Diagnosis of Chronic Musculoskeletal Pain by Using Functional Near-Infrared Spectroscopy and Machine Learning
Abstract
1. Introduction
2. Methods
2.1. Participants
2.2. fNIRS Data Acquisition and Preprocessing
2.3. Brain Network Analysis
2.4. Analysis of the Amplitude of Low-Frequency Fluctuations
2.5. Graph Theory Analysis
2.6. Feature Selection and Classification
3. Results
3.1. Correlation Matrix
3.2. ALFF Results
3.3. Comparison of Functional Network Characteristics
3.4. Classification Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Treede, R.D.; Rief, W.; Barke, A.; Aziz, Q.; Bennett, M.I.; Benoliel, R.; Cohen, M.; Evers, S.; Finnerup, N.B.; First, M.B.; et al. A classification of chronic pain for ICD-11. Pain 2015, 156, 1003–1007. [Google Scholar] [CrossRef]
- Treede, R.D.; Rief, W.; Barke, A.; Aziz, Q.; Bennett, M.I.; Benoliel, R.; Cohen, M.; Evers, S.; Finnerup, N.B.; First, M.B.; et al. Chronic pain as a symptom or a disease: The IASP Classification of Chronic Pain for the International Classification of Diseases (ICD-11). Pain 2019, 160, 19–27. [Google Scholar] [CrossRef]
- Dansie, E.J.; Turk, D.C. Assessment of patients with chronic pain. Br. J. Anaesth. 2013, 111, 19–25. [Google Scholar] [CrossRef]
- Barnett, K.; Mercer, S.W.; Norbury, M.; Watt, G.; Wyke, S.; Guthrie, B. Epidemiology of multimorbidity and implications for health care, research, and medical education: A cross-sectional study. Lancet 2012, 380, 37–43. [Google Scholar] [CrossRef]
- Khera, T.; Rangasamy, V. Cognition and Pain: A Review. Front. Psychol. 2021, 12, 673962. [Google Scholar] [CrossRef]
- Malfliet, A.; Coppieters, I.; Van Wilgen, P.; Kregel, J.; De Pauw, R.; Dolphens, M.; Ickmans, K. Brain changes associated with cognitive and emotional factors in chronic pain: A systematic review. Eur. J. Pain 2017, 21, 769–786. [Google Scholar] [CrossRef]
- Bushnell, M.C.; Ceko, M.; Low, L.A. Cognitive and emotional control of pain and its disruption in chronic pain. Nat. Rev. Neurosci. 2013, 14, 502–511. [Google Scholar] [CrossRef]
