Near-Infrared Blood Vessel Image Segmentation Using Background Subtraction and Improved Mathematical Morphology
Abstract
1. Introduction
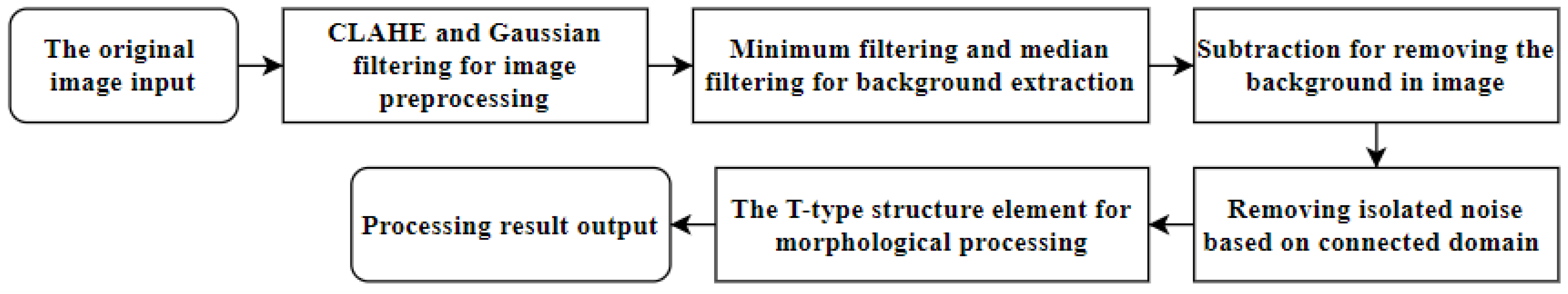
2. Primary Computational Methods
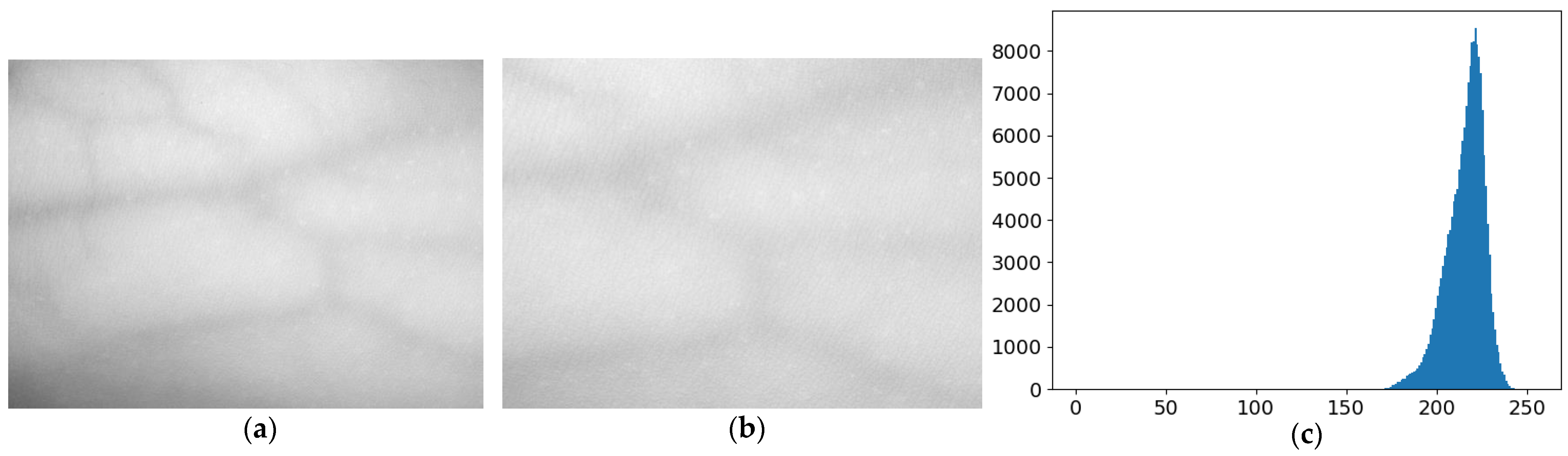
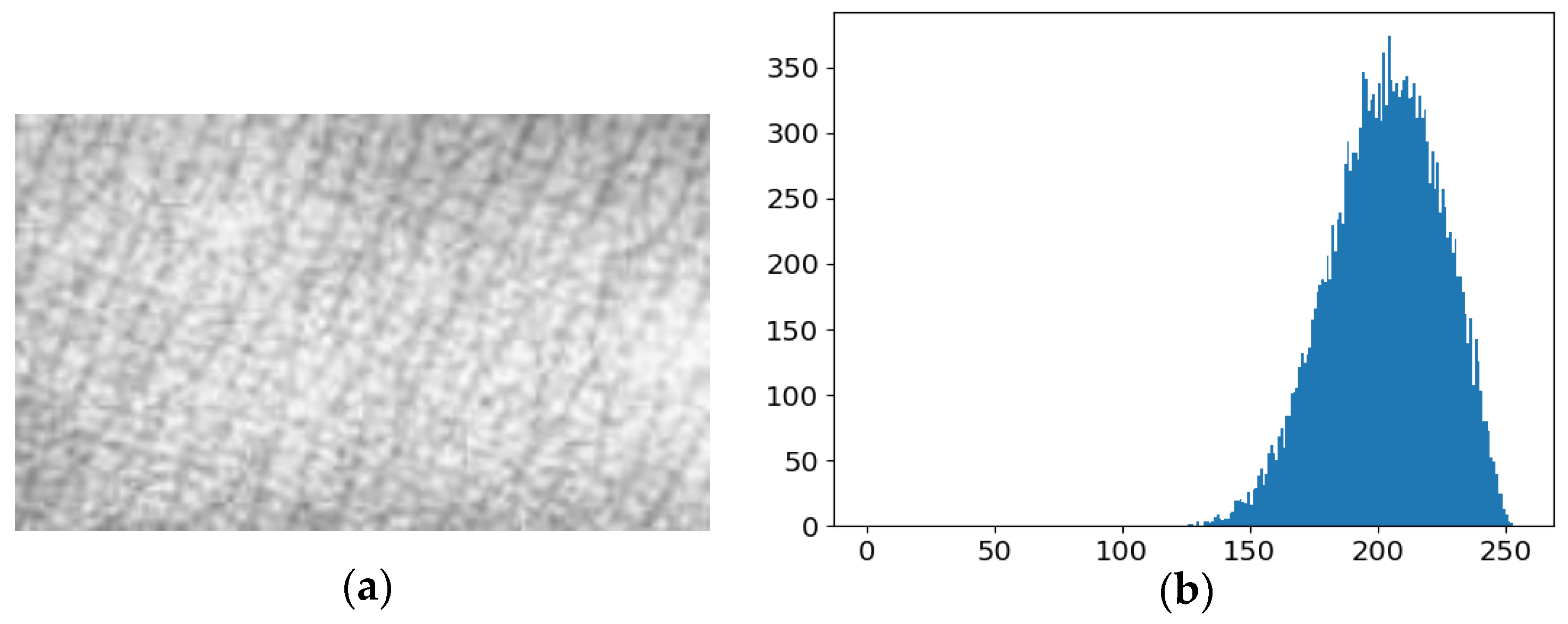
2.1. Dataset Preprocessing and Enhancement
2.2. Vascular Information Extraction and Image Background Subtraction
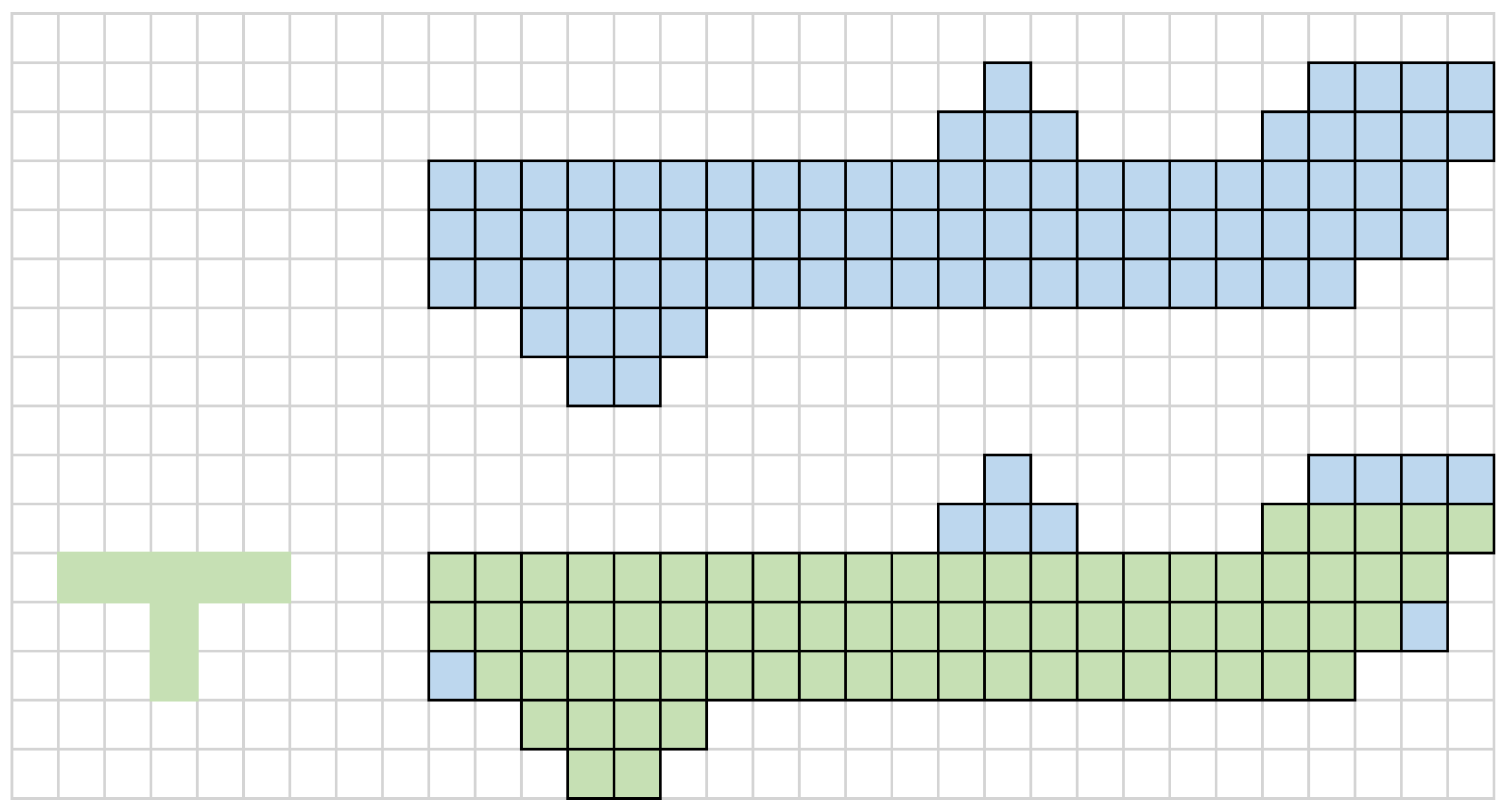
2.3. Image Denoising and Morphological Processing
3. Experiment Results and Evaluations
3.1. Evaluation of Image Enhancement Algorithm
3.2. Evaluation of Noise Reduction Algorithm
3.3. Evaluation of Image Segmentation Algorithm
3.4. Evaluation of Morphological Algorithm
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waitt, C.; Waitt, P.; Pirmohamed, M. Intravenous therapy. Postgrad. Med. J. 2004, 80, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lamperti, M.; Pittiruti, M., II. Difficult peripheral veins: Turn on the lights. Br. J. Anaesth. 2013, 110, 888–891. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, S.; Chatterjee, S.; Katz, N.; Nelson, M.; Goldbaum, M. Detection of blood vessels in retinal images using two-dimensional matched filters. IEEE Trans. Med. Imaging 1989, 8, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; You, J.; Zang, D. Vessel segmentation and width estimation in retinal images using multiscale production of matched filter responses. Expert Syst. Appl. 2012, 39, 7600–7610. [Google Scholar] [CrossRef]
