Classifying Heart-Sound Signals Based on CNN Trained on MelSpectrum and Log-MelSpectrum Features
Abstract
:1. Introduction
2. MelSpectrum and Log-MelSpectrum Features
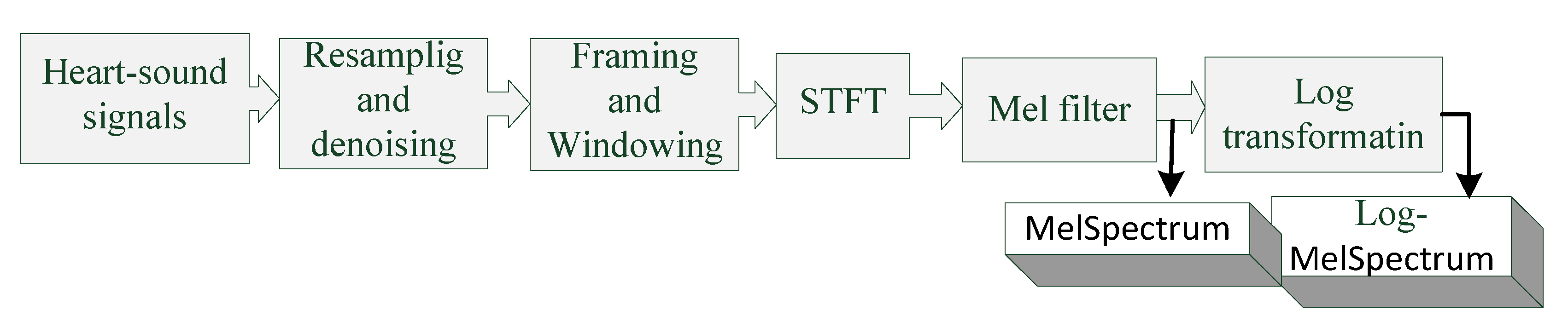
2.1. Extraction of MelSpectrum and Log-MelSpectrum Features
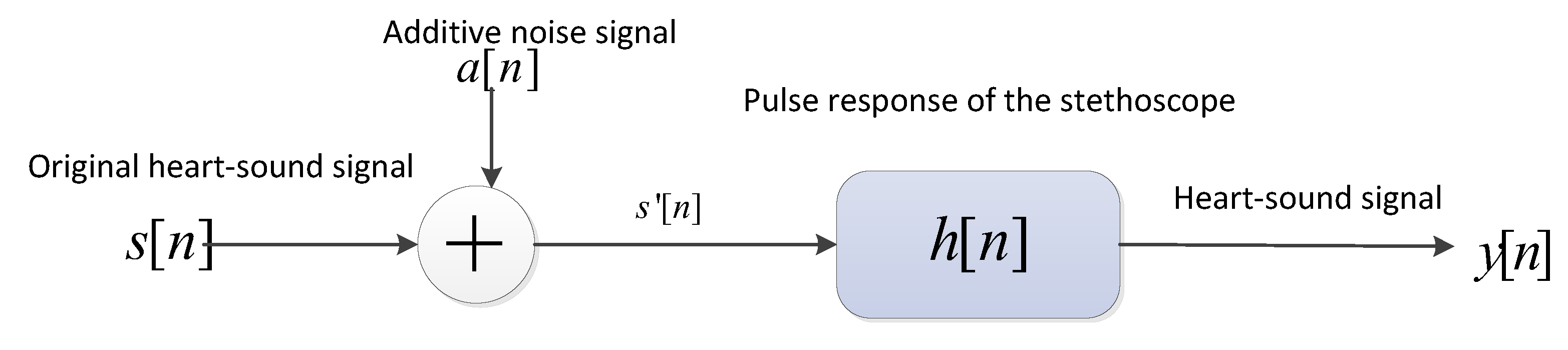
- The heart-sound signals are resampled from 25 Hz to 950 Hz using a Butterworth filter with a sampling frequency of 2000 Hz. The signals are then passed through a Savitzky–Golay filter to improve the smoothness of the time-frequency feature graph and reduce noise interference.
- The filtered signals are framed and windowed using a Hanning window function to fix the signals into a selected frame length.
- Frames are transformed into the periodogram estimate of the power spectrum using STFT.
- Each periodogram estimate is mapped onto the Mel-scale using Mel filters, which consist of several triangular filters. The output of the Mel filter is called the MelSpectrum.
- Logarithmic transformation is applied to the MelSpectrum features to obtain the Log-MelSpectrum.
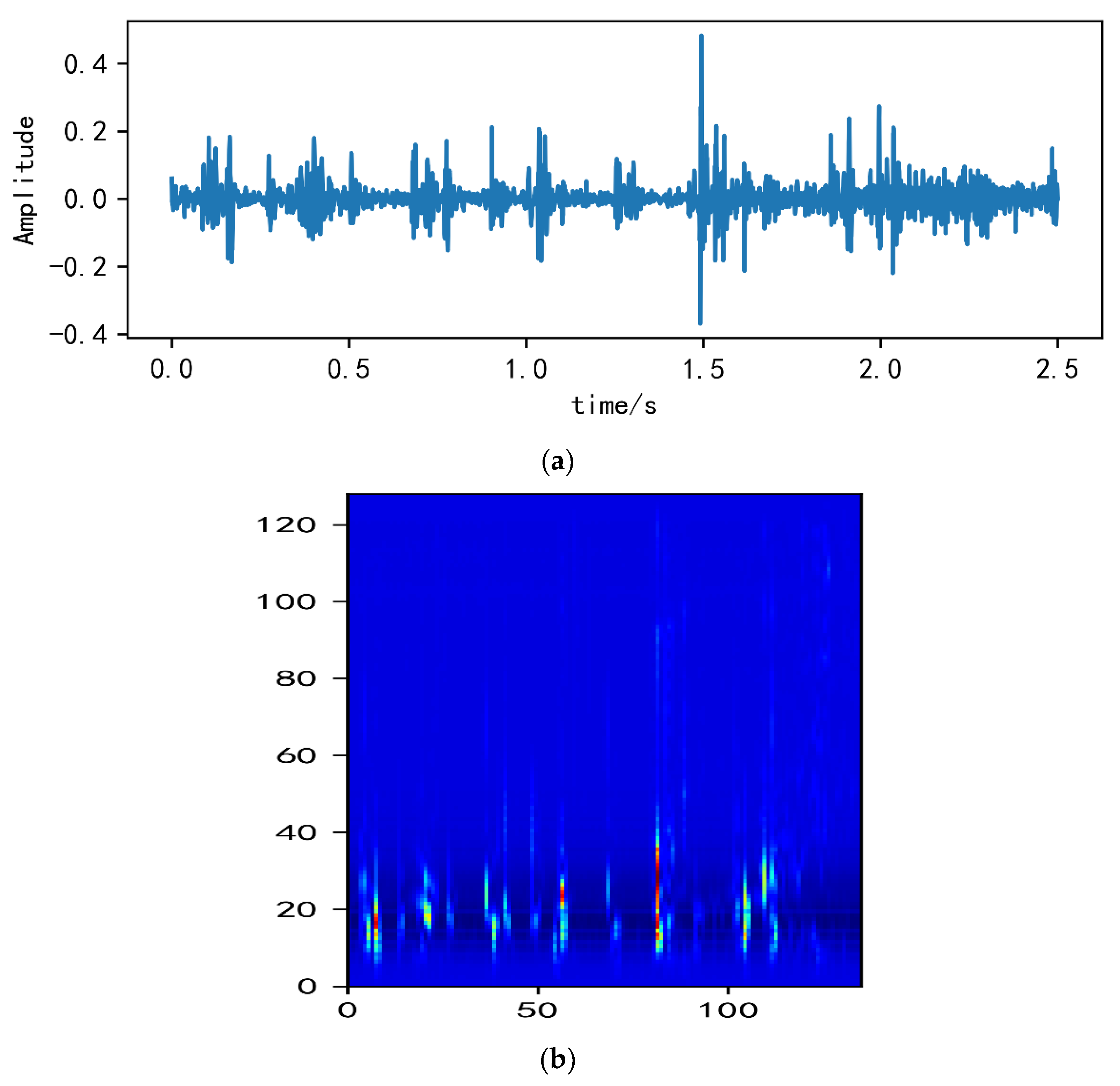
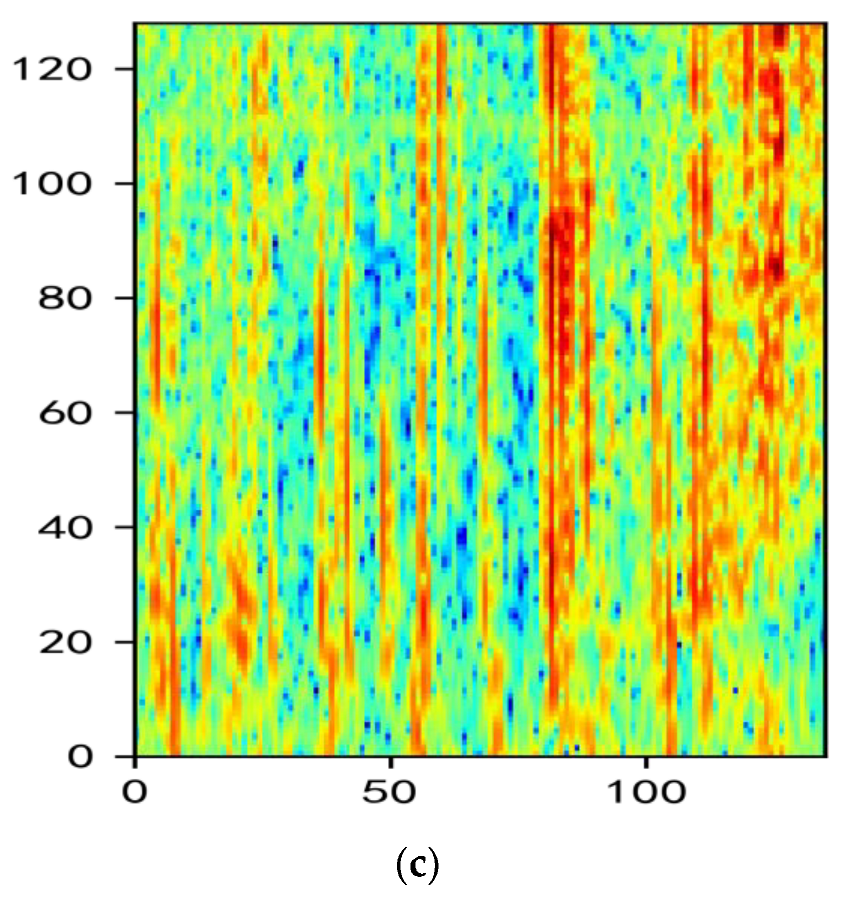
2.2. Analysis of MelSpectrum and Log-MelSpectrum
3. Experiments and Results
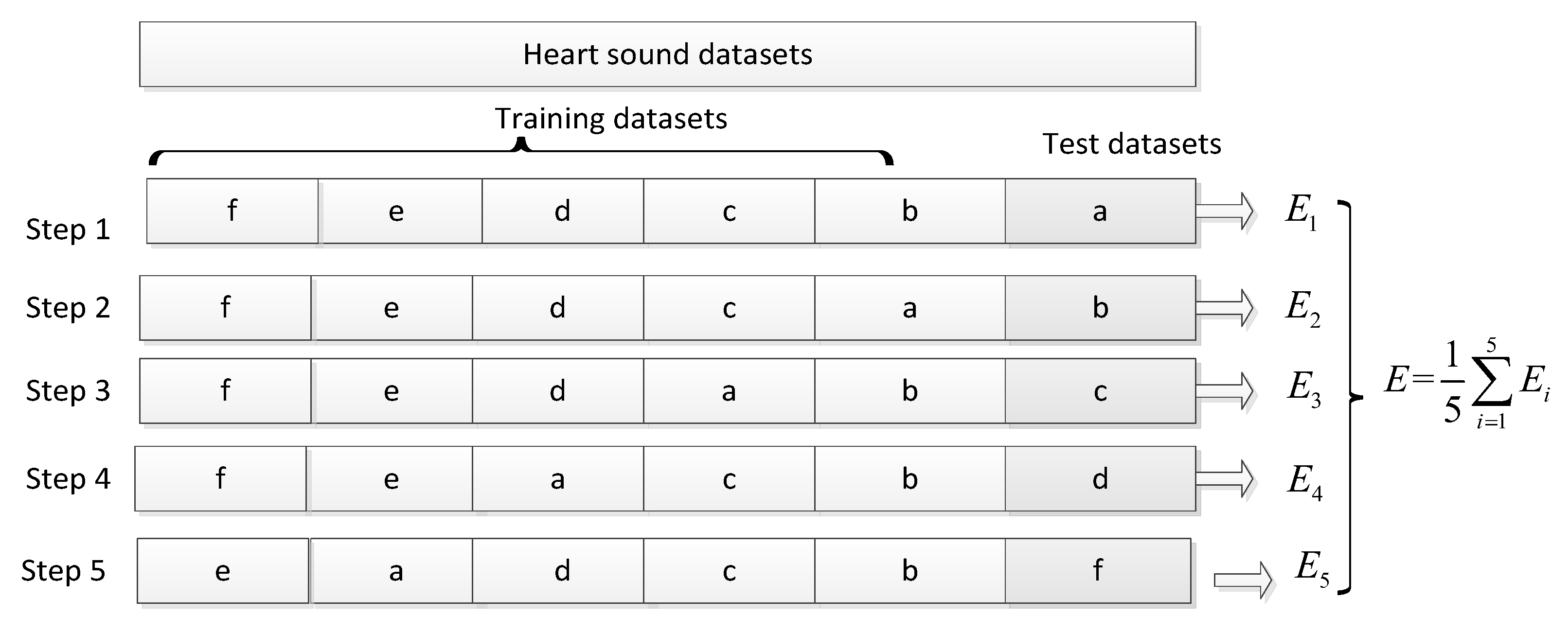
3.1. Heart-Sound Datasets
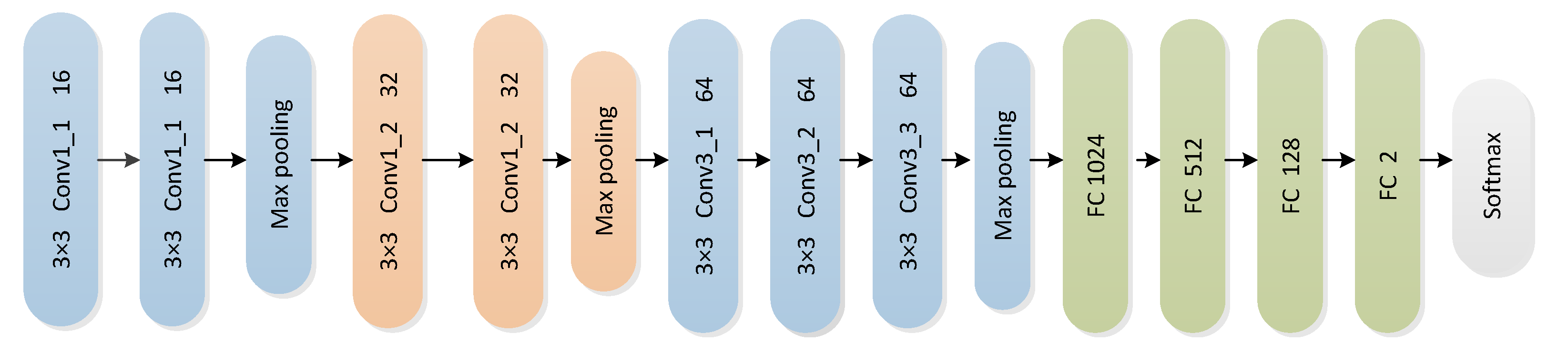
3.2. CNN Architecture
3.3. Experimental Process
3.4. Experimental Results and Analysis
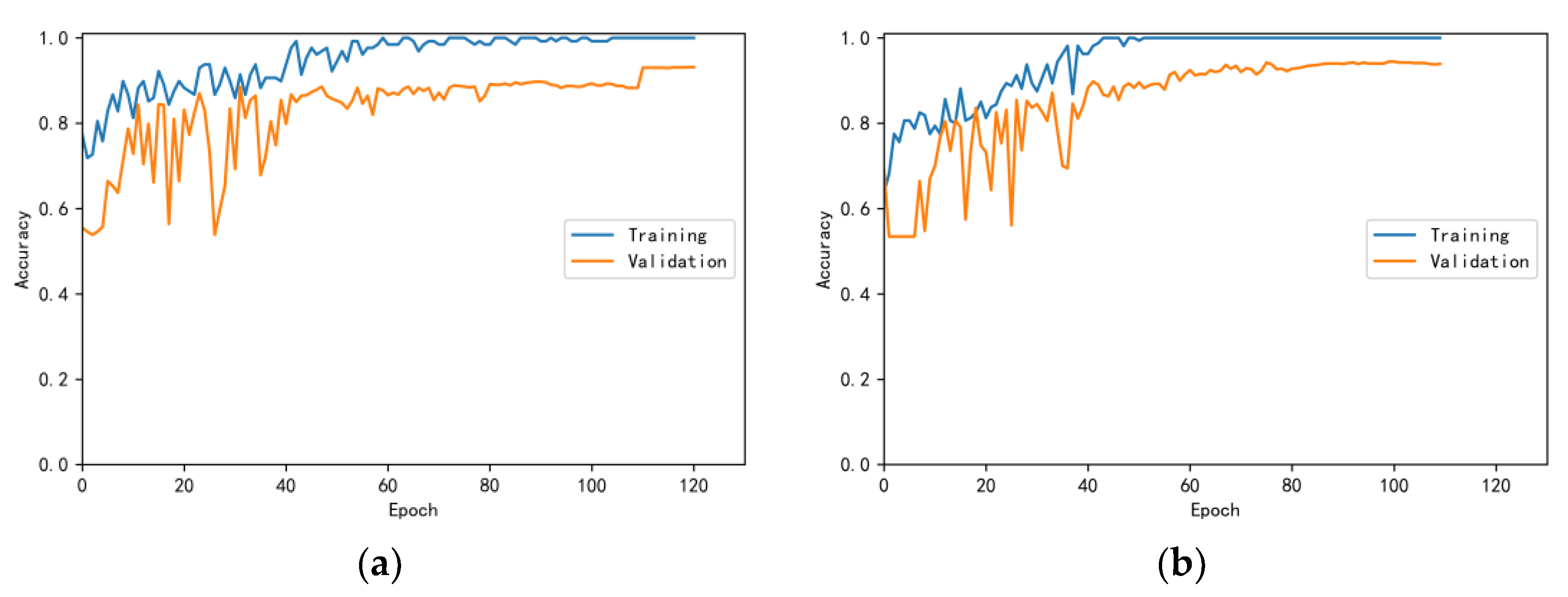
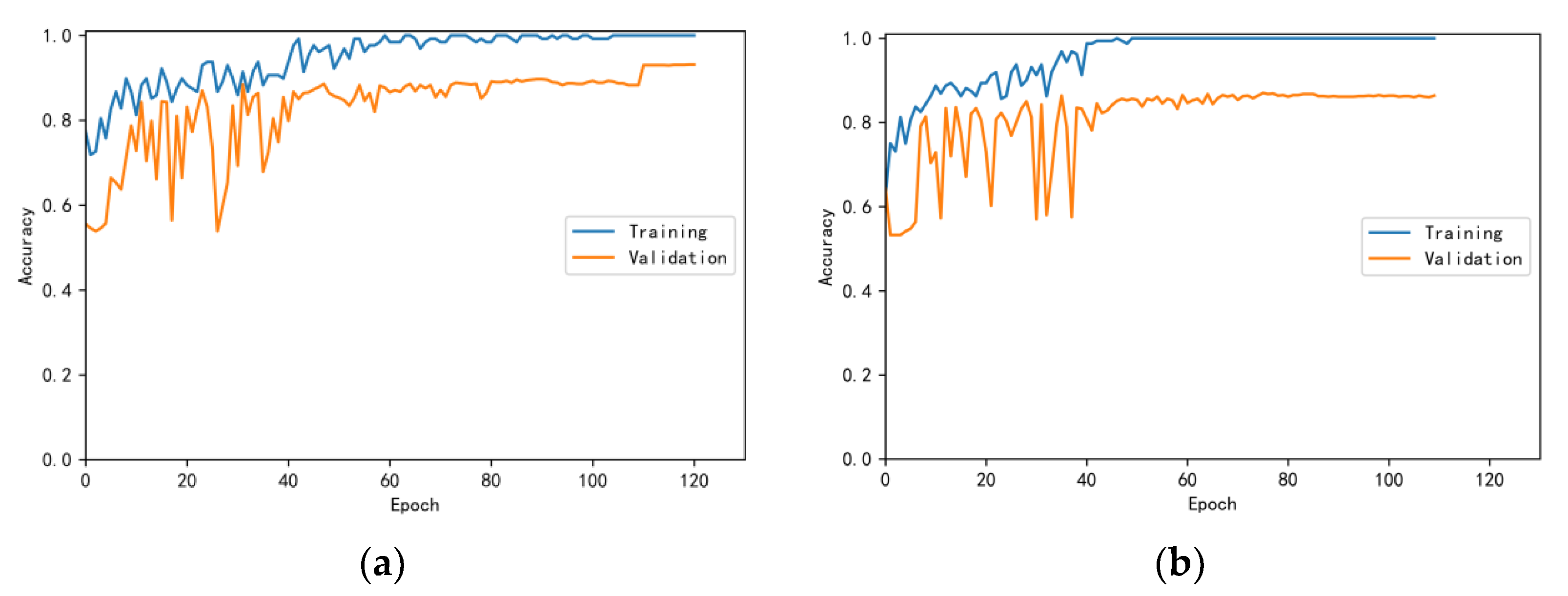
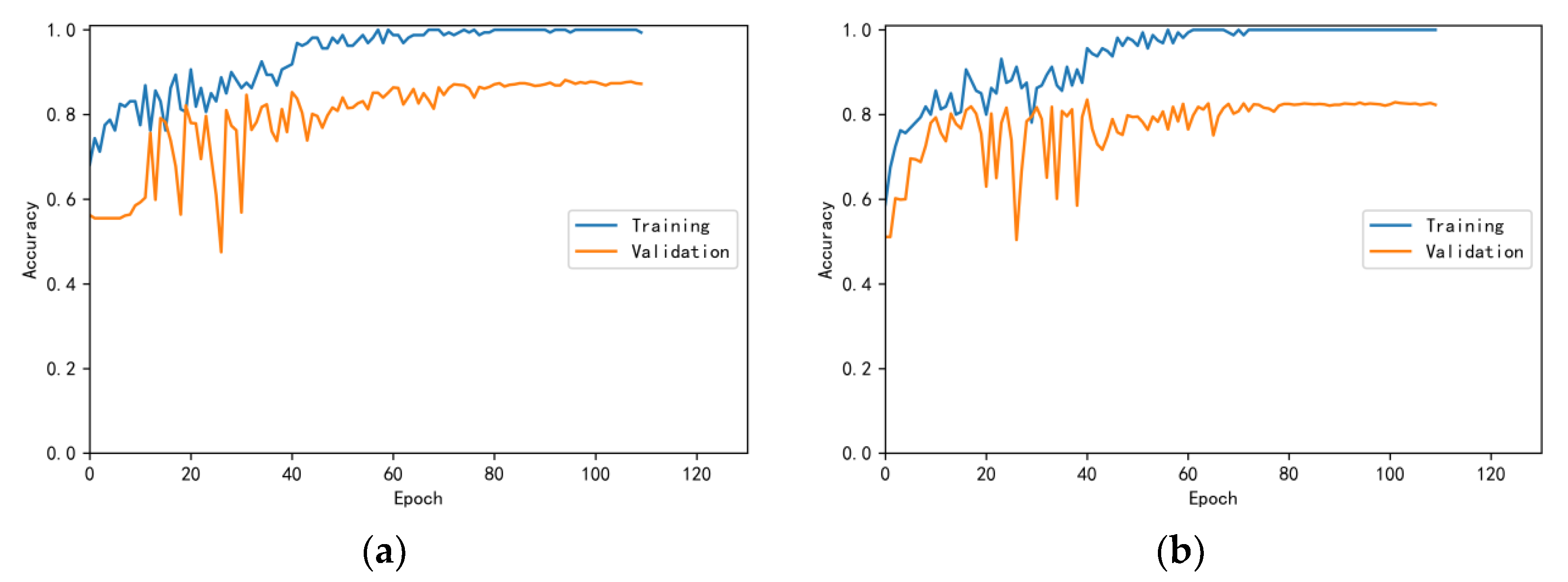
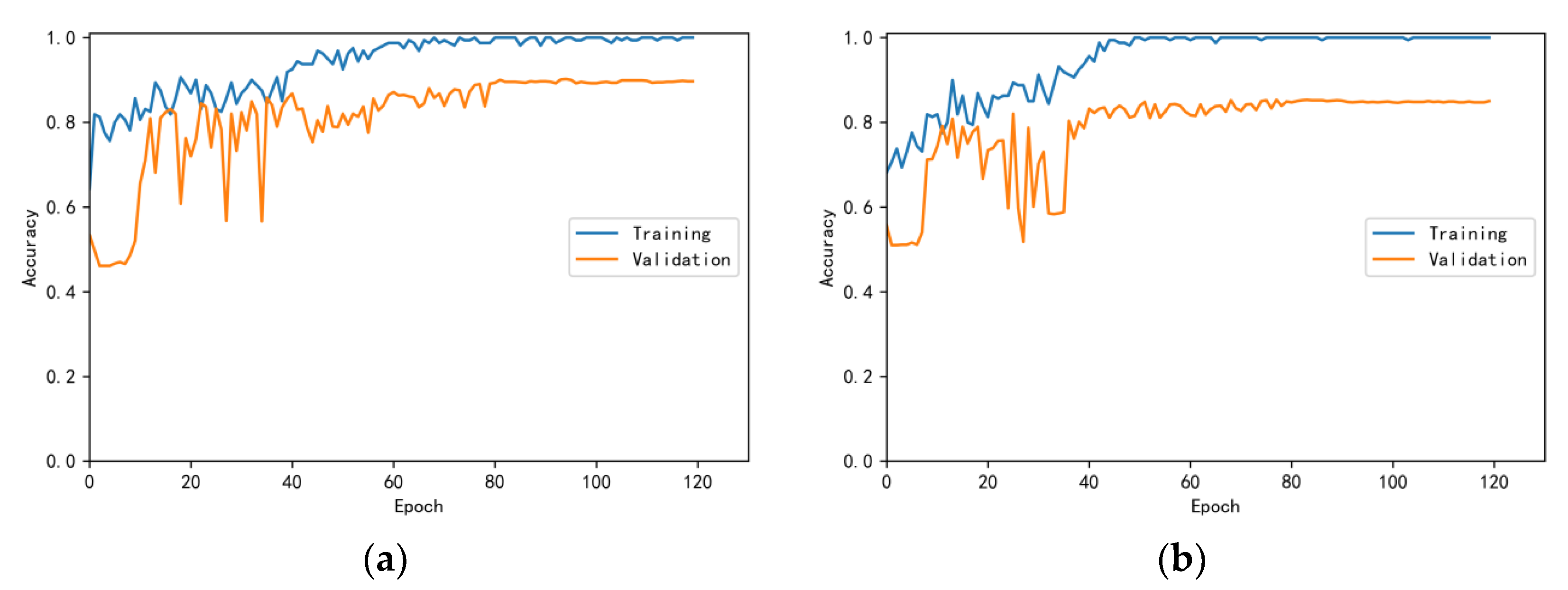
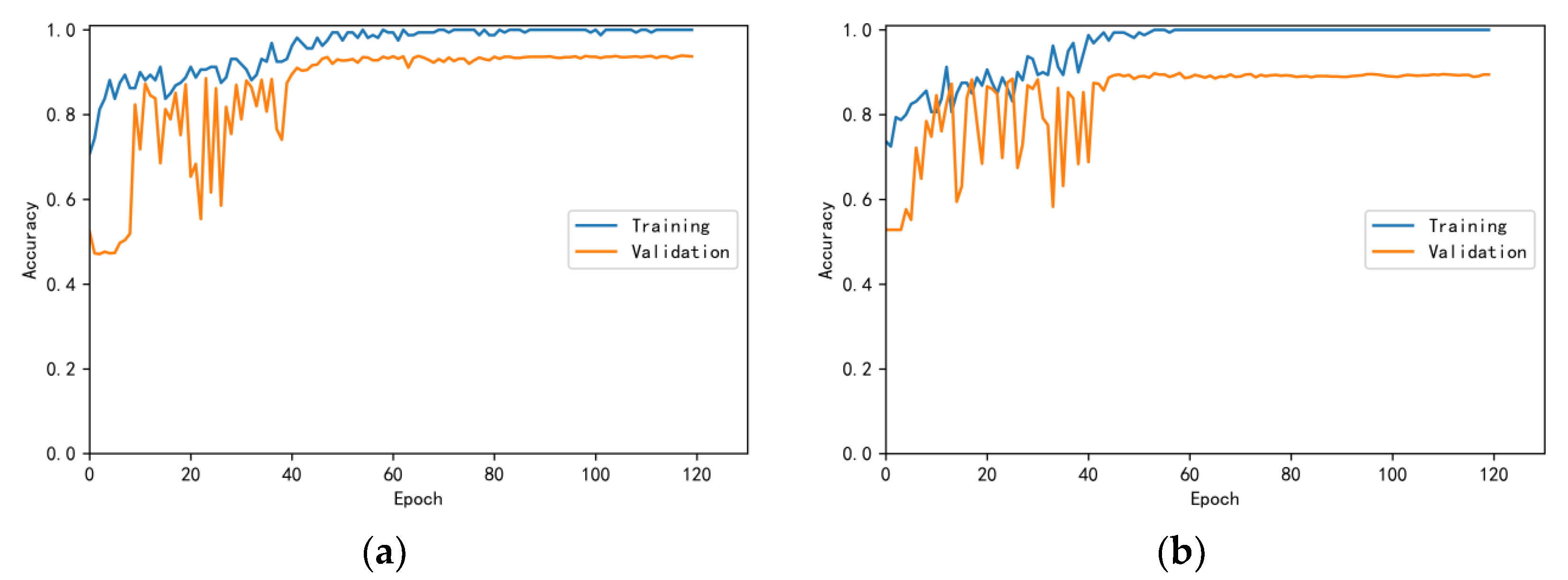