- Archibald, J.; Warner, F.M.; Ortiz, O.; Todd, M.; Jutzeler, C.R. Recent advances in objectifying pain using neuroimaging techniques. J. Neurophysiol. 2018, 120, 387–390. [Google Scholar] [CrossRef]
- Seminowicz, D.A.; Moayedi, M. The Dorsolateral Prefrontal Cortex in Acute and Chronic Pain. J. Pain 2017, 18, 1027–1035. [Google Scholar] [CrossRef]
- Ihara, N.; Wakaizumi, K.; Nishimura, D.; Kato, J.; Yamada, T.; Suzuki, T.; Hashiguchi, S.; Terasawa, Y.; Kosugi, S.; Morisaki, H. Aberrant resting-state functional connectivity of the dorsolateral prefrontal cortex to the anterior insula and its association with fear avoidance belief in chronic neck pain patients. PLoS ONE 2019, 14, e0221023. [Google Scholar] [CrossRef]
- Pfannmöller, J.; Lotze, M. Review on biomarkers in the resting-state networks of chronic pain patients. Brain Cogn. 2019, 131, 4–9. [Google Scholar] [CrossRef]
- De Pauw, R.; Aerts, H.; Siugzdaite, R.; Meeus, M.; Coppieters, I.; Caeyenberghs, K.; Cagnie, B. Hub disruption in patients with chronic neck pain: A graph analytical approach. Pain 2020, 161, 729–741. [Google Scholar] [CrossRef]
- Lenoir, D.; Cagnie, B.; Verhelst, H.; De Pauw, R. Graph Measure Based Connectivity in Chronic Pain Patients: A Systematic Review. Pain Physician 2021, 24, E1037–E1058. [Google Scholar]
- Öztürk, Ö.; Algun, Z.C.; Bombacı, H.; Erdoğan, S.B. Changes in prefrontal cortex activation with exercise in knee osteoarthritis patients with chronic pain: An fNIRS study. J. Clin. Neurosci. Off. J. Neurosurg. Soc. Australas. 2021, 90, 144–151. [Google Scholar] [CrossRef]
- Santana, A.N.; Cifre, I.; de Santana, C.N.; Montoya, P. Using Deep Learning and Resting-State fMRI to Classify Chronic Pain Conditions. Front. Neurosci. 2019, 13, 1313. [Google Scholar] [CrossRef]
- Pinti, P.; Tachtsidis, I.; Hamilton, A.; Hirsch, J.; Aichelburg, C.; Gilbert, S.; Burgess, P.W. The present and future use of functional near-infrared spectroscopy (fNIRS) for cognitive neuroscience. Ann. N. Y. Acad. Sci. 2020, 1464, 5–29. [Google Scholar] [CrossRef]
- Xu, S.Y.; Cheong, L.I.; Zhuang, Y.; Couto, T.A.P.; Yuan, Z. Conducting Concurrent Electroencephalography and Functional Near-Infrared Spectroscopy Recordings with a Flanker Task. J. Vis. Exp. JoVE 2020, 24, e60669. [Google Scholar] [CrossRef]
