Abstract
The tremendous personal and economic burden worldwide caused by low back pain (LBP) has been surging in recent years. While intervertebral disc degeneration (IVDD) is the leading cause of LBP and vast efforts have been made to develop effective therapies, this problem is far from being resolved, as most treatments, such as painkillers and surgeries, mainly focus on relieving the symptoms rather than reversing the cause of IVDD. However, as stem/progenitor cells possess the potential to regenerate IVD, a deeper understanding of the early development and role of these cells could help to improve the effectiveness of stem/progenitor cell therapy in treating LBP. Single-cell RNA sequencing results provide fresh insights into the heterogeneity and development patterns of IVD progenitors; additionally, we compare mesenchymal stromal cells and IVD progenitors to provide a clearer view of the optimal cell source proposed for IVD regeneration.
1. Introduction
Low back pain (LBP) is an increasing social and economic burden on both global governments and individuals [1]. One of the major causes of LBP is the intervertebral disc degeneration (IVDD), which is characterized by the loss and dysfunction of IVD cells and the exhaustion of IVD progenitors [2,3]. The further development of IVDD leads to disc herniation, which exacerbates LBP. Current therapies against IVDD and disc herniation mainly include immobilization, analgesic drugs and surgeries [4]. While these therapies alleviate the symptoms, none reverse the IVD condition; however, developing stem/progenitor therapies could restore the IVD matrix and promote the growth of IVD cells. The IVD consists of three parts that are from distinct embryonic origins: the nucleus pulposus (NP), the annulus fibrosus (AF) and the cartilaginous endplate (CEP). The NP originates from the notochord [5], while the AF and CEP originate from the sclerotome [6]. After a series of cellular transformations with intrinsic regulation, the notochord turns into the NP, and the maturation of cells within the AF and CEP also results in the ablation of multipotency. However, a group of progenitors within the IVD retain their stemness and may play critical roles in future therapies.
Progenitors originating from NP, AF and CEP have all been discovered, and they possess tremendous potential to revive degenerated IVDs [7] by differentiating into corresponding mature IVD cells, giving rise to local mature IVD cells, promoting IVD matrix production and modulating several signaling pathways [8]. Several stem/progenitor cells clinical experiments have already been carried out to treat IVDD. However, current clinical studies tend to focus on the use of traditional mesenchymal stromal cells, such as bone marrow stromal cells (BMSCs) [9]. Although significant progress has been made, not enough studies have been conducted to allow comparisons to be made between IVD progenitors and traditional cells; thus, we are unable to determine which cells are the optimal choice for progenitor therapy [10]. Additionally, the origins and developmental routines of IVD cells have yet to be identified, especially in terms of how to precisely manipulate IVD progenitor differentiation.
2. The Heterogeneity of IVD Cells: Evidence from Single-Cell RNA Sequencing
As each part of the IVD possesses distinct cell compositions, developing a deeper understanding of heterogeneity is a prerequisite for cracking the development code of the IVD cells [10]. As illustrated in Figure 1, using single-cell RNA sequencing techniques, the discovery of novel clustering patterns and progenitor markers has provided fresh insights into the existence and development of IVD progenitors.
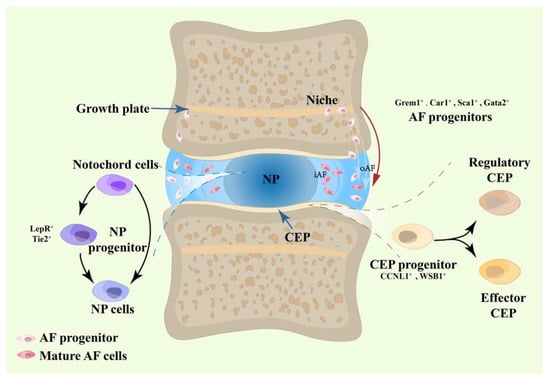
Figure 1.
Evidence from single-cell RNA sequencing to determine the existence of IVD progenitors.
Table 1 summarizes the isolation protocols for the IVD progenitors, which involve mechanically mincing, digesting, expanding and confirming the phenotypes. The age of the donor is a primary factor affecting cellular biology. Tie2+ cells, which were thought to be potential IVD progenitors, have been found to possess much lower viability when they were isolated from older donors, and the viability decreases rapidly after the age of 25 in human donors [11]. Similar conclusions were found in the murine and canine IVD progenitors [12].

Table 1.
Isolation protocols of IVD progenitors from different tissues.
2.1. NP Progenitors
Single-cell RNA sequencing supports the existence of NP progenitors. In a recent study, uniform manifold approximation and projection (UMAP) analysis identified 15 cell populations in human neonatal and adult IVDs, within which a special cluster was termed as NC/NPC because it possessed both the notochord cell (NC) marker SOX4 and the NP cell marker Col2a1, suggesting a transition state in the development process or the presence of an NP progenitor [18]. Transcriptional entropy analysis, which evaluates the extent of stemness, was also implemented in the bovine tail IVD; by using the transcriptional entropy score of the NC cluster (0.89) as a benchmark, the possible progenitor clusters in the NP region reached a score of 0.86, while other clusters achieved lower scores [19]. Although NP progenitors were derived from NCs, it has been found that the NC–NP progenitor–NP route is not the only development route [18].
Single-cell RNA sequencing detected other clustering patterns. NP cells extracted from human NP tissues were classified into six clusters presenting different functions, such as immunomodulation, fibrocartilaginous growth and inflammation. CD70+ and CD82+ NP progenitors have also been found [20], and UTS2R [21] and PDGFRA [22] were identified as progenitor markers in human IVDs. In rat IVDs, stem-like cells expressing MSC markers were observed and termed NP progenitors [23]. Clusters close to notochordal lineages in bovine discs were also discovered and characterized by the pluripotent or progenitor genes KRT15, CD44 and CD55 [24]. Another study that adopted degenerated human NPs as specimens suggested the presence of NP progenitors using the leptin receptor (LepR), which has recently been identified as a stem/progenitor marker [25]. NP progenitors were found to be positive on LepR and displayed a descending trend afterbirth like NCs. Anabolic matrix proteins, such as aggrecan, also generally surrounded LepR+ NP cells [25].
2.2. AF Progenitors
Single-cell RNA sequencing revealed multiple clusters within bovine tail AF, representing various functions, including AF progenitor cells, which were found by single-cell RNA sequencing [24]. The transcriptional entropy analysis found the potential high stemness of cell clusters; the entropy score stayed around 0.85–0.86, compared to 0.89 for NCs and 0.86 for potential NP progenitors. The potential AF progenitor existed only in the outer AF (oAF), as the score of the inner AF (iAF) cells was lower than 0.85 [19]. A group of type-II collagen positive cells found in the AF contributed greatly to IVD development and repairment, presenting a descending trend afterbirth. In addition, the deletion of the type-II collagen gene led to the disruption of the spine pattern, characterized by an apparent reduction in the cartilaginous area and ECM production [26]. Another study using rat AF identified Grem1+ cells as AF progenitors, which was proved by their stemness markers Id1, Cripsld1, Cytl1 and Fos, as well as their high entropy scores [27].