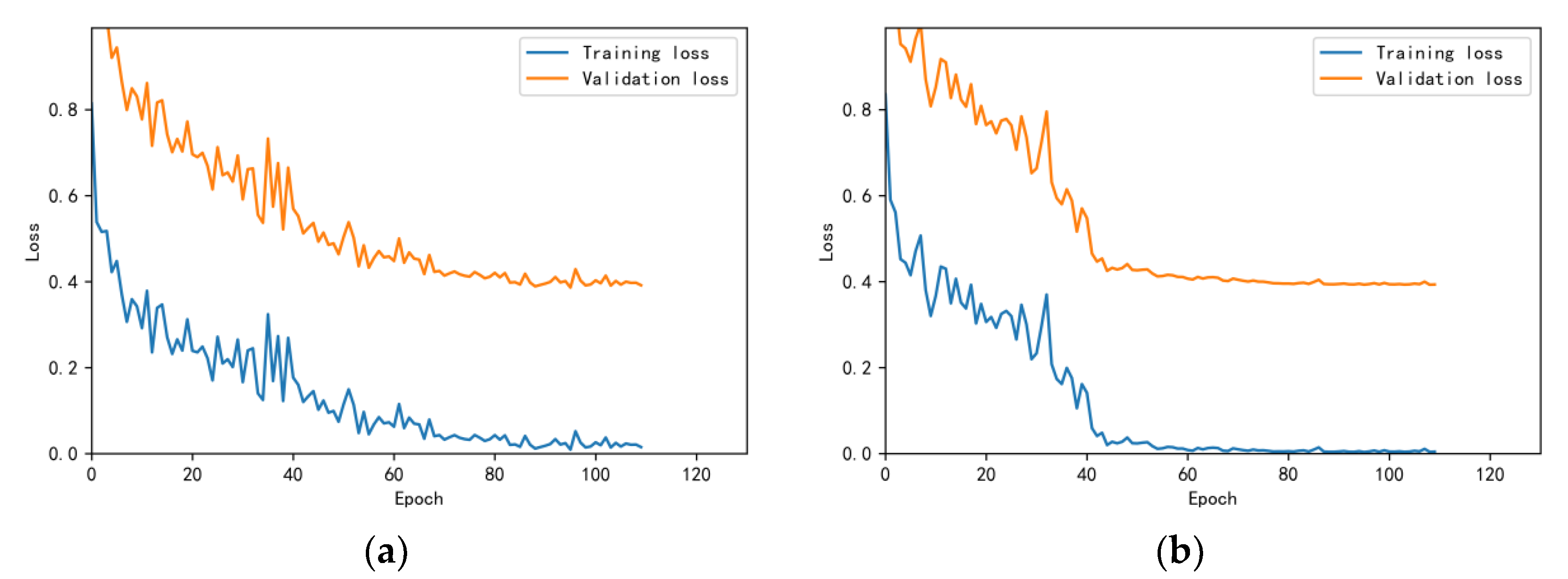
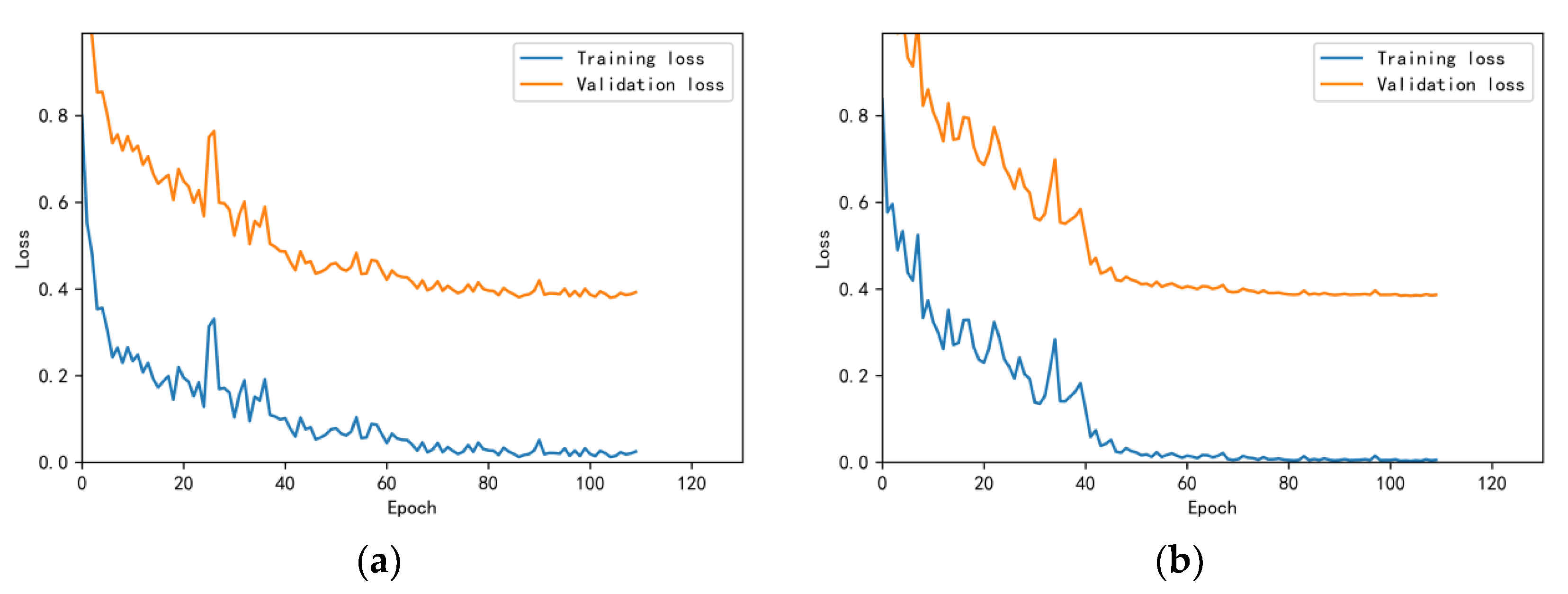
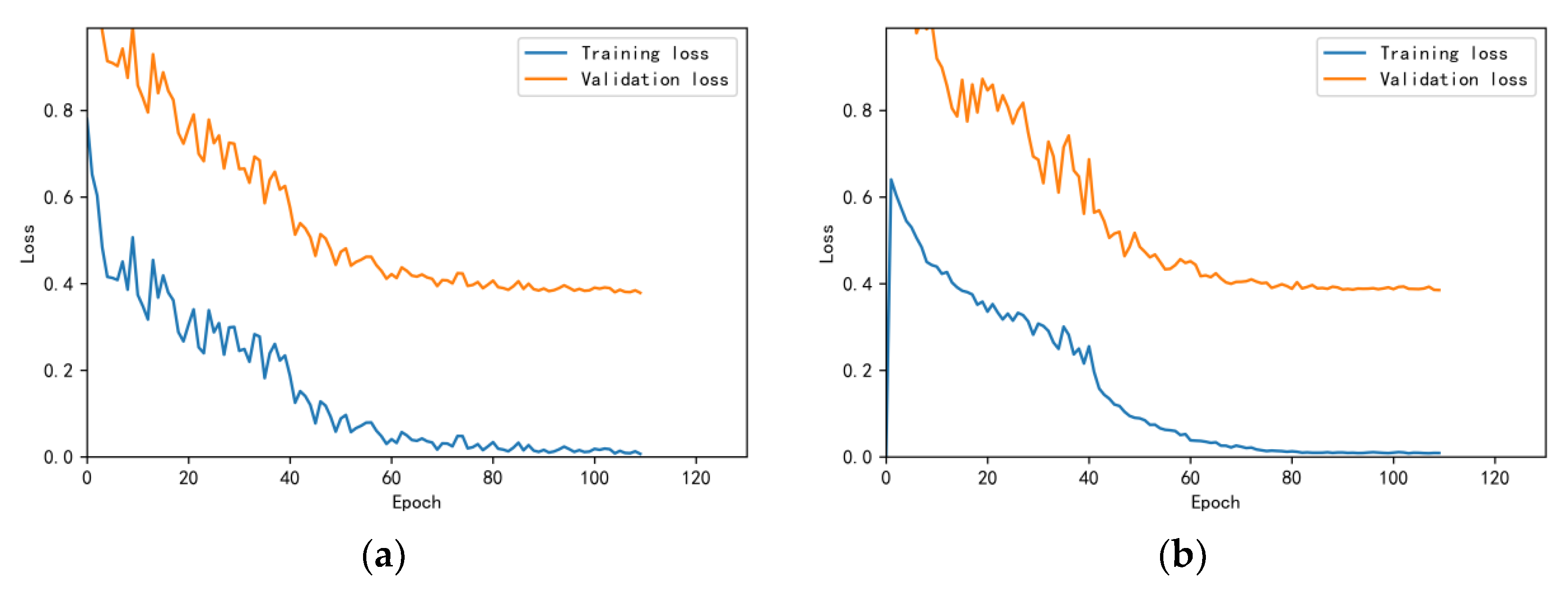
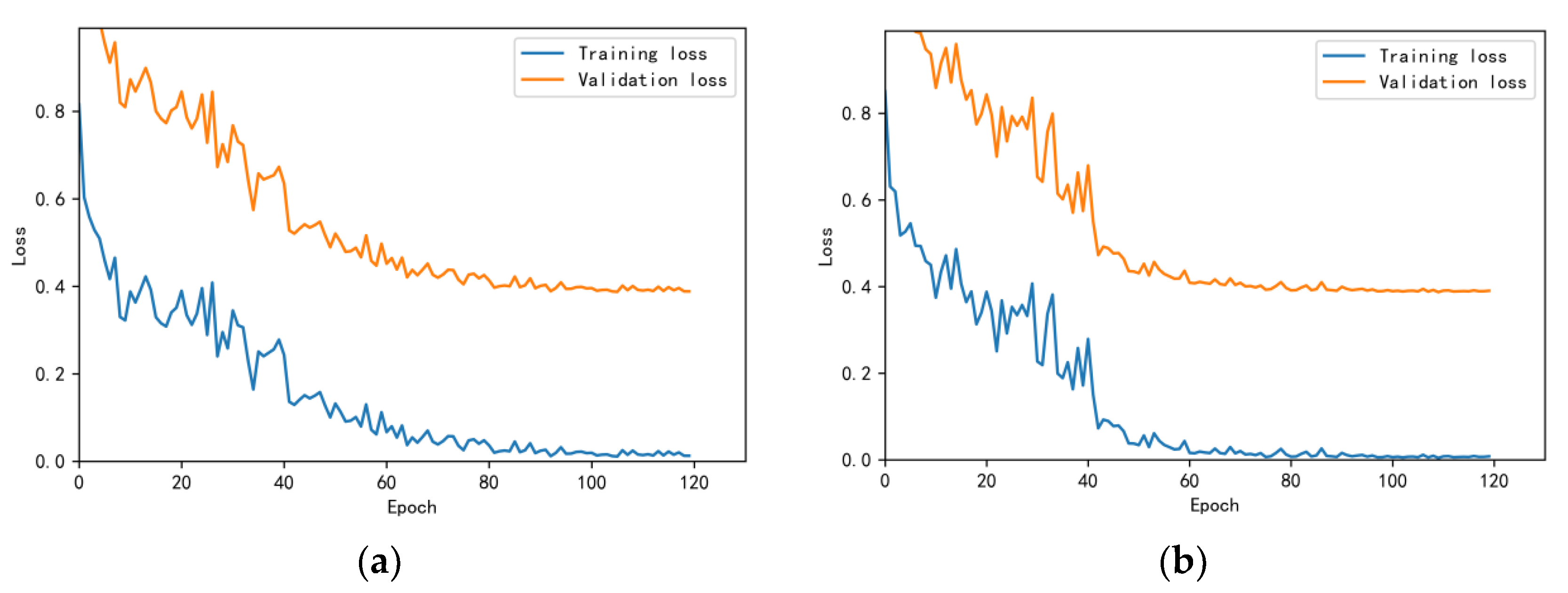
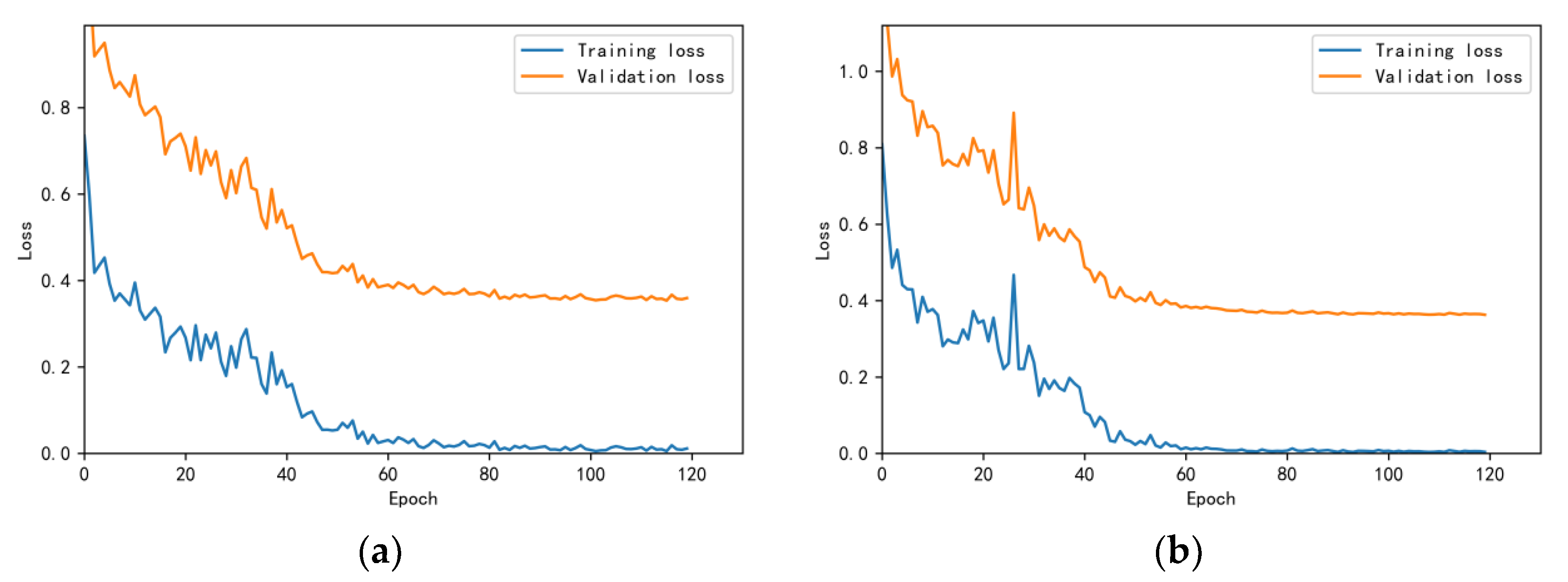
3.4.1. Model Training Results and Analysis
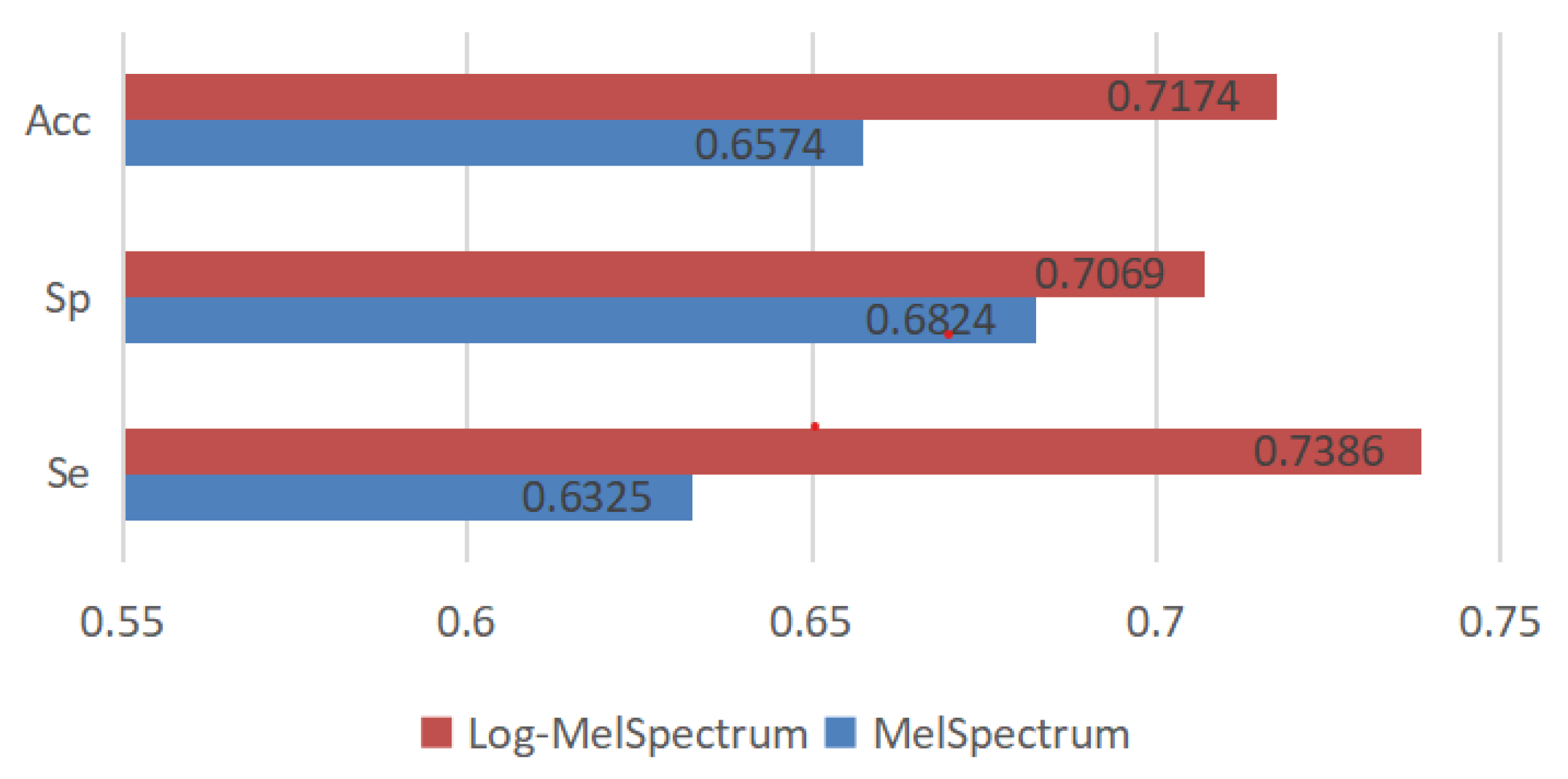
3.4.2. Test Results and Analysis
4. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Cardiovascular Diseases (CVDs) [EB/OL]. Available online: https://www.who.int/zh/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 1 May 2023).
- Liu, C.; Murray, A. Applications of Complexity Analysis in Clinical Heart Failure. In Complexity and Nonlinearity in Cardiovascular Signals; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Baghel, N.; Dutta, M.K.; Burget, R. Automatic diagnosis of multiple cardiac diseases from PCG signals using convolutional neural network. Comput. Methods Programs Biomed. 2020, 197, 105750. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Sun, Q.; Chen, X.; Xie, G.; Wu, H.; Xu, C. Deep Learning Methods for Heart Sounds Classification: A Systematic Review. Entropy 2021, 23, 667. [Google Scholar] [CrossRef] [PubMed]
- Costin, H.; Păsărică, A.; Alexa, I.-D.; Ilie, A.C.; Rotariu, C.; Costin, D. Short-Term Heart Rate Variability using Wrist-Worn Pulse Wave Monitor Compared to a Holter ECG. In Proceedings of the IEEE 6th International Conference on E-Health and Bioengineering–EHB 2017, Sinaia, Romania, 22–24 June 2017; pp. 635–638. [Google Scholar]