- Wilcox, T.; Biondi, M. fNIRS in the developmental sciences. Wiley Interdiscip. Rev. Cogn. Sci. 2015, 6, 263–283. [Google Scholar] [CrossRef]
- Rahman, M.A.; Siddik, A.B.; Ghosh, T.K.; Khanam, F.; Ahmad, M. A Narrative Review on Clinical Applications of fNIRS. J. Digit. Imaging 2020, 33, 1167–1184. [Google Scholar] [CrossRef]
- Yang, D.; Hong, K.S. Quantitative Assessment of Resting-State for Mild Cognitive Impairment Detection: A Functional Near-Infrared Spectroscopy and Deep Learning Approach. J. Alzheimer’s Dis. JAD 2021, 80, 647–663. [Google Scholar] [CrossRef]
- Xu, L.; Liu, Y.; Yu, J.; Li, X.; Yu, X.; Cheng, H.; Li, J. Characterizing autism spectrum disorder by deep learning spontaneous brain activity from functional near-infrared spectroscopy. J. Neurosci. Methods 2020, 331, 108538. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qin, W.; Nan, J.; Li, J.; Yuan, K.; Zhao, L.; Zeng, F.; Sun, J.; Yu, D.; Dong, M.; et al. Gender-related differences in the dysfunctional resting networks of migraine suffers. PLoS ONE 2011, 6, e27049. [Google Scholar] [CrossRef]
- Fishburn, F.A.; Ludlum, R.S.; Vaidya, C.J.; Medvedev, A.V. Temporal Derivative Distribution Repair (TDDR): A motion correction method for fNIRS. NeuroImage 2019, 184, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.-Y.; Lu, F.-M.; Wang, M.-Y.; Hu, Z.-S.; Zhang, J.; Chen, Z.-Y.; Armada-da-Silva, P.A.S.; Yuan, Z. Altered Functional Connectivity in the Motor and Prefrontal Cortex for Children with Down’s Syndrome: An fNIRS Study. Front. Hum. Neurosci. 2020, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Mondloch, C.J.; Ding, X.P.; Short, L.A.; Sun, L.; Lee, K. The neural correlates of the face attractiveness aftereffect: A functional near-infrared spectroscopy (fNIRS) study. NeuroImage 2014, 85 Pt 1, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zhang, Z.; Zhao, C.; Duan, L.; Gong, Y.; Li, Z.; Zhu, C. NIRS-KIT: A MATLAB toolbox for both resting-state and task fNIRS data analysis. Neurophotonics 2021, 8, 010802. [Google Scholar] [CrossRef]