- Hoover, A.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J. Finger-vein image enhancement based on combination of gray-level grouping and circular Gabor filter. In Proceedings of the 2009 International Conference on Information Engineering and Computer Science, Wuhan, China, 19–21 December 2009; pp. 1–4. [Google Scholar]
- Lee, E.C.; Park, K.R. Restoration method of skin scattering blurred vein image for finger vein recognition. Electron. Lett. 2009, 45, 1074–1076. [Google Scholar] [CrossRef]
- Yin, Y.; Adel, M.; Bourennane, S. Retinal vessel segmentation using a probabilistic tracking method. Pattern Recognit. 2012, 45, 1235–1244. [Google Scholar] [CrossRef]
- Liu, B.; Jiang, Q.; Liu, W.; Wang, M.; Zhang, S.; Zhang, X.; Zhang, B.; Yue, Z. A vessel segmentation method for serialized cerebral vascular DSA images based on spatial feature point set of rotating coordinate system. Comput. Methods Programs Biomed. 2018, 161, 55–72. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, S. Adaptive Kalman snake for semiautonomous 3D vessel tracking. Comput. Methods Programs Biomed. 2015, 122, 56–75. [Google Scholar] [CrossRef]
- Song, M.; Civco, D. Road extraction using SVM and image segmentation. Photogramm. Eng. Rem. 2004, 70, 1365–1371. [Google Scholar] [CrossRef]
- Pathak, D.; Krähenbühl, P.; Darrell, T. Constrained convolutional neural networks for weakly supervised segmentation. IEEE Int. Conf. Comput. Vis. 2015, 15, 1796–1804. [Google Scholar]
- Liskowski, P.; Krawiec, K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans. Med. Imaging 2016, 35, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. Lect. Notes Comput. Sci. 2015, 16, 711–720. [Google Scholar]