- Chen, X.; Guo, X.; Zheng, Y.; Lv, C. Heart function grading evaluation based on heart sounds and convolutional neural networks. Phys. Eng. Sci. Med. 2023, 46, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, P.; Rajguru, G.; Datta, D.; Mukhopadhyay, S. Automatic sleep stage classification using time-frequency images of CWT and transfer learning using convolution neural network. Biocybern. Biomed. Eng. 2020, 40, 494–504. [Google Scholar] [CrossRef]
- Khan, M.U.; Samer, S.; Alshehri, M.D.; Baloch, N.K.; Khan, H.; Hussain, F.; Kim, S.W.; Zikria, Y.B. Artificial neural network-based cardiovascular disease prediction using spectral features. Comput. Electr. Eng. 2022, 101, 108094. [Google Scholar] [CrossRef]
- Mei, N.; Wang, H.; Zhang, Y.; Liu, F.; Jiang, X.; Wei, S. Classification of heart sounds based on quality assessment and wavelet scattering transform. Comput. Biol. Med. 2021, 137, 104814. [Google Scholar] [CrossRef]
- Rath, A.; Mishra, D.; Panda, G.; Pal, M. Development and assessment of machine learning based heart disease detection using imbalanced heart sound signal. Biomed. Signal Process. Control 2022, 76, 103730. [Google Scholar] [CrossRef]
- Xiang, M.; Zang, J.; Wang, J.; Wang, H.; Zhou, C.; Bi, R.; Zhang, Z.; Xue, C. Research of heart sound classification using two-dimensional features. Biomed. Signal Process. Control 2023, 79, 104190. [Google Scholar] [CrossRef]
- Grooby, E.; Sitaula, C.; Kwok, T.C.; Sharkey, D.; Marzbanrad, F.; Malhotra, A. Artificial intelligence-driven wearable technologies for neonatal cardiorespiratory monitoring: Part 1 wearable technology. Pediatr. Res. 2023, 93, 413–425. [Google Scholar] [CrossRef]
- Dominguez-Morales, J.P.; Jimenez-Fernandez, A.F.; Dominguez-Morales, M.J.; Jimenez-Moreno, G. Deep Neural Networks for the Recognition and Classification of Heart Murmurs Using Neuromorphic Auditory Sensors. IEEE Trans. Biomed. Circuits Syst. 2018, 12, 24–34. [Google Scholar] [CrossRef]