- Storey, J.D. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B 2002, 64, 479–498. [Google Scholar] [CrossRef]
- Zhou, F.; Gu, L.; Hong, S.; Liu, J.; Jiang, J.; Huang, M.; Zhang, Y.; Gong, H. Altered low-frequency oscillation amplitude of resting state-fMRI in patients with discogenic low-back and leg pain. J. Pain Res. 2018, 11, 165–176. [Google Scholar] [CrossRef]
- Zhang, S.S.; Wu, W.; Yang, J.M.; Wang, C.H. Abnormal Spontaneous Brain Activity in Acute Low-Back Pain Revealed by Resting-State Functional MRI. Am. J. Phys. Med. Rehabil. 2017, 96, 253–259. [Google Scholar] [CrossRef]
- Chen, H.; Qi, G.; Zhang, Y.; Huang, Y.; Zhang, S.; Yang, D.; He, J.; Mu, L.; Zhou, L.; Zeng, M. Altered Dynamic Amplitude of Low-Frequency Fluctuations in Patients with Migraine without Aura. Front. Hum. Neurosci. 2021, 15, 636472. [Google Scholar] [CrossRef]
- Zhang, B.; Jung, M.; Tu, Y.; Gollub, R.; Lang, C.; Ortiz, A.; Park, J.; Wilson, G.; Gerber, J.; Mawla, I.; et al. Identifying brain regions associated with the neuropathology of chronic low back pain: A resting-state amplitude of low-frequency fluctuation study. Br. J. Anaesth. 2019, 123, e303–e311. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Xia, M.; Liao, X.; Evans, A.; He, Y. GRETNA: A graph theoretical network analysis toolbox for imaging connectomics. Front. Hum. Neurosci. 2015, 9, 386. [Google Scholar]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef]
- He, Y.; Chen, Z.J.; Evans, A.C. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb. Cortex 2007, 17, 2407–2419. [Google Scholar] [CrossRef]
- Beleites, C.; Neugebauer, U.; Bocklitz, T.; Krafft, C.; Popp, J. Sample size planning for classification models. Anal. Chim. Acta 2013, 760, 25–33. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E. Pattern Classification; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Hooten, W.M. Chronic Pain and Mental Health Disorders: Shared Neural Mechanisms, Epidemiology, and Treatment. Mayo Clin. Proc. 2016, 91, 955–970. [Google Scholar] [CrossRef]
- Garcia-Larrea, L.; Peyron, R. Pain matrices and neuropathic pain matrices: A review. Pain 2013, 154 (Suppl. S1), S29–S43. [Google Scholar] [CrossRef]