- Sánchez, J.C.G.; Magnusson, M.; Sandborg, M.; Carlsson; Tedgren, Å.; Malusek, A. Segmentation of bones in medical dual-energy computed tomography volumes using the 3D U-Net. Phys. Med. 2020, 69, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Hiasa, Y.; Otake, Y.; Takao, M.; Ogawa, T.; Sugano, N.; Sato, Y. Automated muscle segmentation from clinical CT using Bayesian U-Net for personalized musculoskeletal modeling. IEEE Trans. Med. Imaging 2020, 39, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Perslev, M.; Pai, A.; Runhaar, J.; Igel, C.; Dam, E.B. Cross-cohort automatic knee MRI segmentation with multi-planar U-Nets. J. Magn. Reason. Imaging 2022, 55, 1650–1663. [Google Scholar] [CrossRef]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted Res-UNet for high-quality retina vessel segmentation. In Proceedings of the 2018 IEEE 9th International Conference on Information Technology in Medicine and Education, Hangzhou, China, 19–21 October 2018; pp. 327–331. [Google Scholar]
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent residual U-Net for medical image segmentation. J. Med. Imaging 2018, 16, 228–233. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, Y.; Bian, K.; Tuo, H.; Song, L. LadderNet: Knowledge transfer based viewpoint prediction in 360° video. In Proceedings of the 2019 IEEE International Conference on Acoustics, Speech and Signal Processing, Brighton, UK, 12–17 May 2019; pp. 1657–1661. [Google Scholar]
- Ning, Z.; Zhong, S.; Feng, Q.; Chen, W.; Zhang, Y. SMU-Net: Saliency-guided morphology-aware U-Net for breast lesion segmentation in ultrasound image. IEEE Trans. Med. Imaging 2021, 41, 476–490. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.; Tian, Z.; Geng, W. Near-infrared vascular image segmentation using improved level set method. Infrared Phys. Technol. 2023, 133, 104678. [Google Scholar] [CrossRef]