- Costin, H.-N.; Sanei, S. Intelligent Biosignal Processing in Wearable and Implantable Sensors. Biosensors 2022, 12, 396. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, F.; Tang, S.; Xiong, W. A Review of Computer-Aided Heart Sound Detection Techniques. BioMed Res. Int. 2020, 2020, 5846191. [Google Scholar] [CrossRef]
- Nogueira, D.M.; Ferreira, C.A.; Gomes, E.F.; Jorge, A.M. Classifying Heart Sounds Using Images of Motifs, MFCC and Temporal Features. J. Med. Syst. 2019, 43, 168. [Google Scholar] [CrossRef] [PubMed]
- Soeta, Y.; Bito, Y. Detection of features of prosthetic cardiac valve sound by spectrogram analysis. Appl. Acoust. 2015, 89, 28–33. [Google Scholar] [CrossRef]
- Abduh, Z.; Nehary, E.A.; Wahed, M.A.; Kadah, Y.M. Classification of Heart Sounds Using Fractional Fourier Transform Based Mel-Frequency Spectral Coefficients and Stacked Autoencoder Deep Neural Network. J. Med. Imaging Health Inf. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Maknickas, V.; Maknickas, A. Recognition of normal abnormal phonocardiographic signals using deep convolutional neural networks and mel-frequency spectral coefcients. Physiol. Meas. 2017, 38, 1671–1684. [Google Scholar] [CrossRef]
- Chen, Y.; Su, B.; Zeng, W.; Yuan, C.; Ji, B. Abnormal heart sound detection from unsegmented phonocardiogram using deep features and shallow classifiers. Multimed Tools Appl. 2023. [Google Scholar] [CrossRef]
- Chen, W.; Sun, Q.; Xu, C. A Novel Deep Learning Neural Network System for Imbalanced Heart Sounds Classification. J. Mech. Med. Biol. 2021, 2150064. [Google Scholar] [CrossRef]