- Ossipov, M.H.; Dussor, G.O.; Porreca, F. Central modulation of pain. J. Clin. Investig. 2010, 120, 3779–3787. [Google Scholar] [CrossRef]
- Shaver, T.K.; Ozga, J.E.; Zhu, B.; Anderson, K.G.; Martens, K.M.; Vonder Haar, C. Long-term deficits in risky decision-making after traumatic brain injury on a rat analog of the Iowa gambling task. Brain Res. 2019, 1704, 103–113. [Google Scholar] [CrossRef]
- Barke, A.; Baudewig, J.; Schmidt-Samoa, C.; Dechent, P.; Kröner-Herwig, B. Neural correlates of fear of movement in high and low fear-avoidant chronic low back pain patients: An event-related fMRI study. Pain 2012, 153, 540–552. [Google Scholar] [CrossRef]
- Mazza, S.; Frot, M.; Rey, A.E. A comprehensive literature review of chronic pain and memory. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 87, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Brighina, F.; Piazza, A.; Vitello, G.; Aloisio, A.; Palermo, A.; Daniele, O.; Fierro, B. rTMS of the prefrontal cortex in the treatment of chronic migraine: A pilot study. J. Neurol Sci. 2004, 227, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Rajain, M.; Bhatia, R.; Tripathi, M.; Kumar, N.; Mathur, R. Low-Frequency Repetitive Transcranial Magnetic Stimulation for Chronic Tension-Type Headache: A Randomized Controlled Study. Cureus 2023, 15, e34922. [Google Scholar] [CrossRef]
- Lamm, C.; Decety, J.; Singer, T. Meta-analytic evidence for common and distinct neural networks associated with directly experienced pain and empathy for pain. NeuroImage 2011, 54, 2492–2502. [Google Scholar] [CrossRef] [PubMed]
- Ta Dinh, S.; Nickel, M.M.; Tiemann, L.; May, E.S.; Heitmann, H.; Hohn, V.D.; Edenharter, G.; Utpadel-Fischler, D.; Tolle, T.R.; Sauseng, P.; et al. Brain dysfunction in chronic pain patients assessed by resting-state electroencephalography. Pain 2019, 160, 2751–2765. [Google Scholar] [CrossRef] [PubMed]
- DeSouza, D.D.; Woldeamanuel, Y.W.; Sanjanwala, B.M.; Bissell, D.A.; Bishop, J.H.; Peretz, A.; Cowan, R.P. Altered structural brain network topology in chronic migraine. Brain Struct. Funct. 2020, 225, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Rubinov, M.; Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage 2010, 52, 1059–1069. [Google Scholar] [CrossRef]