- Alwazzan, M.J.; Ismael, M.A.; Ahmed, A.N. A hybrid algorithm to enhance colour Retinal fundus images using a Wiener filter and CLAHE. J. Digit. Imaging 2021, 34, 750–759. [Google Scholar] [CrossRef]
- Arpah, B.A.S.; Taib, M.N.; Khalid, N.E.; Taib, H. Variations of adaptive histogram equalization (AHE) analysis on intra-oral dental radiograph. In Proceedings of the 2015 IEEE 6th Control and System Graduate Research Colloquium, Shah Alam, Malaysia, 10–11 August 2015; pp. 87–92. [Google Scholar]
- Liu, X.W.; Liu, C.Y. An optional Gauss filter image denoising method based on difference image fast fuzzy clustering. Appl. Mech. Mater. 2013, 414, 1348–1352. [Google Scholar] [CrossRef]
- Portilla, J.; Strela, V.; Wainwright, M.J.; Simoncelli, E.P. Image denoising using scale mixtures of Gaussians in the wavelet domain. IEEE Trans. Image Process. 2003, 12, 1338–1351. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wang, Y.; Cao, W.; Yang, J. Improved adaptive median filtering for structured light image denoising. In Proceedings of the 2019 7th International Conference on Information, Communication and Networks, Macao, China, 24–26 April 2019; pp. 146–149. [Google Scholar]
- Cheng, H.; Jiang, X.; Sun, Y.; Wang, J. Color image segmentation: Advances and prospects. Pattern Recognit. 2001, 34, 2259–2281. [Google Scholar] [CrossRef]
- Xiaosong, S.; Yongjie, H.; Yongge, L. Text on Oracle rubbing segmentation method based on connected domain. In Proceedings of the 2016 IEEE Advanced Information Management, Communicates, Electronic and Automation Control Conference, Xi’an, China, 3–5 October 2016; pp. 414–418. [Google Scholar]