- Deng, M.; Meng, T.; Cao, J.; Wang, S.; Zhang, J.; Fan, H. Heart sound classification based on improved MFCC features and convolutional recurrent neural networks. Neural Netw. 2020, 130, 22–32. [Google Scholar] [CrossRef]
- Wu, J.M.-T.; Tsai, M.-H.; Huang, Y.Z.; Islam, S.H.; Hassan, M.M.; Alelaiwi, A.; Fortino, G. Applying an ensemble convolutional neural network with Savitzky–Golay filter to construct a phonocardiogram prediction model. Appl. Soft Comput. 2019, 78, 29–40. [Google Scholar] [CrossRef]
- Noman, F.; Ting, C.-M.; Salleh, S.-H.; Ombao, H. Short-segment heart sound classification Using an ensemble of deep con-volutional neural networks. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1318–1322. [Google Scholar]
- Nilanon, T.; Yao, J.; Hao, J.; Purushotham, S. Normal/abnormal heart sound recordings classification using convolutional neural network. In Proceedings of the Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; pp. 585–588. [Google Scholar]
- Yıldırım, M. Automatic classification and diagnosis of heart valve diseases using heart sounds with MFCC and proposed deep model. Concurr. Comput. Pract. Exp. 2022, 34, e7232. [Google Scholar] [CrossRef]
- Abdollahpur, M.; Ghaffari, A.; Ghiasi, S.; Mollakazemi, M. Detection of pathological heart sounds. Physiol. Meas. 2017, 38, 1616–1630. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Huang, J.; Li, Y.; Gui, G. Design and application of a Laconic heart sound neural networ. IEEE Access 2019, 7, 124417–124425. [Google Scholar] [CrossRef]