- Zhu, J.; Qian, Y.; Zhang, B.; Li, X.; Bai, Y.; Li, X.; Yu, Y. Abnormal synchronization of functional and structural networks in schizophrenia. Brain Imaging Behav. 2020, 14, 2232–2241. [Google Scholar] [CrossRef]
- Van den Heuvel, M.P.; Sporns, O. Rich-club organization of the human connectome. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 15775–15786. [Google Scholar] [CrossRef]
- Bullmore, E.; Sporns, O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009, 10, 186–198. [Google Scholar] [CrossRef]
- Fernandez Rojas, R.; Huang, X.; Ou, K.L. A Machine Learning Approach for the Identification of a Biomarker of Human Pain using fNIRS. Sci. Rep. 2019, 9, 5645. [Google Scholar] [CrossRef] [PubMed]
- Bagarinao, E.; Johnson, K.A.; Martucci, K.T.; Ichesco, E.; Farmer, M.A.; Labus, J.; Ness, T.J.; Harris, R.; Deutsch, G.; Apkarian, V.A.; et al. Preliminary structural MRI based brain classification of chronic pelvic pain: A MAPP network study. Pain 2014, 155, 2502–2509. [Google Scholar] [CrossRef] [PubMed]
- Labus, J.S.; Van Horn, J.D.; Gupta, A.; Alaverdyan, M.; Torgerson, C.; Ashe-McNalley, C.; Irimia, A.; Hong, J.Y.; Naliboff, B.; Tillisch, K.; et al. Multivariate morphological brain signatures predict patients with chronic abdominal pain from healthy control subjects. Pain 2015, 156, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Ung, H.; Brown, J.E.; Johnson, K.A.; Younger, J.; Hush, J.; Mackey, S. Multivariate classification of structural MRI data detects chronic low back pain. Cereb. Cortex 2014, 24, 1037–1044. [Google Scholar] [CrossRef]
- Shen, W.; Tu, Y.; Gollub, R.L.; Ortiz, A.; Napadow, V.; Yu, S.; Wilson, G.; Park, J.; Lang, C.; Jung, M.; et al. Visual network alterations in brain functional connectivity in chronic low back pain: A resting state functional connectivity and machine learning study. NeuroImage Clin. 2019, 22, 101775. [Google Scholar] [CrossRef]