The temporal and developmental patterns of AF cells were also discovered in mice models. AF progenitor cells from mice IVD were divided into five clusters based on the molecular signature, such as Car1, Adgrg1 and Cnmd. The earliest form of the AF progenitors lay in the stem cell niche adjacent to the epiphyseal plate, and they migrated through a specific route to the AF and differentiated into chondrocyte-like AF and fibroblast-like AF progenitors. Both types of progenitors developed into corresponding mature AF cells and migrated simultaneously to the inner and outer AF, respectively [17]. Notably, increasing evidence showed that AF progenitor cells exist in the outer AF (oAF) zone rather than the inner AF (iAF), as the stem/progenitor marker LepR was primarily aggregated in the oAF [25]. Moreover, bovine oAF cells had higher scores on entropy analysis [19], and type-II collagen positive cells were more intense in the oAF area [26].
2.3. CEP Progenitors
CEP progenitors were found to be spindle-shaped and positive in CD29, CD44, CD73, CD90 and CD105 but negative in CD34, CD45, CD11b, CD19 and HLA-DR [28,29]; novel biomarkers such as CCNL1 and WSB1 were also found to be positive in CEP progenitors [8]. Previous studies have shown that CEP progenitors have osteogenic, adipogenic and chondrogenic potential [28,30].
In a single-cell RNA sequencing utilizing human IVD data, three clusters of chondrocytes were found: homeostatic, regulatory and effector. Trajectory analysis predicted that the homeostatic chondrocytes, which express CCNL1, WSB1 and MSCs markers, were located in the root area in place of CEP progenitors; then, they became regulatory and effector chondrocytes that were responsible for IVDD and bone/cartilage growth, respectively [8].
3. Key Pathways in the Early Development of IVD Progenitors
Cells within the IVD possess unique developmental trajectories. During embryogenesis, the early stage of the human embryo forms the axial mesoderm and the paraxial mesoderm. The axial mesoderm then becomes the notochord, whose cells are currently thought to have a multilineage differentiation ability and maintain disc homeostasis. Notochordal cells transform into NP cells and offer migration and differentiation signals to transform the paraxial mesoderm-originated sclerotome into the AF [31]. As mentioned previously, mature discs also comprised various functional clusters that demonstrate distinct molecular signatures and prospective functions, and they derived from the corresponding IVD progenitor cells. Although mechanisms for curing IVDD have been proposed [32], many studies have solely focused on helping IVD progenitor cells survive longer in the harsh environment within the degenerated IVD or on activating their therapeutic potential [33]. However, it is important to understand the mechanisms responsible for early differentiation [32]. It is also crucial to use these mechanisms to promote the more precisely directed differentiation of IVD progenitors to reduce degeneration.
As summarises in Figure 2, notochord formation involves a series of intrinsic signals, including BMP, WNT and Activin/Nodal. By activating the WNT pathway via GSK3β, CHIR definitively induced differentiation towards mesendoderm progenitor cells (MSPCs), but it was not enough to maintain their notochordal fate, even with Activin/Nodal. The subsequent transfection of the notochordal typical gene NOTO in MSPCs created and maintained a stable notochordal cell population, which was in accordance with a previous study [34]. Sequencing results coincided with this, as the prolonged expression of mesendoderm genes persisted only in NOTO-transfected cells. Additional functional enrichment analysis highlighted notochord development, which further validated this finding [31]. This finding supports the vital impression of the WNT and BMP pathways in the formation of NC cells.
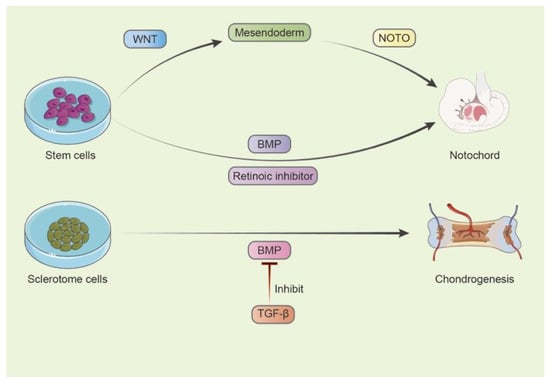
Figure 2.
Key pathways controlling progenitor development.
The notochord marker gene NOTO transfection in mesendoderm progenitor cells was also found to induce distinct expression genes [31]. Upregulated genes can be divided into two clusters. The maximum expression level of one cluster appeared on day 3 post-transfection and degraded on day 7, while the other peaked on day 7, including the pivotal notochord markers sonic hedgehog (Shh) and FoxA1 [31]. In another study, LAFC-induced notochord differentiation from hESCs demonstrated the decreased stemness markers Nanog and Sox-2 at day 6, which represented an effective commitment of mesodermal lineage [34]. A conclusion can be made that, in notochord induction, approximately one week might be the key point of success.
Another study combined BMP and the retinoic acid inhibitor LDN/AGN/FGF (LAF treatment) to initiate NC differentiation, marked by an increased expression of NOTO and FOXA2 mRNA levels, which could also be enhanced by CHIR. Single-cell RNA sequencing revealed that, after LAFC (LAF + CHIR) treatment, human ESCs developed NP-like cell clusters [34]. The ablation of the type-II collagen gene in mice embryos also resulted in calcified vertebrae, short limbs and rapid death after birth, and postnatal deletion disrupted AF and NP cells and ECM formation. This experiment highlighted the importance of the type-II collagen gene, suggesting that it might be the controlling gene of progenitor function [26].
For sclerotome development, chondrogenesis and fibrogenesis require varied intercellular signals. TGF-β increased the expression of the BMP antagonist Noggin and thus inhibited the BMP/Smad pathway, promoting chondrogenesis. Sclerotome cells were induced into chondrocytes by signaling molecules from the BMP family; however, this process could be inhibited by TGF-β1. Additionally, BMP/Noggin enhanced Sox-9/4 and Scx expression. On the other hand, exogenous BMP inhibitors, such as Gremlin, suppressed chondrogenesis, but not enough to initiate the fibrotic differentiation of sclerotome cells [35]. This finding indicates the lineage differentiation of the sclerotome and ways to optimize progenitor/stem cell therapy.
In short, single-cell RNA sequencing not only provided fresh information about the heterogeneity within the IVDs but also implied that important pathways (such as BMP, WNT and retinoic acid) control the development of IVD progenitors (Figure 2).