- Haralick, R.M.; Sternberg, S.R.; Zhuang, X. Image analysis using mathematical morphology. IEEE Trans. Pattern Anal. Mach. Intell. 1987, 9, 532–550. [Google Scholar] [CrossRef] [PubMed]
- Sargun; Rana, S.B. Performance evaluation of HE, AHE and fuzzy image enhancement. Int. J. Comput. Appl. 2015, 122, 14–19. [Google Scholar] [CrossRef]
- Zhang, G.; Sun, D.; Yan, P.; Zhao, H.; Li, Z. A LDCT image contrast enhancement algorithm based on single-scale Retinex theory. In Proceedings of the 2008 International Conference on Computational Intelligence for Modelling Control & Automation, Vienna, Austria, 10–12 December 2008; pp. 1282–1287. [Google Scholar]
- Yu, H.; Li, X.; Lou, Q.; Yan, L. Underwater image enhancement based on color-line model and homomorphic filtering. Signal Image Video P 2021, 16, 83–91. [Google Scholar] [CrossRef]
- Chen, B.; Tseng, Y.; Yin, J. Gaussian-adaptive bilateral filter. IEEE Signal Process. Lett. 2020, 27, 1670–1674. [Google Scholar] [CrossRef]
- Erkan, U.; Gökrem, L.; Enginoğlu, S. Different applied median filter in salt and pepper noise. Comput. Electr. Eng. 2018, 70, 789–798. [Google Scholar] [CrossRef]
- Fu, B.; Xiong, X.; Sun, G. An efficient mean filter algorithm. In Proceedings of the 2011 IEEE/ICME International Conference on Complex Medical Engineering, Harbin, China, 22–25 May 2011; pp. 466–470. [Google Scholar]
- Yang., J.; Fan, J.; Ai, D.; Zheng, Y.; Tang, S.; Wang, Y. Local statistics and non-local mean filter for speckle noise reduction in medical ultrasound image. Neural Comput. 2016, 195, 88–95. [Google Scholar] [CrossRef]