- You, B.; Wen, H.; Jackson, T. Identifying resting state differences salient for resilience to chronic pain based on machine learning multivariate pattern analysis. Psychophysiology 2021, 58, e13921. [Google Scholar] [CrossRef]
- Rogachov, A.; Cheng, J.C.; Hemington, K.S.; Bosma, R.L.; Kim, J.A.; Osborne, N.R.; Inman, R.D.; Davis, K.D. Abnormal Low-Frequency Oscillations Reflect Trait-Like Pain Ratings in Chronic Pain Patients Revealed through a Machine Learning Approach. J. Neurosci. Off. J. Soc. Neurosci. 2018, 38, 7293–7302. [Google Scholar] [CrossRef]
- Mano, H.; Kotecha, G.; Leibnitz, K.; Matsubara, T.; Sprenger, C.; Nakae, A.; Shenker, N.; Shibata, M.; Voon, V.; Yoshida, W.; et al. Classification and characterisation of brain network changes in chronic back pain: A multicenter study. Wellcome Open Res. 2018, 3, 19. [Google Scholar] [CrossRef]
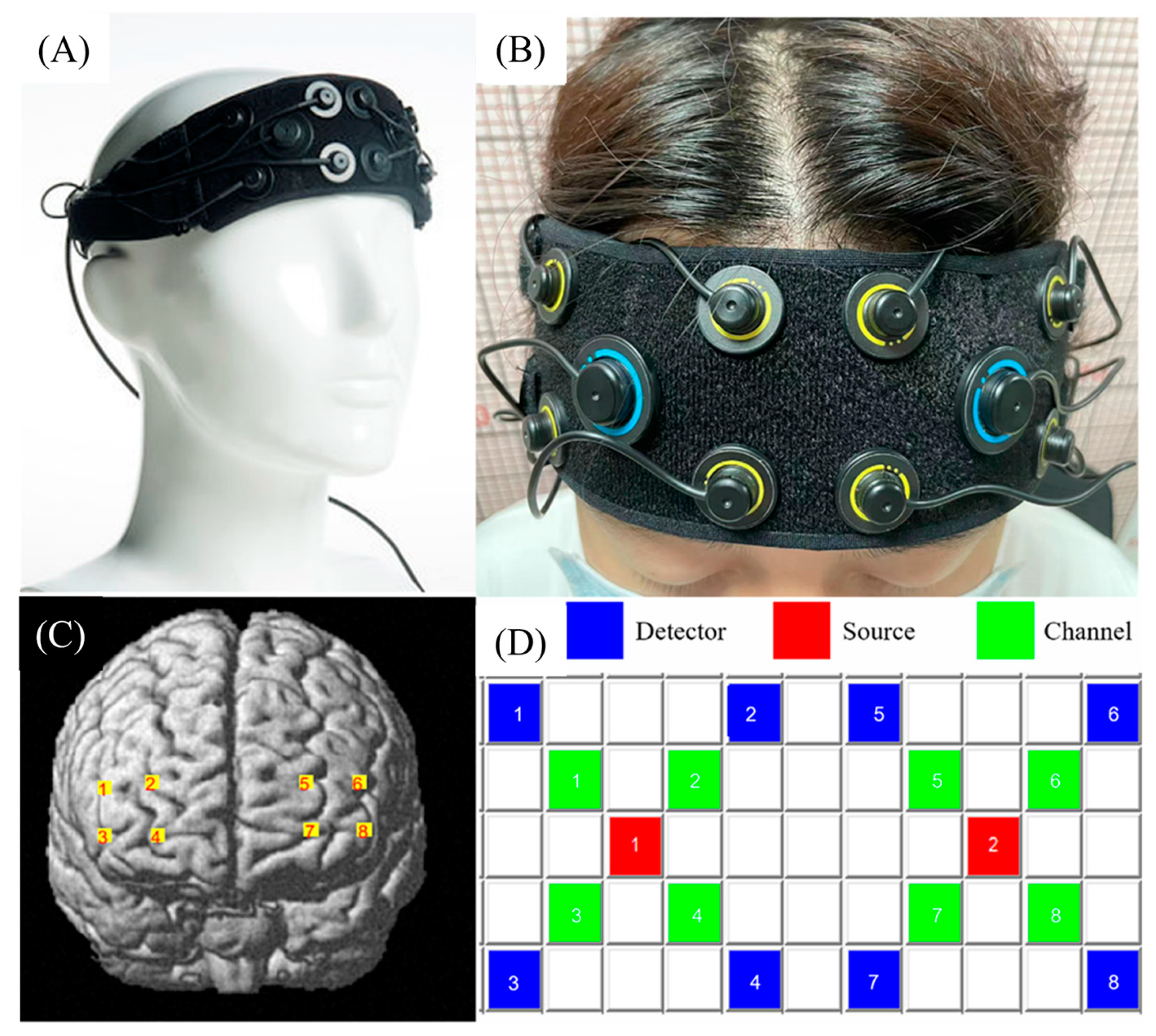
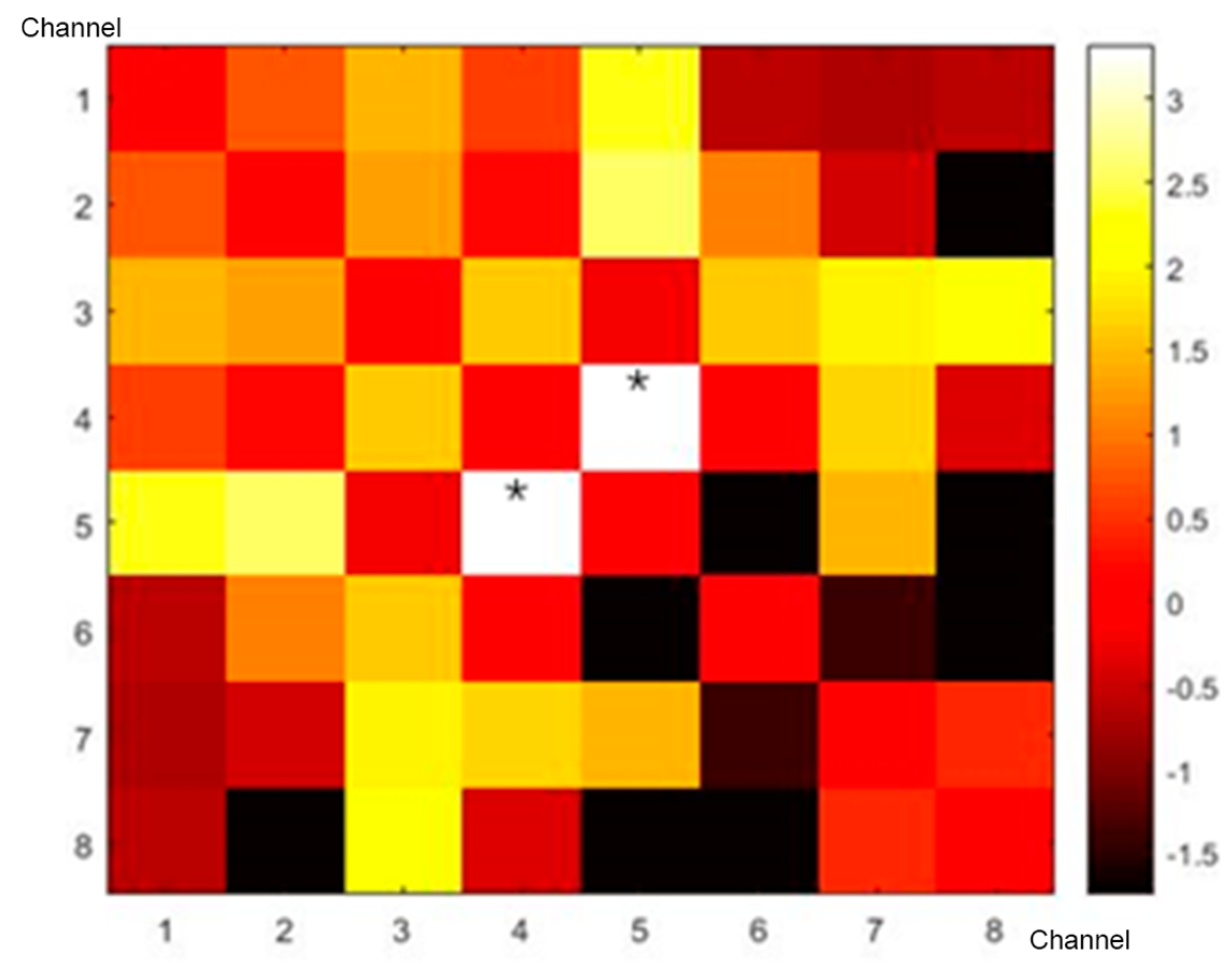
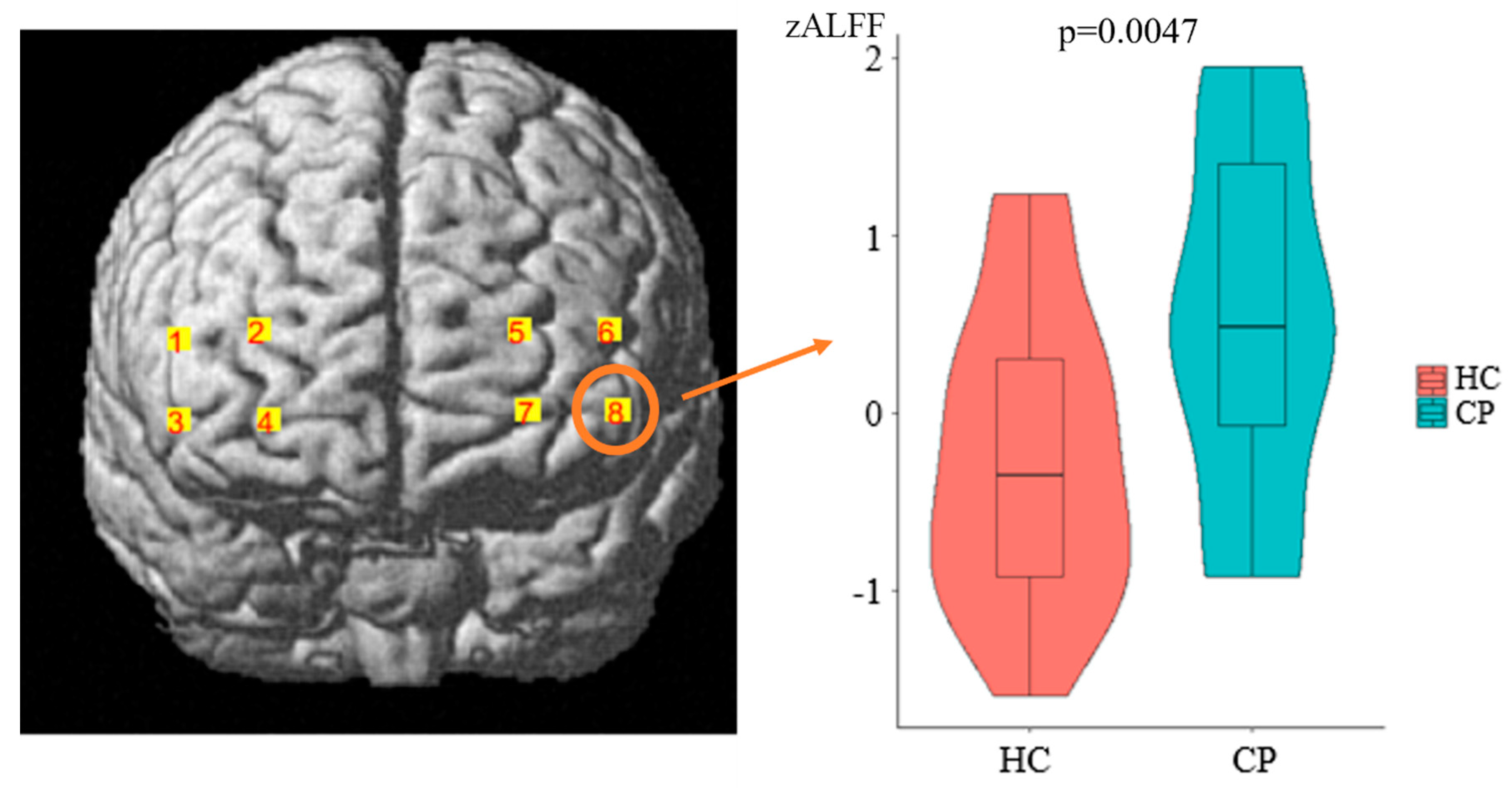
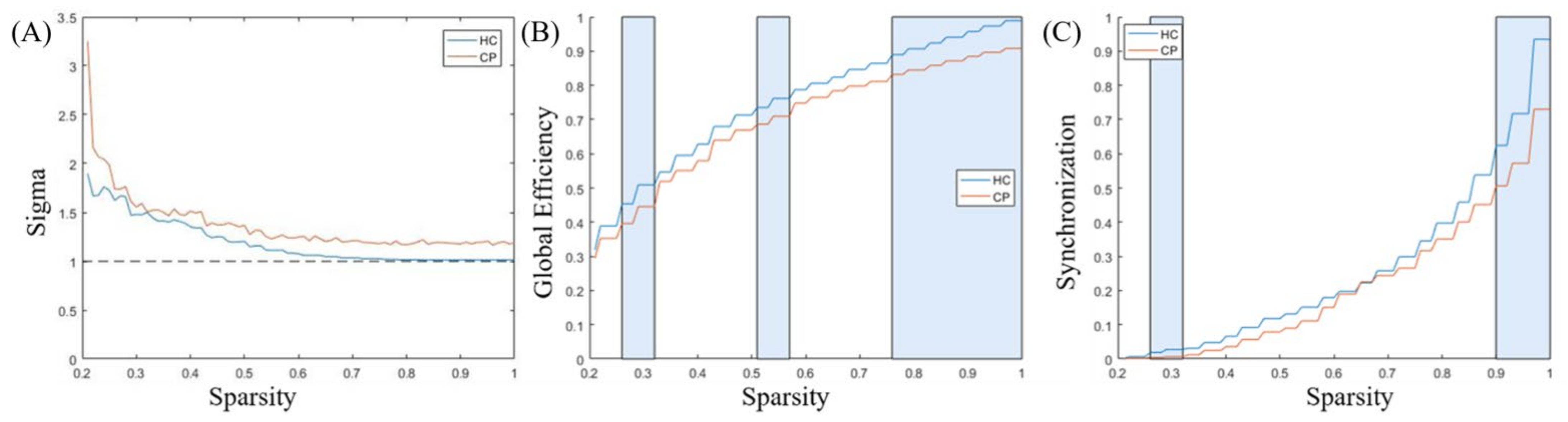
| Channel Numbers | MNI | Anatomical Label | Percentage of Overlap | ||
|---|---|---|---|---|---|
| X | Y | Z | |||
| 1 | 51 | 46.33 | 16.33 | 45—pars triangularis Broca’s area; 46—Dorsolateral prefrontal cortex; | 60.517% 39.483% |
| 2 | 31.67 | 65 | 17 | 10—Frontopolar area; 46—Dorsolateral prefrontal cortex; | 80.989% 19.011% |
| 3 | 50.33 | 51.67 | −1.33 | 45—pars triangularis Broca’s area; 46—Dorsolateral prefrontal cortex; 47—Inferior prefrontal gyrus; | 2.9197% 96.35% 0.72993% |
| 4 | 30.67 | 68.33 | −1.67 | 10—Frontopolar area; 11—Orbitofrontal area; | 36.093% 63.907% |
| 5 | −25.67 | 66.67 | 17.67 | 10—Frontopolar area; 46—Dorsolateral prefrontal cortex; | 86.716% 13.284% |
| 6 | −46 | 49.33 | 17.33 | 45—pars triangularis Broca’s area; 46—Dorsolateral prefrontal cortex; | 41.985% 58.015% |
| 7 | −27.67 | 67.33 | 0.67 | 10—Frontopolar area; 11—Orbitofrontal area; | 51.495% 48.505% |
| 8 | −47.67 | 51.67 | −0.67 | 10—Frontopolar area; 45—pars triangularis Broca’s area; 46—Dorsolateral prefrontal cortex; | 2.2642% 3.7736% 93.962% |
| Selected Measurement | HC (Mean ± Standard Deviation) | Pain Patients (Mean ± Standard Deviation) | t Value | p Values |