4. Characteristic of Disc Progenitor Cells Compared to Traditional MSCs
Bone marrow stromal cells (BMSCs) demonstrate a typical spindle-shaped fibriform outline, positive for CD29, CD90, CD105 and CD146 and negative for CD34 and CD45 [36]. BMSCs can differentiate into various IVD cell lineages, such as NP cells [37,38] and AF cells [38]. They also preserve osteogenic [36] and chondrogenic [39] capabilities. An ex vivo experiment demonstrated that BMSCs can ameliorate IVDD by preserving NP and AF cells [37,40] and can enhance matrix regeneration [38,41]. Coculturing NP cells with BMSCs led to decreased levels of type-II collagen and MMP-13, as well as increased levels of type-I collagen and aggrecan; this process was also named ECM remodeling [42]. The interaction between cells may be attributed to complex immunomodulation involving TGF-β [43]. Its therapeutic potential has been proven in rabbit [41,44], canine [39], porcine [45], sheep [46] and rat [47] models.
Umbilical cord stem cells (UCSCs) were positive for CD29, CD44, CD73, CD90, CD105 and CD166, and negative for CD11b, CD14, CD34, CD45, CD79 and HLA-DR [48,49,50]. UCSCs also possessed multilineage differentiation potential. Various in vitro studies have demonstrated that UCSCs can differentiate into all three lineages of cells: osteocytes, adipocytes and chondrocytes [49,51]. Experiment also showed that UCSCs could promote osteogenesis [48] and NP-like differentiation [52,53]. In one study, cells from Wharton’s jelly, which displayed stem cell markers, enhanced matrix production and revived degenerate NP cells [52]. In another study utilizing UCSCs, purified exosomes improved NP cell viability by adjusting the methyltransferase METTL14 [54]. The UCSCs regenerated bony connections between vertebrae, and their repairing effect extended to the AF [55], thus accelerating cartilaginous regeneration [51]. In vivo studies conducted on rabbits [53,55] and rats [51] verified the therapeutic potential of UCSCs. However, another in vivo experiment on rat models demonstrated a less satisfying outcome of the regeneration ability of human UCSCs, which may be due to heterogeneity [50].
Cytometry results revealed that adipose-derived stem cells (ADSCs) were positive for CD73, CD105, CD44 and Sca-1 [56] and negative for CD34, CD11b and CD45 [57]. NP-like [57] and adipocyte differentiation can be induced in ADSCs. Compared to BMSCs, ADSCs exhibited a higher potential for differentiating into NP-like cells, as shown by both genetic and mRNA analysis [56]. Staining also revealed the osteogenic and chondrogenic potential of ADSCs [58]. In addition, ADSCs were found to proliferate faster than BMSCs in both 3D and 2D cultures [56]. However, another study found that unstimulated ADSCs and BMSCs had similar proliferation abilities, which may be ascribed to different test methods. ADSCs migrated to NP-rich regions and induced a higher cell density of Tie2+ NP progenitors in an ex vivo degenerated ovine disc [59]. Transplanting ADSCs also revived degenerated chondrocytes and promoted endogenous repair, possibly by enhancing chondrogenic cytokines [60]. Except for the enhancement and differentiation of local cells, these multipotent stem cells were capable of restoring the extracellular matrix within the IVDs by improving the production of glycosaminoglycan (GAG) and proteoglycan [56,59]. ADSCs also exerted immunomodulatory effects, as they could produce more anti-inflammatory cytokines under inflammatory conditions [58]. The conditioned medium of ADSCs and extracellular vesicles also reduced inflammation of the AF area and exerted a protective influence [61]. Rat [56,62], sheep [63], mice [57] and rabbit [64] in vivo models were used to indicate the therapeutic potential of ADSCs.
As shown in Table 2, the comparisons of the characteristics of IVD progenitors and MSCs are summarized. NP progenitors are fibroblast-like bipolar spindle cells that form a whirlpool array in monolayer cultures. They were positive for CD24, CD73, CD90, Tie2 [65] and CD44 and negative for CD29, CD45 [66], CD11b, CD14, CD34 and HLA-DR. Another experiment found them positive for CD29 and CD105, but with much lower amounts than UCSCs [49]. Some studies have suggested dividing NP progenitors into two groups based on their levels of MSC markers expression [67], of which aging was the major cause. The novel markers PDGFRA [8] and UTS2R [21] were also discovered. Cells harvested from human degenerated IVDs could induce osteogenesis in certain media [68], and staining revealed that calcium deposition and lipid drops could be found within those cells [49]. Chondrogenesis was also detected [69]. NP progenitors were able to produce type-II collagen and aggrecan [66], reducing the matrix loss caused by punctures [70]. Various experiments proved the multilineage differentiation ability of NP progenitors [69]. Gene analysis conducted in a recent study revealed that NP progenitors may play a key role in extracellular matrix regeneration, mineral deposition, ossification, cartilage repair and immunomodulatory reactions such as T/B cell activities [8]. The same study also suggested a complex cell-to-cell signaling cascade involving multiple immune pathways, such as SPP1 and MIF. In addition, NP progenitors produce growth factors and exert a possible supporting influence on both themselves and surrounding cells [8]. However, animal tests are far from abundant, as in vivo experiments have only been conducted on rats [70,71]. When NP progenitors were transplanted into degenerated rat IVDs, they survived in a harsh environment and facilitated ECM restoration [72,73] by increasing proteoglycan and type-II collagen and damping MMP13 expression. An increased water content and elastic modulus were also found in punctured NP injected with exogenous NP progenitors [73]. This finding supported the imaging results indicating that the disc height was better preserved after transplantation. Studies have also utilized NP progenitors pre-conditioned by biomaterials [73] or combined with scaffolds [74] for potential future applications, but none of these studies tested the migration ability of NP progenitors. A group of angiopoietin-1 receptor (Tie2)-positive NP cells was identified as potential key markers of NP progenitors and for future therapies [75]. Recent studies have found that Tie2+ NP progenitors decreased after IVDD induced by injury [59] and displayed an age-related pattern. In an in vitro study utilizing human IVDs, the majority of NP progenitors that highly expressed Tie2 were from donors below 20 years old, while NP progenitors from donors above 25 years old demonstrated much lower Tie2 expression [67]. Additionally, the loss of Tie2+ NP progenitors could be rejuvenated by MSC transplantation [76]. When human normal NP (NNP) and degenerated NP (DNP) were compared by single-cell RNA sequencing, five types of chondrocytes were found. Chondrocytes 1, staying in the starting position of the development trajectory, were dominant in NNP. Chondrocytes 2, 4 and 5 presented the activities of calcification inhibition, inflammation and calcifying, respectively. Chondrocytes 5 were thought to be key cells leading to NP degeneration, while chondrocytes 2 may play a role in delaying NP degeneration. In short, compared to NNP, DNP contained more chondrocytes that were in the later positions of the development trajectory, and chondrocytes responsible for NP degeneration, pain and inflammation increased in DNP [77]. In a study performed to compare BMSCs and NP progenitors, no significant difference was detected in the expansion ability of the two types of cells [78]. Additional evidence illustrates that UCSCs possessed an extensively higher proliferation ability than NP progenitors [49].

Table 2.
Comparisons of the characteristics of MSCs and disc progenitor cells.