- Malladi, S.R.; Ram, S.; Rodríguez, J.J. Image denoising using superpixel-based PCA. IEEE Trans. Multimedia 2021, 23, 2297–2309. [Google Scholar] [CrossRef]
- Yan, Z.; Chen, G.; Xu, W.; Yang, C.; Lu, Y. Study of an image autofocus method based on power threshold function wavelet reconstruction and a quality evaluation algorithm. Appl. Opt. 2018, 33, 9714–9721. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Wu, M.; Liu, K. Multiple levels perceptual noise backed visual information fidelity for picture quality assessment. In Proceedings of the 2022 International Symposium on Intelligent Signal Processing and Communication Systems, Penang, Malaysia, 22–25 November 2022; pp. 1–4. [Google Scholar]
- Kim, D.; Lee, S.; Hong, W.; Lee, H.; Jeon, S.; Han, S.; Nam, J. Image segmentation for FIB-SEM serial sectioning of a Si/C–graphite composite anode microstructure based on preprocessing and global thresholding. Microsc. Microanal. 2019, 25, 1139–1154. [Google Scholar] [CrossRef] [PubMed]
- Hasikin, K.; Isa, N.A.; Mohamed, M.; Suandi, S.A.; Kadri, N.A. A new region-based adaptive thresholding for sperm motility segmentation. Malays. J. Comput. Sci. 2017, 29, 272–286. [Google Scholar] [CrossRef]
- Wong, A.K.C.; Sahoo, P.K. A gray-level threshold selection method based on maximum entropy principle. IEEE Trans. Syst. Man Cybern. Syst. 1989, 19, 866–871. [Google Scholar] [CrossRef]
- Fu, Z.L. Image thresholding method—A generalization of Otsu’s method. Comput. Appl. 2000, 20, 37–39. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. Syst. 2007, 9, 62–66. [Google Scholar] [CrossRef]