- Rubin, J.; Abreu, R.; Ganguli, A.; Nelaturi, S.; Matei, I.; Sricharan, K. Classifying heart sound recordings using deep convolutional neural networks and mel-frequency cepstral coefficients. In Proceedings of the 2016 Computing in Cardiology Conference (CinC), Vancouver, BC, Canada, 11–14 September 2016; pp. 813–816. [Google Scholar]
- Springer, D.B.; Tarassenko, L.; Clifford, G.D. Logistic Regression-HSMM-Based Heart Sound Segmentation. IEEE Trans. Biomed. Eng. 2016, 63, 822. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Lin, W.W.; Huang, J.H. Heart Sound Classification Using Deep Learning Techniques Based on Log-mel Spectrogram. Circuits Syst. Signal Process. 2023, 42, 344–360. [Google Scholar] [CrossRef]
- Li, F.; Zhang, Z.; Wang, L.; Liu, W. Heart sound classification based on improved mel-frequency spectral coefficients and deep residual learning. Front Physiol. 2022, 13, 1084420. [Google Scholar] [CrossRef]
- Liu, C.; Springer, D.; Li, Q.; Moody, B.; Juan, R.A.; Chorro, F.J.; Castells, F.; Roig, J.M.; Silva, I.; Johnson, A.E.W.; et al. An open access database for the evaluation of heart sound algorithms. Physiol. Meas. 2016, 37, 2181. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the ICLR, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
| Parameter | Value | Description |
|---|---|---|
| Low and high frequency | 25 Hz to 950 Hz | The band frequency of Butterworth filter |
| Window_function | Hanning | Window function selected in FFT operation |
| Hop_length | 60 | Number of samples between consecutive frames |
| Sampling_frequency | 2000 Hz | Sampling frequency |
| N-FFT | 512 | The length of FFT operation |
| Window_size | 240 | Frame length in FFT operation |