When referring to AF progenitor cells, different descriptions of their outlines, such as cobblestone-like and pancake-like, were used due to different cell sources and medium ingredients. CD29, CD44, CD69, CD105, Gata2 and Tnfaip3 were found to be positive, while CD34 and CD45 were negative. Three stemness markers, Oct4, SSEA4 and nucleostemin, were also discovered [17,79,80,81]. In addition, Scx was identified as one of the earliest markers of tendon progenitors, which were identical to AF progenitors in the early stages of development [82]. Multi-differentiation ability was also observed in AF progenitors, as adipocytes, osteocytes and chondrocytes can all be induced in appropriate cultures [80]. AF progenitors could also express type-I, II collagen and aggrecan, with biochemical tests showing accordant results [83]. Evidence indicating that chondroid matrix restoration is parallel to the reorientation and reestablishment of fibers in AF lamellae supports this finding [17]. However, no recent in vivo animal studies nor experiments have been conducted on impaired IVD models.
CEP progenitors, which could differentiate into NP cells, could be enhanced by exosomes via the HIF1-α and TGF-β pathways [84]. Moreover, the exosomes of CEP progenitors could transport Sphk2 to NP cells and inhibit apoptosis [30]. When CEP progenitors were in the same coculture system, activating ERK1/2 and Akt pathways enhanced NP cell growth [29]. In vivo studies have verified the regeneration potential in rat [28,30] models. However, in a study that simultaneously covered NP, AF and CEP progenitors, the proliferation ability measured by both the cell growth curve and colony forming displayed no significant difference among them. In the study, CEP progenitors demonstrated the most powerful invasion ability over the other two cells [7], and they regenerated impaired IVD; this effect may have come from the exosomes produced by CEP progenitors, which penetrated the AF and migrated into NP cells [30]. They also restored the disc height and hydration in rat tail nucleotomy models and increased ECM protein levels, such as aggrecan and type-II collagen [85]. The same study revealed that the injected CEP progenitors preserved endogenous NP progenitors and exerted anti-inflammatory and anti-catabolic effects in impaired IVDs.
With certain stimulation, dermal fibroblasts originating from induced pluripotent stem cells (iPSCs) can differentiate into primitive streak (PS) mesoderm cells and then into NCs; these NCs maintain their phenotypes for at least 8 weeks and exert protective effects both in vitro and in vivo [86]. Human embryonic stem cells (hESCs) can also be induced to a similar notochord-NP cell lineage as iPSCs [87]. These findings were supported by transcriptomic similarities between the induced and native NP tissues and between the differentiating trajectories of iPSCs and hESCs [87]. In vivo studies conducted in rats showed that iPSCs-derived mature NP cells exerted a similar protective effect to induced NCs, possibly by replacing endogenous NP tissue spatially and functionally and preventing CEP destruction [88]. Because of their pluripotency, hESCs and iPSCs possessed greater differentiation ability than IVD progenitors and MSCs. However, the safety of hESCs and iPSCs still requires further validation.
5. Conclusions
Recent evidence from single-cell RNA sequencing provides more solid proof of the existence of IVD progenitors and the heterogeneity within the IVD. The early development of IVD progenitors is controlled by signaling pathways such as Wnt, BMP and retinoic acid. In addition, IVD progenitors may have advantages over MSCs because of their similarity with endogenous tissues, but the evidence for determining the optimal cell source for IVD regeneration is still lacking. In summary, progenitor cell-based therapy holds significant potential in repairing the degenerated IVD, and well-designed experiments are necessary to verify their therapeutic ability.
Author Contributions
Conceptualization, Y.-D.Z. and Y.-C.H.; methodology, Y.-D.Z.; software, none; validation, Y.-D.Z.; formal analysis, Y.-D.Z.; writing—original draft preparation, Y.-D.Z.; writing—review and editing, Y.-D.Z., J.-L.L. and Y.-C.H.; visualization, Y.-D.Z.; supervision, W.-S.L. and Y.-C.H.; project administration, W.-S.L.; funding acquisition, Y.-C.H. and W.-S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Guangdong Basic and Applied Basic Research Foundation (No. 2021A1515220086) and the Shenzhen Science and Technology Programs (No. JCYJ20190809182213535 & No. GJHZ20210705142543019).
Institutional Review Board Statement
Not relevant to this study.
Informed Consent Statement
Not relevant to this study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Katz, J.N. Lumbar disc disorders and low-back pain: Socioeconomic factors and consequences. J. Bone Jt. Surg. 2006, 88 (Suppl. S2), 21–24. [Google Scholar] [CrossRef]
- Freemont, A.J.; Peacock, T.E.; Goupille, P.; Hoyland, J.A.; O’Brien, J.; Jayson, M.I. Nerve ingrowth into diseased intervertebral disc in chronic back pain. Lancet 1997, 350, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Kepler, C.K.; Markova, D.Z.; Dibra, F.; Yadla, S.; Vaccaro, A.R.; Risbud, M.V.; Albert, T.J.; Anderson, D.G. Expression and relationship of proinflammatory chemokine RANTES/CCL5 and cytokine IL-1β in painful human intervertebral discs. Spine 2013, 38, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.A.; Williams, N.H.; Sutton, A.J.; Burton, K.; Din, N.U.; Matar, H.E.; Hendry, M.; Phillips, C.J.; Nafees, S.; Fitzsimmons, D.; et al. Comparative clinical effectiveness of management strategies for sciatica: Systematic review and network meta-analyses. Spine J. 2015, 15, 1461–1477. [Google Scholar] [CrossRef] [PubMed]