- Eelbode, T.; Bertels, J.; Berman, M.; Vandermeulen, D.; Maes, F.; Bisschops, R.; Blaschko, M.B. Optimization for medical image segmentation: Theory and practice when evaluating with Dice score or Jaccard index. IEEE Trans. Med. Imaging 2020, 39, 3679–3690. [Google Scholar] [CrossRef]
- Moccia, S.; Momi, E.D.; Hadji, S.E.; Mattos, L.S. Blood vessel segmentation algorithms—Review of methods, datasets and evaluation metrics. Comput. Methods Programs Biomed. 2018, 158, 71–91. [Google Scholar] [CrossRef]
- Ng, H.P.; Ong, S.H.; Foong, K.W.; Goh, P.; Nowinski, W.L. Medical image segmentation using K-Means clustering and improved watershed algorithm. In Proceedings of the 2006 IEEE Southwest Symposium on Image Analysis and Interpretation, Denver, CO, USA, 26–28 March 2006; pp. 61–65. [Google Scholar]
- Roerdink, J.B.T.M.; Meijster, A. The watershed transform: Definitions, algorithms and parallelization strategies. Fund. Inform. 2000, 41, 187–228. [Google Scholar] [CrossRef]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Med. Imaging 2001, 10, 266–277. [Google Scholar] [CrossRef]
- Li, C.; Kao, C.-Y.; Gore, J.C.; Ding, Z. Implicit active contours driven by local binary fitting energy. In Proceedings of the 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007; pp. 1–7. [Google Scholar]


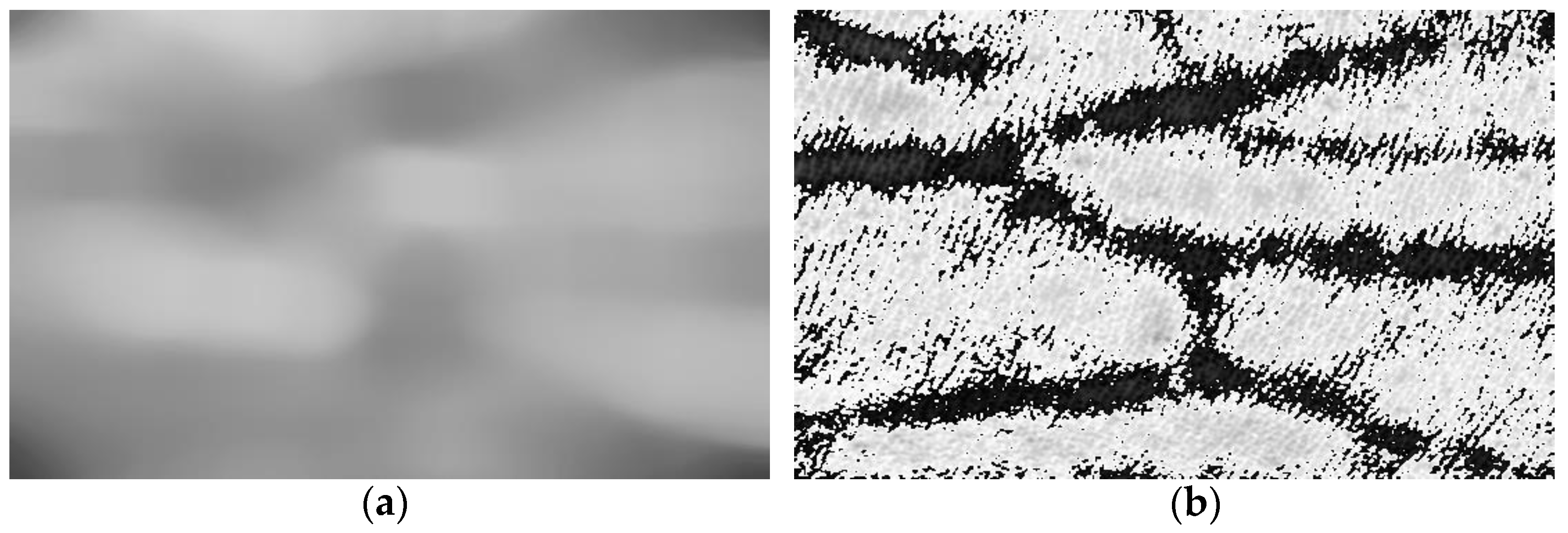




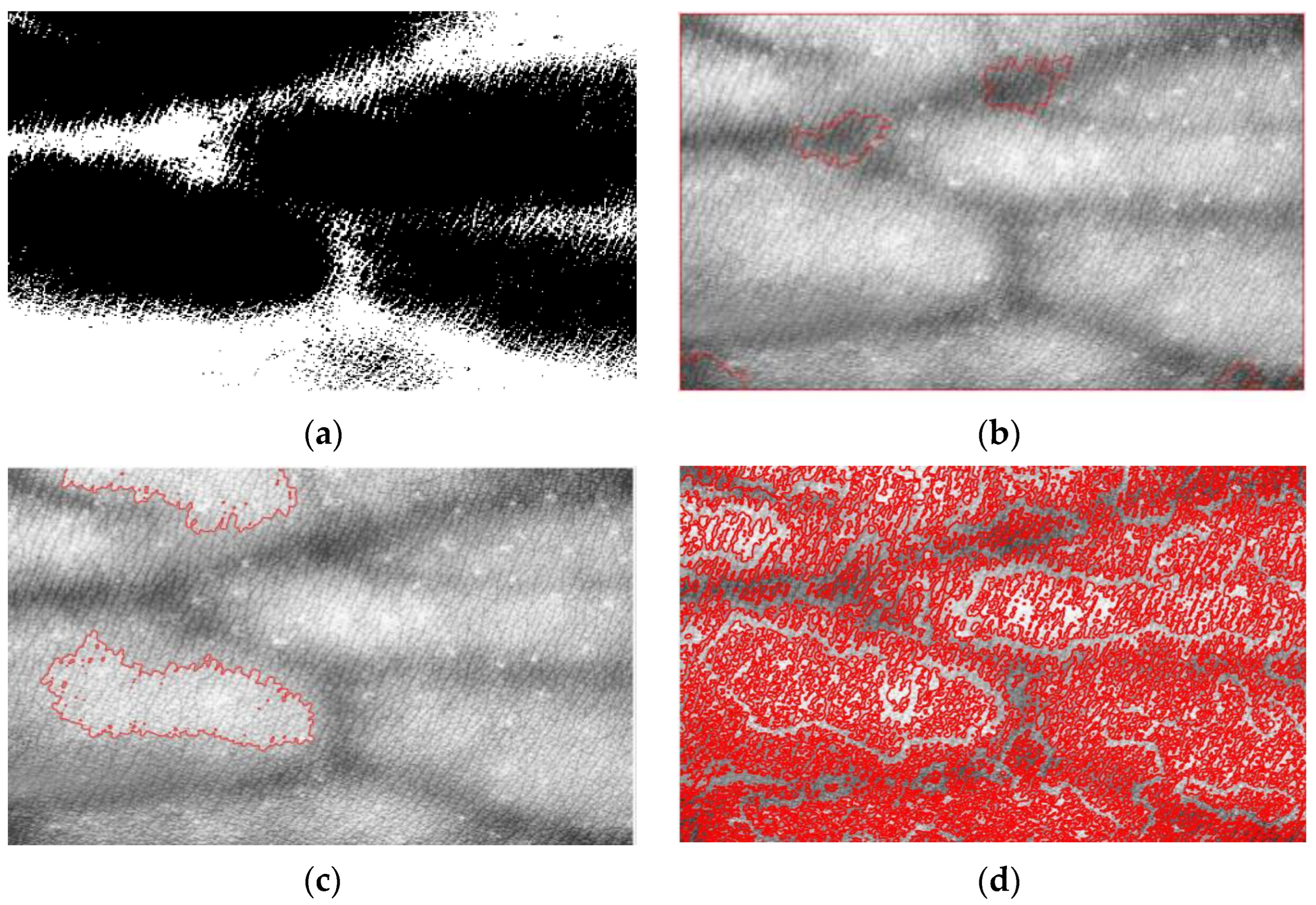
| Name of Filtering Algorithm | Energy | Brenner | VIF |
|---|---|---|---|
| Bilateral filtering | 0.111113 | 0.149405 | 0.0229357 |
| Median filtering | 0.142327 | 0.185371 | 0.0171459 |
| Mean filtering | 0.542328 | 0.717005 | 0.228890 |
| Non-local means denoising | 0.166525 | 0.200364 | 0.192109 |
| Bandpass filtering | 0 | 0 | 6.829285 × 10−5 |
| Gaussian filtering (Our method) | 0.579129 | 0.788554 | 0.314532 |