| Sample_size | 2.5 s | The length of heart sound signals selected from the start position the cardiac cycle |
| Mel_filters | 128 | The number of Mel filters |
| Subset | Normal Recordings | Abnormal Recordings | Account for Total Datasets | Acquisition Equipment |
|---|---|---|---|---|
| a | 117 | 292 | 12.62% | Welch Allyn Meditron |
| b | 386 | 104 | 15.12% | 3 M Littmann E4000 |
| c | 7 | 24 | 0.96% | AUDIOSCOPE |
| d | 27 | 28 | 1.70% | Infral Corp. Prototype |
| e | 1958 | 183 | 66.08% | MLT201/Piezo, 3 M Littmann |
| f | 80 | 34 | 3.52% | JABES |
| Total | 2575 | 665 | 100% |
| Parameter | Step 1 | Step 2 | Step 3 | Step 4 | Step 5 |
|---|---|---|---|---|---|
| Training datasets/Test datasets | b, c, d, e, f/a | a, c, d, e, f/b | a, b, d, e, f/c | a, b, c, e, f/d | a, b, c, d, e/f |
| Learning rate | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 |
| Epoch_size | 25 | 22 | 26 | 26 | 25 |
| Epoch | 110 | 110 | 110 | 120 | 120 |
| BatchSize | 160 | 160 | 160 | 160 | 160 |
| Optimiser | Adam | Adam | Adam | Adam | Adam |
| Loss function | Cross-Entropy | Cross-Entropy | Cross-Entropy | Cross-Entropy | Cross-Entropy |
| Input | Dataset- a | Dataset- b | Dataset- c | Dataset- d | Dataset- f | Meana Ccuracy+ Variance |
|---|---|---|---|---|---|---|
| Log- MelSpectrum | 97.0% | 93.0% | 87.3% | 87.7% | 93.7% | 91.74 + 3.72 |
| MelSpectrum | 93.9% | 86.4% | 82.3% | 85% | 89.5% | 87.42 + 3.99 |
| Input Feature | Se | Sp | MAcc |
|---|---|---|---|
| Performance of the model based on test dataset-a | |||
| MelSpectrum | 0.6267 | 0.5299 | 0.5783 |
| Log-MelSpectrum | 0.5582 | 0.7949 | 0.6765 |
| Performance of the model based on test dataset-b | |||