- Cornejo, M.C.; Cho, S.K.; Giannarelli, C.; Iatridis, J.C.; Purmessur, D. Soluble factors from the notochordal-rich intervertebral disc inhibit endothelial cell invasion and vessel formation in the presence and absence of pro-inflammatory cytokines. Osteoarthr. Cartil. 2015, 23, 487–496. [Google Scholar] [CrossRef]
- Brand-Saberi, B.; Christ, B. Evolution and development of distinct cell lineages derived from somites. Curr. Top. Dev. Biol. 2000, 48, 1–42. [Google Scholar]
- Liu, S.; Liang, H.; Lee, S.M.; Li, Z.; Zhang, J.; Fei, Q. Isolation and identification of stem cells from degenerated human inter-vertebral discs and their migration characteristics. Acta Biochim. Biophys. Sin. 2017, 49, 101–109. [Google Scholar] [CrossRef]
- Li, W.; Zhang, S.; Zhao, Y.; Wang, D.; Shi, Q.; Ding, Z.; Wang, Y.; Gao, B.; Yan, M. Revealing the Key MSCs Niches and Pathogenic Genes in Influencing CEP Homeostasis: A Conjoint Analysis of Single-Cell and WGCNA. Front. Immunol. 2022, 13, 933721. [Google Scholar] [CrossRef]
- Sakai, D.; Andersson, G.B. Stem cell therapy for intervertebral disc regeneration: Obstacles and solutions. Nat. Rev. Rheumatol. 2015, 11, 243–256. [Google Scholar] [CrossRef]
- Lyu, F.J.; Cheung, K.M.; Zheng, Z.; Wang, H.; Sakai, D.; Leung, V.Y. IVD progenitor cells: A new horizon for understanding disc homeostasis and repair. Nat. Rev. Rheumatol. 2019, 15, 102–112. [Google Scholar] [CrossRef]
- Sakai, D.; Schol, J.; Bach, F.C.; Tekari, A.; Sagawa, N.; Nakamura, Y.; Chan, S.C.W.; Nakai, T.; Creemers, L.B.; Frauchiger, D.A.; et al. Successful fishing for nucleus pulposus progenitor cells of the intervertebral disc across species. JOR Spine 2018, 1, e1018. [Google Scholar] [CrossRef]
- Bach, F.C.; Zhang, Y.; Miranda-Bedate, A.; Verdonschot, L.C.; Bergknut, N.; Creemers, L.B.; Ito, K.; Sakai, D.; Chan, D.; Meij, B.P.; et al. Increased caveolin-1 in intervertebral disc degeneration facilitates repair. Arthritis Res. Ther. 2016, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, S.; Ma, K.; He, R.; Xiong, L.; Hu, Y.; Deng, X.; Yang, A.; Ma, X.; Shao, Z. Comparison of different methods for the isolation and purification of rat nucleus pulposus-derived mesenchymal stem cells. Connect. Tissue Res. 2020, 61, 426–434. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, C.; Li, Y.; Xu, H. Mechanism of the Mitogen-Activated Protein Kinases/Mammalian Target of Rapamycin Pathway in the Process of Cartilage Endplate Stem Cell Degeneration Induced by Tension Load. Glob. Spine J. 2022; ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Sun, C.; Zou, F.; Wang, H.; Lu, F.; Song, J.; Liu, S.; Xia, X.; Jiang, J.; Ma, X. Carbohydrate sulfotransferase 3 (CHST3) overexpression promotes cartilage endplate-derived stem cells (CESCs) to regulate molecular mechanisms related to repair of intervertebral disc degeneration by rat nucleus pulposus. J. Cell. Mol. Med. 2021, 25, 6006–6017. [Google Scholar] [CrossRef] [PubMed]
- Chu, G.; Zhang, W.; Zhou, P.; Yuan, Z.; Zhu, C.; Wang, H.; Li, J.; Zhou, F.; Yang, Q.; Yang, H.; et al. Substrate Topography Regulates Differentiation of Annulus Fibrosus-Derived Stem Cells via CAV1-YAP-Mediated Mecha-notransduction. ACS Biomater. Sci. Eng. 2021, 7, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, D.; Luo, B.; Wang, D.; Jia, H.; Peng, P.; Shang, Q.; Mao, J.; Gao, C.; Peng, Y.; et al. Decoding the annulus fibrosus cell atlas by scRNA-seq to develop an inducible composite hydrogel: A novel strategy for disc reconstruction. Bioact. Mater. 2022, 14, 350–363. [Google Scholar] [CrossRef]
- Jiang, W.; Glaeser, J.D.; Salehi, K.; Kaneda, G.; Mathkar, P.; Wagner, A.; Ho, R.; Sheyn, D. Single-cell atlas unveils cellular heterogeneity and novel markers in human neonatal and adult intervertebral discs. iScience 2022, 25, 104504. [Google Scholar] [CrossRef]
- Panebianco, C.J.; Dave, A.; Charytonowicz, D.; Sebra, R.; Iatridis, J.C. Single-cell RNA-sequencing atlas of bovine caudal in-tervertebral discs: Discovery of heterogeneous cell populations with distinct roles in homeostasis. FASEB J. 2021, 35, e21919. [Google Scholar] [CrossRef]
- Ling, Z.; Liu, Y.; Wang, Z.; Zhang, Z.; Chen, B.; Yang, J.; Zeng, B.; Gao, Y.; Jiang, C.; Huang, Y.; et al. Single-Cell RNA-Seq Analysis Reveals Macrophage Involved in the Progression of Human Intervertebral Disc Degeneration. Front. Cell Dev. Biol. 2021, 9, 833420. [Google Scholar] [CrossRef]
- Gao, B.; Jiang, B.; Xing, W.; Xie, Z.; Luo, Z.; Zou, W. Discovery and Application of Postnatal Nucleus Pulposus Progenitors Essential for Intervertebral Disc Homeostasis and Degeneration. Adv. Sci. 2022, 9, e2104888. [Google Scholar] [CrossRef]
- Gan, Y.; He, J.; Zhu, J.; Xu, Z.; Wang, Z.; Yan, J.; Hu, O.; Bai, Z.; Chen, L.; Xie, Y.; et al. Spatially defined single-cell transcriptional profiling characterizes diverse chondrocyte subtypes and nucleus pulposus pro-genitors in human intervertebral discs. Bone Res. 2021, 9, 37. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Y.; Huang, L.; Shi, K.; Wang, J.; Zhu, C.; Li, L.; Zhang, L.; Feng, G.; Liu, L.; et al. Novel biomarkers of intervertebral disc cells and evidence of stem cells in the intervertebral disc. Osteoarthr. Cartil. 2021, 29, 389–401. [Google Scholar] [CrossRef]
- Calió, M.; Gantenbein, B.; Egli, M.; Poveda, L.; Ille, F. The Cellular Composition of Bovine Coccygeal Intervertebral Discs: A Comprehensive Single-Cell RNAseq Analysis. Int. J. Mol. Sci. 2021, 22, 4917. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Yin, J.; Xu, X.; Fan, J.; Wang, D.; Zheng, C.; Lu, W.; Cheng, P.; Sun, J.; Wang, D.; et al. Leptin receptor-expressing cells represent a distinct subpopulation of notochord-derived cells and are essential for disc ho-moeostasis. J. Orthop. Transl. 2020, 21, 91–99. [Google Scholar]