|---|---|---|---|---|
| Network Efficiency | 0.6596 ± 0.1093 | 0.7528 ± 0.0855 | −3.1684 | 0.0027 |
| Nodal Local Efficiency_5 | 0.5703 ± 0.3846 | 0.8632 ± 0.2392 | −2.9788 | 0.0045 |
| Nodal Cluster Efficiency 5 | 0.5129 ± 0.3675 | 0.8035 ± 0.2646 | −2.9981 | 0.0043 |
| Local Efficiency of Nodal 8 | 0.6031 ± 0.4249 | 0.8784 ± 0.1535 | −2.7089 | 0.0093 |
| Community Index of Nodal 7 | 1.7419 ± 0.6308 | 1.1053 ± 0.8753 | 2.9849 | 0.0045 |
| Clustering coefficient | 0.5920 ± 0.1174 | 0.6829 ± 0.1024 | −2.7854 | 0.0076 |
| Efficiency of Nodal 7 | 0.6859 ± 0.1328 | 0.4743 ± 0.3383 | 3.1267 | 0.0030 |
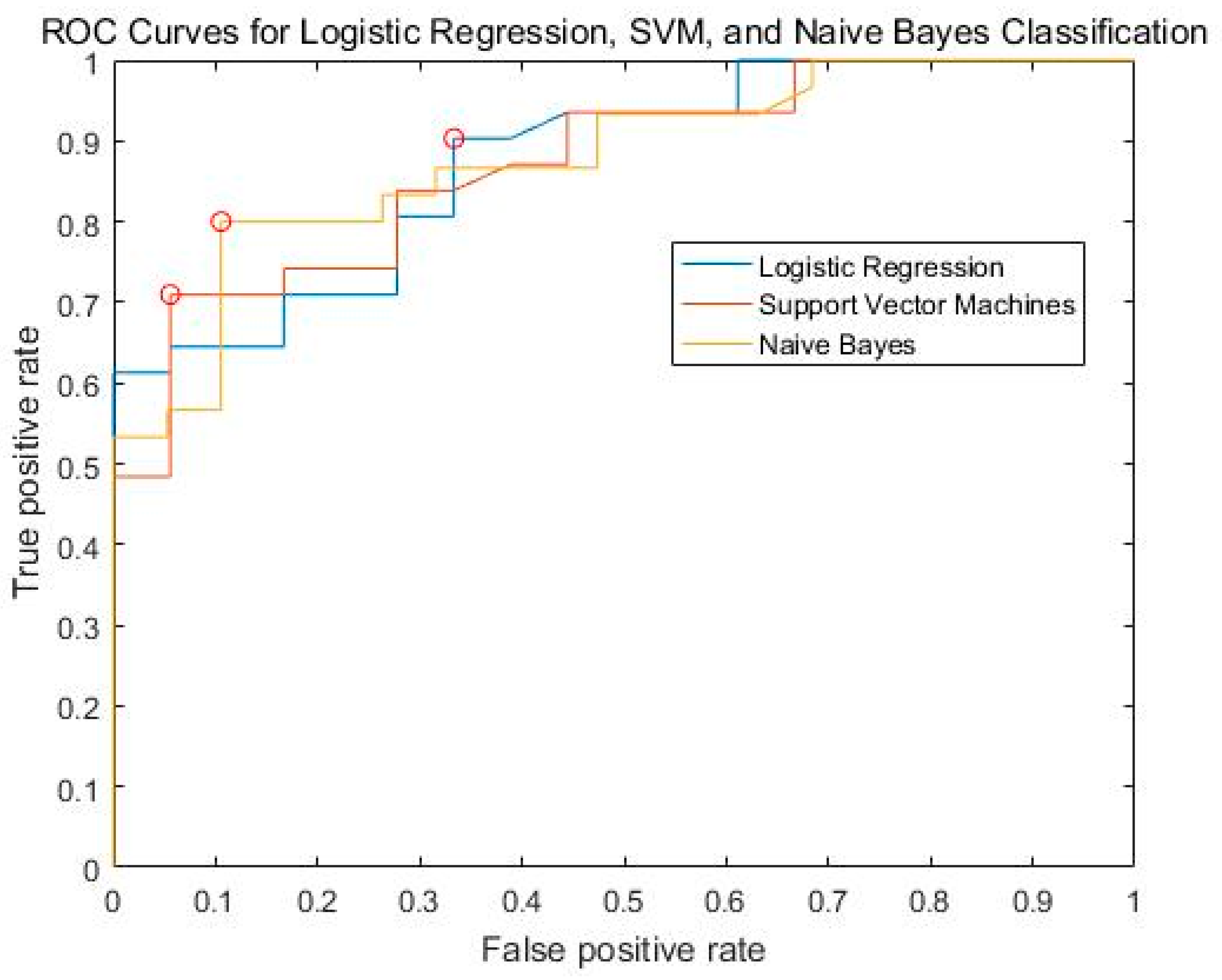
| Learning Model | Accuracy | Precise | Recall | F1 Score | AUC |
|---|---|---|---|---|---|
| SVM (linear) | 0.7559 | 0.7517 | 0.9135 | 0.8229 | 0.8719 |
| Logistic Regression | 0.7898 | 0.7418 | 0.9135 | 0.7131 | 0.8754 |
| naïve Bayes | 0.7755 | 0.7297 | 0.7269 | 0.7279 | 0.8781 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.; Tang, W.; Yang, J.; Lin, X.; Du, M.; Chen, X.; Yuan, Z.; Zhang, Z.; Chen, Z. Diagnosis of Chronic Musculoskeletal Pain by Using Functional Near-Infrared Spectroscopy and Machine Learning. Bioengineering 2023, 10, 669. https://doi.org/10.3390/bioengineering10060669
Zeng X, Tang W, Yang J, Lin X, Du M, Chen X, Yuan Z, Zhang Z, Chen Z. Diagnosis of Chronic Musculoskeletal Pain by Using Functional Near-Infrared Spectroscopy and Machine Learning. Bioengineering. 2023; 10(6):669. https://doi.org/10.3390/bioengineering10060669
Chicago/Turabian StyleZeng, Xinglin, Wen Tang, Jiajia Yang, Xiange Lin, Meng Du, Xueli Chen, Zhen Yuan, Zhou Zhang, and Zhiyi Chen. 2023. "Diagnosis of Chronic Musculoskeletal Pain by Using Functional Near-Infrared Spectroscopy and Machine Learning" Bioengineering 10, no. 6: 669. https://doi.org/10.3390/bioengineering10060669
APA StyleZeng, X., Tang, W., Yang, J., Lin, X., Du, M., Chen, X., Yuan, Z., Zhang, Z., & Chen, Z. (2023). Diagnosis of Chronic Musculoskeletal Pain by Using Functional Near-Infrared Spectroscopy and Machine Learning. Bioengineering, 10(6), 669. https://doi.org/10.3390/bioengineering10060669









