 | 97.69 | 97.73 | 97.66 | 97.69 | 12.65 | 12.74 | 26 | 0.75 | 0.81 | 0.78 | |
| EfficientNet-B1 | 95.46 | 97.37 | 97.40 | 97.34 | 97.37 | 9.81 | 3.37 | 23 | 0.63 | 0.65 | 0.64 | |
| DeepLabv3+ | ResNet50 | 96.25 | 97.97 | 98.02 | 97.93 | 97.97 | 26.68 | 9.2 | 26 | 0.81 | 0.86 | 0.83 |
| MobileNet v2 | 95.22 | 97.36 | 97.42 | 97.32 | 97.37 | 4.38 | 1.52 | 27 | 0.75 | 0.82 | 0.78 | |
| EfficientNet-B1 | 95.93 | 97.75 | 97.83 | 97.70 | 97.76 | 7.41 | 0.56 | 23 | 0.72 | 0.72 | 0.72 | |
| PAN | ResNet50 | 96.06 | 97.87 | 98.00 | 97.78 | 97.89 | 8.71 | 24.26 | 26 | 0.77 | 0.82 | 0.80 |
| MobileNet v2 | 95.17 | 97.27 | 97.50 | 97.14 | 97.31 | 2.42 | 0.79 | 26 | 0.72 | 0.77 | 0.75 | |
| EfficientNet-B1 | 91.83 | 95.09 | 98.86 | 92.71 | 95.52 | 6.6 | 0.09 | 22 | 0.52 | 0.33 | 0.43 | |
| LinkNet | ResNet50 | 96.31 | 98.03 | 98.03 | 98.03 | 98.03 | 31.18 | 10.77 | 26 | 0.81 | 0.86 | 0.83 |
| MobileNet v2 | 95.11 | 97.32 | 97.33 | 97.32 | 97.32 | 4.32 | 0.94 | 26 | 0.71 | 0.77 | 0.74 | |
| EfficientNet-B1 | 96.32 | 98.04 | 98.08 | 98.00 | 98.04 | 3.67 | 0.19 | 22 | 0.76 | 0.72 | 0.74 | |
| MA-Net | ResNet50 | 96.34 | 98.03 | 98.06 | 98.01 | 98.03 | 147.44 | 18.64 | 21 | 0.54 | 0.55 | 0.55 |
| MobileNet v2 | 96.23 | 97.98 | 98.00 | 97.96 | 97.98 | 48.89 | 5.27 | 24 | 0.74 | 0.76 | 0.75 | |
| EfficientNet-B1 | 96.57 | 98.15 | 98.16 | 98.14 | 98.15 | 11.6 | 2.41 | 21 | 0.74 | 0.70 | 0.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, T.; Qin, Y.Y.; Wong, P.K.; Ren, H.; Wong, C.H.; Yao, L.; Hu, Y.; Chan, C.I.; Gao, S.; Chan, P.P. Semantic Segmentation of Gastric Polyps in Endoscopic Images Based on Convolutional Neural Networks and an Integrated Evaluation Approach. Bioengineering 2023, 10, 806. https://doi.org/10.3390/bioengineering10070806
Yan T, Qin YY, Wong PK, Ren H, Wong CH, Yao L, Hu Y, Chan CI, Gao S, Chan PP. Semantic Segmentation of Gastric Polyps in Endoscopic Images Based on Convolutional Neural Networks and an Integrated Evaluation Approach. Bioengineering. 2023; 10(7):806. https://doi.org/10.3390/bioengineering10070806
Chicago/Turabian StyleYan, Tao, Ye Ying Qin, Pak Kin Wong, Hao Ren, Chi Hong Wong, Liang Yao, Ying Hu, Cheok I Chan, Shan Gao, and Pui Pun Chan. 2023. "Semantic Segmentation of Gastric Polyps in Endoscopic Images Based on Convolutional Neural Networks and an Integrated Evaluation Approach" Bioengineering 10, no. 7: 806. https://doi.org/10.3390/bioengineering10070806
APA StyleYan, T., Qin, Y. Y., Wong, P. K., Ren, H., Wong, C. H., Yao, L., Hu, Y., Chan, C. I., Gao, S., & Chan, P. P. (2023). Semantic Segmentation of Gastric Polyps in Endoscopic Images Based on Convolutional Neural Networks and an Integrated Evaluation Approach. Bioengineering, 10(7), 806. https://doi.org/10.3390/bioengineering10070806