- Li, X.; Yang, S.; Qin, L.; Yang, S. Type II collagen-positive embryonic progenitors are the major contributors to spine and in-tervertebral disc development and repair. Stem Cells Transl. Med. 2021, 10, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wang, H.; Zhang, W.; Mao, H.; Li, B. Single-cell RNA sequencing reveals resident progenitor and vasculariza-tion-associated cell subpopulations in rat annulus fibrosus. J. Orthop. Transl. 2023, 38, 256–267. [Google Scholar]
- Luo, L.; Jian, X.; Sun, H.; Qin, J.; Wang, Y.; Zhang, J.; Shen, Z.; Yang, D.; Li, C.; Zhao, P.; et al. Cartilage endplate stem cells inhibit intervertebral disc degeneration by releasing exosomes to nucleus pulposus cells to activate Akt/autophagy. Stem Cells 2021, 39, 467–481. [Google Scholar] [CrossRef]
- He, Z.; Jia, M.; Yu, Y.; Yuan, C.; Wang, J. Roles of SDF-1/CXCR4 axis in cartilage endplate stem cells mediated promotion of nucleus pulposus cells proliferation. Biochem. Biophys. Res. Commun. 2018, 506, 94–101. [Google Scholar] [CrossRef]
- Luo, L.; Gong, J.; Wang, Z.; Liu, Y.; Cao, J.; Qin, J.; Zuo, R.; Zhang, H.; Wang, S.; Zhao, P.; et al. Injectable cartilage matrix hydrogel loaded with cartilage endplate stem cells engineered to release exosomes for non-invasive treatment of intervertebral disc degeneration. Bioact. Mater. 2022, 15, 29–43. [Google Scholar] [CrossRef]
- Colombier, P.; Halgand, B.; Chédeville, C.; Chariau, C.; François-Campion, V.; Kilens, S.; Vedrenne, N.; Clouet, J.; David, L.; Guicheux, J.; et al. NOTO Transcription Factor Directs Human Induced Pluripotent Stem Cell-Derived Mesendoderm Progenitors to a Notochordal Fate. Cells 2020, 9, 509. [Google Scholar] [CrossRef] [PubMed]
- Binch, A.L.A.; Fitzgerald, J.C.; Growney, E.A.; Barry, F. Cell-based strategies for IVD repair: Clinical progress and translational obstacles. Nat. Rev. Rheumatol. 2021, 17, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Urban, J.P.; Luk, K.D. Intervertebral disc regeneration: Do nutrients lead the way? Nat. Rev. Rheumatol. 2014, 10, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Hernandez, M.E.; Khan, N.M.; Trochez, C.M.; Yoon, T.; Maye, P.; Presciutti, S.M.; Gibson, G.; Drissi, H. Derivation of notochordal cells from human embryonic stem cells reveals unique regulatory networks by single cell-transcriptomics. J. Cell. Physiol. 2020, 235, 5241–5255. [Google Scholar] [CrossRef]
- Ban, G.I.; Williams, S.; Serra, R. Antagonism of BMP signaling is insufficient to induce fibrous differentiation in primary scle-rotome. Exp. Cell Res. 2019, 378, 11–20. [Google Scholar] [CrossRef]
- Lee, Y.C.; Chan, Y.H.; Hsieh, S.C.; Lew, W.Z.; Feng, S.W. Comparing the Osteogenic Potentials and Bone Regeneration Ca-pacities of Bone Marrow and Dental Pulp Mesenchymal Stem Cells in a Rabbit Calvarial Bone Defect Model. Int. J. Mol. Sci. 2019, 20, 5015. [Google Scholar] [CrossRef]
- Frauchiger, D.A.; Heeb, S.R.; May, R.D.; Wöltje, M.; Benneker, L.M.; Gantenbein, B. Differentiation of MSC and annulus fi-brosus cells on genetically engineered silk fleece-membrane-composites enriched for GDF-6 or TGF-β3. J. Orthop. Res. 2018, 36, 1324–1333. [Google Scholar] [CrossRef]
- Sun, B.; Lian, M.; Han, Y.; Mo, X.; Jiang, W.; Qiao, Z.; Dai, K. A 3D-Bioprinted dual growth factor-releasing intervertebral disc scaffold induces nucleus pulposus and annulus fibrosus re-construction. Bioact. Mater. 2021, 6, 179–190. [Google Scholar] [CrossRef]
- Steffen, F.; Bertolo, A.; Affentranger, R.; Ferguson, S.J.; Stoyanov, J. Treatment of Naturally Degenerated Canine Lumbosacral Intervertebral Discs with Autologous Mesenchymal Stromal Cells and Collagen Microcarriers: A Prospective Clinical Study. Cell Transplant. 2019, 28, 201–211. [Google Scholar] [CrossRef]
- Liao, Z.; Luo, R.; Li, G.; Song, Y.; Zhan, S.; Zhao, K.; Hua, W.; Zhang, Y.; Wu, X.; Yang, C. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics 2019, 9, 4084–4100. [Google Scholar] [CrossRef]
- Feng, G.; Zhang, Z.; Dang, M.; Rambhia, K.J.; Ma, P.X. Nanofibrous spongy microspheres to deliver rabbit mesenchymal stem cells and anti-miR-199a to regenerate nucleus pulposus and prevent calcification. Biomaterials 2020, 256, 120213. [Google Scholar] [CrossRef]
- Wang, Z.; Ding, X.; Cao, F.; Zhang, X.; Wu, J. Bone Mesenchymal Stem Cells Promote Extracellular Matrix Remodeling of Degenerated Nucleus Pulposus Cells via the miR-101-3p/EIF4G2 Axis. Front. Bioeng. Biotechnol. 2021, 9, 642502. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, T.P.; Jakub, G.; Harasymczuk, J.; Jagodziński, P.P. Transforming growth factor β mediates communication of co-cultured human nucleus pulposus cells and mesenchymal stem cells. J. Orthop. Res. 2018, 36, 3023–3032. [Google Scholar] [CrossRef] [PubMed]
- Chiang, E.R.; Ma, H.L.; Wang, J.P.; Chang, M.C.; Liu, C.L.; Chen, T.H.; Hung, S.C. Use of Allogeneic Hypoxic Mesenchymal Stem Cells for Treating Disc Degeneration in Rabbits. J. Orthop. Res. 2019, 37, 1440–1450. [Google Scholar] [CrossRef]
- Omlor, G.W.; Lorenz, S.; Nerlich, A.G.; Guehring, T.; Richter, W. Disc cell therapy with bone-marrow-derived autologous mesenchymal stromal cells in a large porcine disc degeneration model. Eur. Spine J. 2018, 27, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Ukeba, D.; Yamada, K.; Suyama, T.; Lebl, D.R.; Tsujimoto, T.; Nonoyama, T.; Sugino, H.; Iwasaki, N.; Watanabe, M.; Matsuzaki, Y.; et al. Combination of ultra-purified stem cells with an in situ-forming bioresorbable gel enhances intervertebral disc regeneration. EBioMedicine 2022, 76, 103845. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y.; Deng, G.; Ma, J.; Huang, X.; Yu, J.; Xi, Y.; Ye, X. Transplantation of Hypoxic-Preconditioned Bone Mesenchymal Stem Cells Retards Intervertebral Disc Degeneration via En-hancing Implanted Cell Survival and Migration in Rats. Stem Cells Int. 2018, 2018, 7564159. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, H.; Tang, X.; Feng, R.; Yao, G.; Chen, W.; Li, W.; Liang, J.; Feng, X.; Sun, L. Mesenchymal Stem Cells Promote the Osteogenesis in Collagen-Induced Arthritic Mice through the Inhibition of TNF-α. Stem Cells Int. 2018, 2018, 4069032. [Google Scholar] [CrossRef]