| MelSpectrum | 0.5714 | 0.9482 | 0.7598 |
| Log-MelSpectrum | 0.7143 | 0.9508 | 0.8325 |
| Performance of the model based on test dataset-c | |||
| MelSpectrum | 0.8333 | 0.5714 | 0.7024 |
| Log-MelSpectrum | 0.875 | 0.5714 | 0.7232 |
| Performance of the model based on test dataset-d | |||
| MelSpectrum | 0.5714 | 0.6293 | 0.6005 |
| Log-MelSpectrum | 0.7857 | 0.5926 | 0.6892 |
| Performance of the model based on test dataset-f | |||
| MelSpectrum | 0.5588 | 0.7333 | 0.6461 |
| Log-MelSpectrum | 0.7059 | 0.625 | 0.6654 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Zhou, Z.; Bao, J.; Wang, C.; Chen, H.; Xu, C.; Xie, G.; Shen, H.; Wu, H. Classifying Heart-Sound Signals Based on CNN Trained on MelSpectrum and Log-MelSpectrum Features. Bioengineering 2023, 10, 645. https://doi.org/10.3390/bioengineering10060645
Chen W, Zhou Z, Bao J, Wang C, Chen H, Xu C, Xie G, Shen H, Wu H. Classifying Heart-Sound Signals Based on CNN Trained on MelSpectrum and Log-MelSpectrum Features. Bioengineering. 2023; 10(6):645. https://doi.org/10.3390/bioengineering10060645
Chicago/Turabian StyleChen, Wei, Zixuan Zhou, Junze Bao, Chengniu Wang, Hanqing Chen, Chen Xu, Gangcai Xie, Hongmin Shen, and Huiqun Wu. 2023. "Classifying Heart-Sound Signals Based on CNN Trained on MelSpectrum and Log-MelSpectrum Features" Bioengineering 10, no. 6: 645. https://doi.org/10.3390/bioengineering10060645
APA StyleChen, W., Zhou, Z., Bao, J., Wang, C., Chen, H., Xu, C., Xie, G., Shen, H., & Wu, H. (2023). Classifying Heart-Sound Signals Based on CNN Trained on MelSpectrum and Log-MelSpectrum Features. Bioengineering, 10(6), 645. https://doi.org/10.3390/bioengineering10060645