| Name of Filtering Algorithm | Dice | Acc | Sen |
|---|---|---|---|
| Threshold segmentation method based on region growth | 0.66151 | 0.570808 | 0.668102 |
| Threshold method based on regional growth | 0.670982 | 0.489470 | 0.811824 |
| Entropy threshold method | 0.662919 | 0.456471 | 0.8487629 |
| OTSU threshold segmentation algorithm | 0.670982 | 0.489471 | 0.811824 |
| Histogram-based technique for threshold segmentation | 0.457068 | 0.657206 | 0.319836 |
| The algorithm in this paper | 0.697933 | 0.443855 | 0.938922 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Liu, H.; Li, Q.; Tian, Z.; Li, Y.; Geng, W.; Wang, S. Near-Infrared Blood Vessel Image Segmentation Using Background Subtraction and Improved Mathematical Morphology. Bioengineering 2023, 10, 726. https://doi.org/10.3390/bioengineering10060726
Li L, Liu H, Li Q, Tian Z, Li Y, Geng W, Wang S. Near-Infrared Blood Vessel Image Segmentation Using Background Subtraction and Improved Mathematical Morphology. Bioengineering. 2023; 10(6):726. https://doi.org/10.3390/bioengineering10060726
Chicago/Turabian StyleLi, Ling, Haoting Liu, Qing Li, Zhen Tian, Yajie Li, Wenjia Geng, and Song Wang. 2023. "Near-Infrared Blood Vessel Image Segmentation Using Background Subtraction and Improved Mathematical Morphology" Bioengineering 10, no. 6: 726. https://doi.org/10.3390/bioengineering10060726
APA StyleLi, L., Liu, H., Li, Q., Tian, Z., Li, Y., Geng, W., & Wang, S. (2023). Near-Infrared Blood Vessel Image Segmentation Using Background Subtraction and Improved Mathematical Morphology. Bioengineering, 10(6), 726. https://doi.org/10.3390/bioengineering10060726