- Wu, H.; Zeng, X.; Yu, J.; Shang, Y.; Tu, M.; Cheang, L.H.; Zhang, J. Comparison of nucleus pulposus stem/progenitor cells isolated from degenerated intervertebral discs with umbilical cord de-rived mesenchymal stem cells. Exp. Cell Res. 2017, 361, 324–332. [Google Scholar] [CrossRef]
- Matta, A.; Karim, M.Z.; Gerami, H.; Benigno, B.; Erwin, W.M. A comparative study of mesenchymal stem cell transplantation and NTG-101 molecular therapy to treat degenerative disc disease. Sci. Rep. 2021, 11, 14804. [Google Scholar] [CrossRef]
- Khalid, S.; Ekram, S.; Salim, A.; Chaudhry, G.R.; Khan, I. Transcription regulators differentiate mesenchymal stem cells into chondroprogenitors, and their in vivo implantation regenerated the intervertebral disc degeneration. World J. Stem Cells 2022, 14, 163–182. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, Y.; Gao, L.; Jiang, S.; Ruan, D. Human Wharton’s Jelly Cells Activate Degenerative Nucleus Pulposus Cells In Vitro. Tissue Eng. Part A 2018, 24, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Perez-Cruet, M.; Beeravolu, N.; McKee, C.; Brougham, J.; Khan, I.; Bakshi, S. Potential of Human Nucleus Pulposus-Like Cells Derived From Umbilical Cord to Treat Degenerative Disc Disease. Neurosurgery 2019, 84, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Ekram, S.; Khalid, S.; Bashir, I.; Salim, A.; Khan, I. Human umbilical cord-derived mesenchymal stem cells and their chon-droprogenitor derivatives reduced pain and inflammation signaling and promote regeneration in a rat intervertebral disc de-generation model. Mol. Cell. Biochem. 2021, 476, 3191–3205. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, Z.; Li, R.; Tian, J.; Sun, G.; Li, L.; Wu, D.; Ding, S.; Zhou, C. A novel biomimetic scaffold with hUCMSCs for lumbar fusion. J. Mater. Chem. B 2017, 5, 5996–6007. [Google Scholar] [CrossRef]
- Dai, X.; Guan, Y.; Zhang, Z.; Xiong, Y.; Liu, C.; Li, H.; Liu, B. Comparison of the differentiation abilities of bone marrow-derived mesenchymal stem cells and adipose-derived mesenchymal stem cells toward nucleus pulposus-like cells in three-dimensional culture. Exp. Ther. Med. 2021, 22, 1018. [Google Scholar] [CrossRef]
- Xiao, L.; Xu, S.J.; Liu, C.; Wang, J.; Hu, B.; Xu, H.G. Sod2 and catalase improve pathological conditions of intervertebral disc degeneration by modifying human adipose-derived mesenchymal stem cells. Life Sci. 2021, 267, 118929. [Google Scholar] [CrossRef] [PubMed]
- Borem, R.; Madeline, A.; Bowman, M.; Gill, S.; Tokish, J.; Mercuri, J. Differential Effector Response of Amnion- and Adi-pose-Derived Mesenchymal Stem Cells to Inflammation; Implications for Intradiscal Therapy. J. Orthop. Res. 2019, 37, 2445–2456. [Google Scholar] [CrossRef]
- Frapin, L.; Clouet, J.; Chédeville, C.; Moraru, C.; Samarut, E.; Henry, N.; André, M.; Bord, E.; Halgand, B.; Lesoeur, J.; et al. Controlled release of biological factors for endogenous progenitor cell migration and intervertebral disc extracellular matrix remodelling. Biomaterials 2020, 253, 120107. [Google Scholar] [CrossRef]
- Bello, A.B.; Kim, Y.; Park, S.; Muttigi, M.S.; Kim, J.; Park, H.; Lee, S. Matrilin3/TGFβ3 gelatin microparticles promote chondrogenesis, prevent hypertrophy, and induce paracrine release in MSC spheroid for disc regeneration. NPJ Regen. Med. 2021, 6, 50. [Google Scholar] [CrossRef]
- González-Cubero, E.; González-Fernández, M.L.; Olivera, E.R.; Villar-Suárez, V. Extracellular vesicle and soluble fractions of adipose tissue-derived mesenchymal stem cells secretome induce inflammatory cytokines modulation in an in vitro model of discogenic pain. Spine J. 2022, 22, 1222–1234. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Li, D.; Wang, C.; Xia, K.; Wang, J.; Zhou, X.; Ying, L.; Shu, J.; Huang, X.; Xu, H.; et al. Injectable kartogenin and apocynin loaded micelle enhances the alleviation of intervertebral disc degeneration by adi-pose-derived stem cell. Bioact. Mater. 2021, 6, 3568–3579. [Google Scholar] [CrossRef]
- Friedmann, A.; Baertel, A.; Schmitt, C.; Ludtka, C.; Milosevic, J.; Meisel, H.J.; Goehre, F.; Schwan, S. Intervertebral Disc Regeneration Injection of a Cell-Loaded Collagen Hydrogel in a Sheep Model. Int. J. Mol. Sci. 2021, 22, 4248. [Google Scholar] [CrossRef]
- Muttigi, M.S.; Kim, B.J.; Kumar, H.; Park, S.; Choi, U.Y.; Han, I.; Park, H.; Lee, S.H. Efficacy of matrilin-3-primed adipose-derived mesenchymal stem cell spheroids in a rabbit model of disc degeneration. Stem Cell Res. Ther. 2020, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guerrero, J.; Croft, A.S.; Albers, C.E.; Häckel, S.; Gantenbein, B. Spheroid-Like Cultures for Expanding Angiopoietin Receptor-1 (aka. Tie2) Positive Cells from the Human Intervertebral Disc. Int. J. Mol. Sci. 2020, 21, 9423. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Wang, R.; Shi, Q.; Yuan, M.; Jin, M.; Li, D. Umbilical cord mesenchymal stem cell conditioned medium restored the expression of collagen II and aggrecan in nucleus pulposus mesenchymal stem cells exposed to high glucose. J. Bone Miner. Metab. 2019, 37, 455–466. [Google Scholar] [CrossRef]
- Wu, H.; Shang, Y.; Yu, J.; Zeng, X.; Lin, J.; Tu, M.; Cheang, L.H.; Zhang, J. Regenerative potential of human nucleus pulposus resident stem/progenitor cells declines with ageing and intervertebral disc degeneration. Int. J. Mol. Med. 2018, 42, 2193–2202. [Google Scholar] [CrossRef]
- Brown, S.J.; Turner, S.A.; Balain, B.S.; Davidson, N.T.; Roberts, S. Is Osteogenic Differentiation of Human Nucleus Pulposus Cells a Possibility for Biological Spinal Fusion? Cartilage 2020, 11, 181–191. [Google Scholar] [CrossRef]
- Zeng, X.; Lin, J.; Wu, H.; Yu, J.; Tu, M.; Cheang, L.H.; Zhang, J. Effect of Conditioned Medium from Human Umbilical Cord-Derived Mesenchymal Stromal Cells on Rejuvenation of Nucleus Pulposus Derived Stem/Progenitor Cells from Degenerated Intervertebral Disc. Int. J. Stem Cells 2020, 13, 257–267. [Google Scholar] [CrossRef]
- Liu, Z.; Zheng, Z.; Qi, J.; Wang, J.; Zhou, Q.; Hu, F.; Liang, J.; Li, C.; Zhang, W.; Zhang, X. CD24 identifies nucleus pulposus progenitors/notochordal cells for disc regeneration. J. Biol. Eng. 2018, 12, 35. [Google Scholar] [CrossRef]
- Ying, J.W.; Wen, T.Y.; Pei, S.S.; Su, L.H.; Ruan, D.K. Stromal cell-derived factor-1α promotes recruitment and differentiation of nucleus pulposus-derived stem cells. World J. Stem Cells 2019, 11, 196–211. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Wang, Z.; Cui, M.; Liu, S.; Wu, W.; Chen, M.; Wu, Y.; Qu, Y.; Lin, H.; Chen, S.; et al. HIF1A Alleviates compression-induced apoptosis of nucleus pulposus derived stem cells via upregulating autophagy. Autophagy 2021, 17, 3338–3360. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.S.; Li, D.D.; Wang, C.G.; Ying, L.W.; Wang, J.K.; Yang, B.; Shu, J.W.; Huang, X.P.; Zhang, Y.A.; Yu, C.; et al. An esterase-responsive ibuprofen nano-micelle pre-modified embryo derived nucleus pulposus progenitor cells promote the regeneration of intervertebral disc degeneration. Bioact. Mater. 2023, 21, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Nan, L.P.; Zhou, S.F.; Liu, Y.; Wang, Z.Y.; Wang, J.C.; Feng, X.M.; Zhang, L. Injectable Hydrogel Combined with Nucleus Pulposus-Derived Mesenchymal Stem Cells for the Treatment of Degenerative Intervertebral Disc in Rats. Stem Cells Int. 2019, 2019, 8496025. [Google Scholar] [CrossRef]
- Sakai, D.; Nakamura, Y.; Nakai, T.; Mishima, T.; Kato, S.; Grad, S.; Alini, M.; Risbud, M.V.; Chan, D.; Cheah, K.S.; et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat. Commun. 2012, 3, 1264. [Google Scholar] [CrossRef]
- Li, X.C.; Wang, M.S.; Liu, W.; Zhong, C.F.; Deng, G.B.; Luo, S.J.; Huang, C.M. Co-culturing nucleus pulposus mesenchymal stem cells with notochordal cell-rich nucleus pulposus explants attenuates tumor necrosis factor-α-induced senescence. Stem Cell Res. Ther. 2018, 9, 171. [Google Scholar] [CrossRef]
- Li, Z.; Ye, D.; Dai, L.; Xu, Y.; Wu, H.; Luo, W.; Liu, Y.; Yao, X.; Wang, P.; Miao, H.; et al. Single-Cell RNA Sequencing Reveals the Difference in Human Normal and Degenerative Nucleus Pulposus Tissue Profiles and Cellular Interactions. Front. Cell Dev. Biol. 2022, 10, 910626. [Google Scholar] [CrossRef]
- Blanco, J.F.; Graciani, I.F.; Sanchez-Guijo, F.M.; Muntión, S.; Hernandez-Campo, P.; Santamaria, C.; Carrancio, S.; Barbado, M.V.; Cruz, G.; Gutierrez-Cosío, S.; et al. Isolation and characterization of mesenchymal stromal cells from human degenerated nucleus pulposus: Comparison with bone marrow mesenchymal stromal cells from the same subjects. Spine 2010, 35, 2259–2265. [Google Scholar] [CrossRef]
- Han, Y.; Yuan, F.; Deng, C.; He, F.; Zhang, Y.; Shen, H.; Chen, Z.; Qian, L. Metformin decreases LPS-induced inflammatory response in rabbit annulus fibrosus stem/progenitor cells by blocking HMGB1 release. Aging 2019, 11, 10252–10265. [Google Scholar] [CrossRef]
- Gao, C.; Ning, B.; Sang, C.; Zhang, Y. Rapamycin prevents the intervertebral disc degeneration via inhibiting differentiation and senescence of annulus fibrosus cells. Aging 2018, 10, 131–143. [Google Scholar] [CrossRef]
- Guo, Q.; Zhou, P.; Li, B. Identification and Characterizations of Annulus Fibrosus-Derived Stem Cells. Methods Mol Biol. 2018, 1842, 207–216. [Google Scholar]
- Kaji, D.A.; Montero, A.M.; Patel, R.; Huang, A.H. Transcriptional profiling of mESC-derived tendon and fibrocartilage cell fate switch. Nat. Commun. 2021, 12, 4208. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Chu, G.; Yuan, Z.; Wang, H.; Zhang, W.; Mao, Y.; Zhu, X.; Chen, W.; Yang, H.; Li, B. Regulation of differentiation of annulus fibrosus-derived stem cells using heterogeneous electrospun fibrous scaffolds. J. Orthop. Transl. 2021, 26, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Gong, J.; Zhang, H.; Qin, J.; Li, C.; Zhang, J.; Tang, Y.; Zhang, Y.; Chen, J.; Zhou, Y.; et al. Cartilage Endplate Stem Cells Transdifferentiate into Nucleus Pulposus Cells via Autocrine Exosomes. Front. Cell Dev. Biol. 2021, 9, 648201. [Google Scholar]
- Bhujel, B.; Yang, S.S.; Kim, H.R.; Kim, S.B.; Min, B.H.; Choi, B.H.; Han, I. An Injectable Engineered Cartilage Gel Improves Intervertebral Disc Repair in a Rat Nucleotomy Model. Int. J. Mol. Sci. 2023, 24, 3146. [Google Scholar] [CrossRef]
- Sheyn, D.; Ben-David, S.; Tawackoli, W.; Zhou, Z.; Salehi, K.; Bez, M.; De Mel, S.; Chan, V.; Roth, J.; Avalos, P.; et al. Human iPSCs can be differentiated into notochordal cells that reduce intervertebral disc degeneration in a porcine model. Theranostics 2019, 9, 7506–7524. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Chen, P.; Ma, C.Y.; Li, C.; Au, T.Y.K.; Tam, V.; Peng, Y.; Wu, R.; Cheung, K.M.C.; et al. Directed Differentiation of Notochord-like and Nucleus Pulposus-like Cells Using Human Pluripotent Stem Cells. Cell Rep. 2020, 30, 2791–2806.e5. [Google Scholar] [CrossRef]
- Kamatani, T.; Hagizawa, H.; Yarimitsu, S.; Morioka, M.; Koyamatsu, S.; Sugimoto, M.; Kodama, J.; Yamane, J.; Ishiguro, H.; Shichino, S.; et al. Human iPS cell-derived cartilaginous tissue spatially and functionally replaces nucleus pulposus. Biomaterials 2022, 284, 121491. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).