Biomedical Relation Extraction Using Dependency Graph and Decoder-Enhanced Transformer Model
Abstract
1. Introduction
2. Related Works
3. Preliminaries
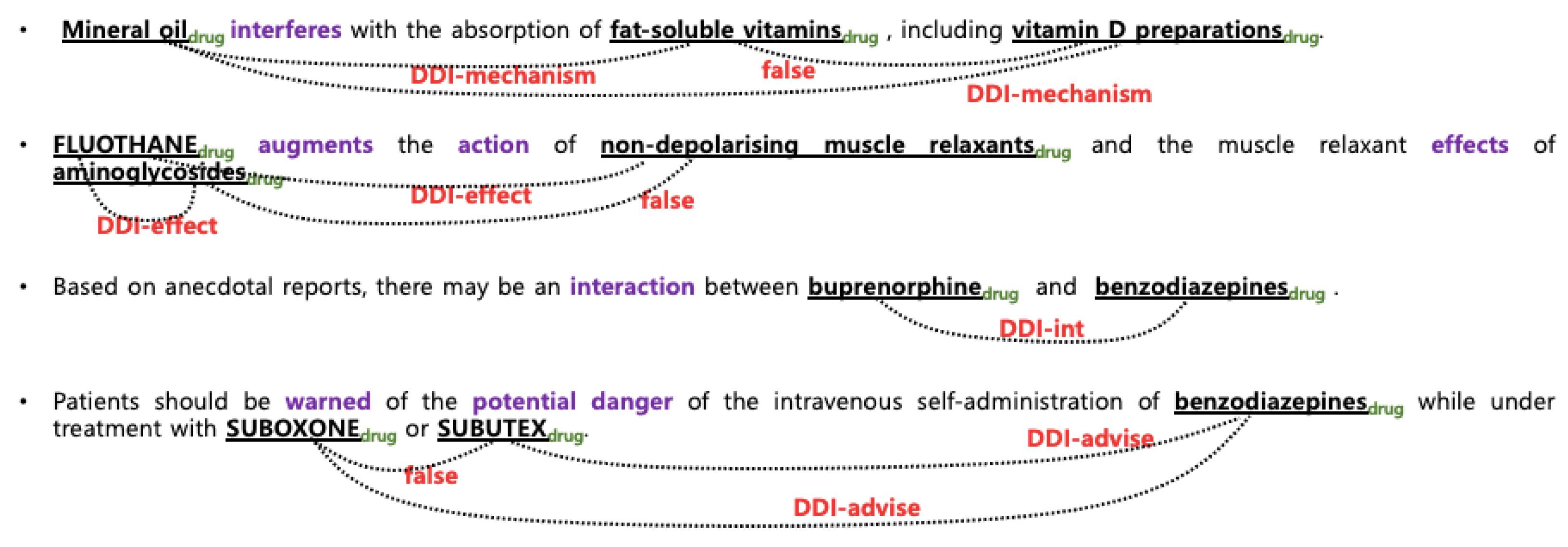
3.1. Data Sets and Target Relations
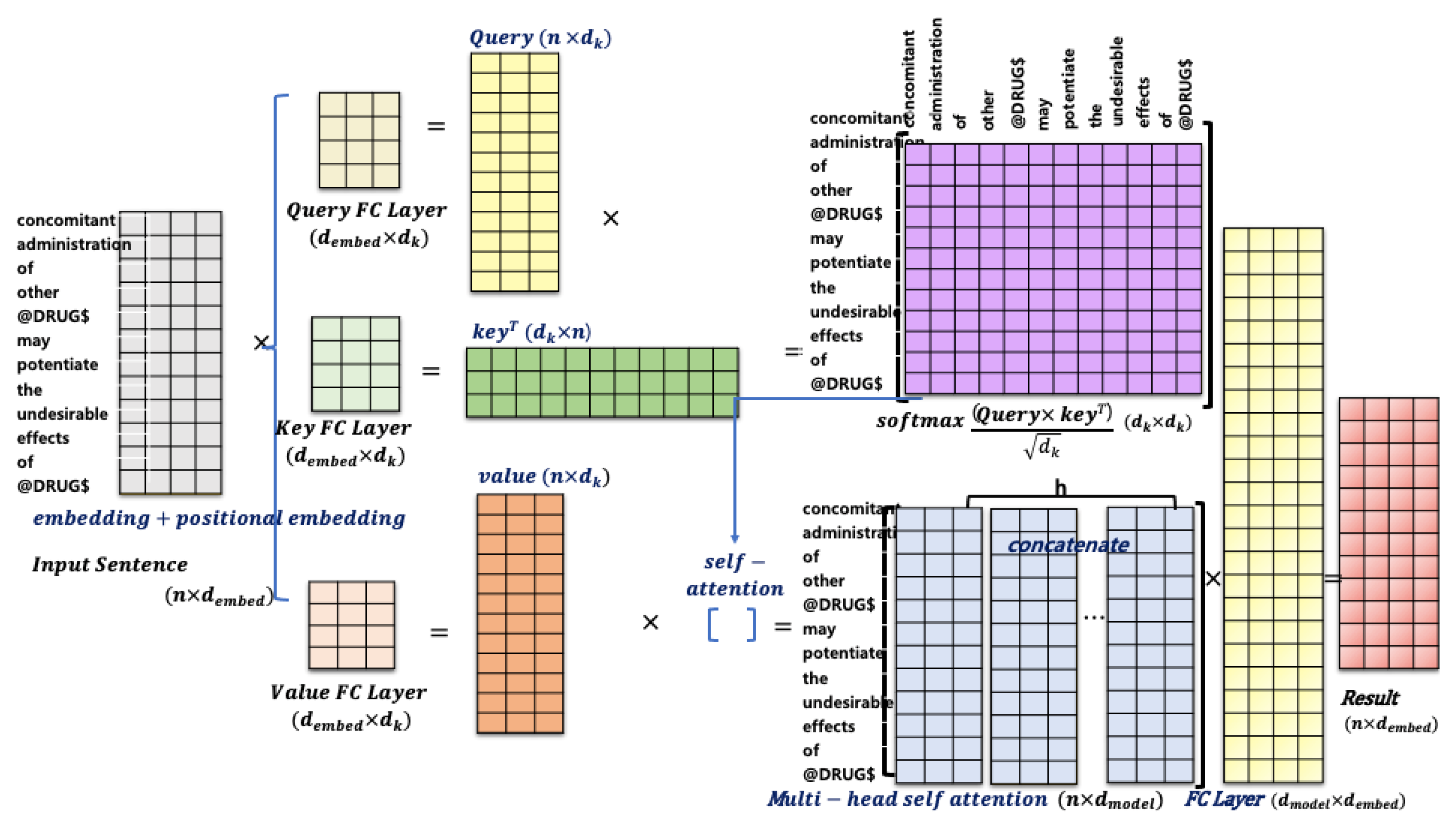
3.2. Transformer and Attention
4. Methods
4.1. Baseline Methods
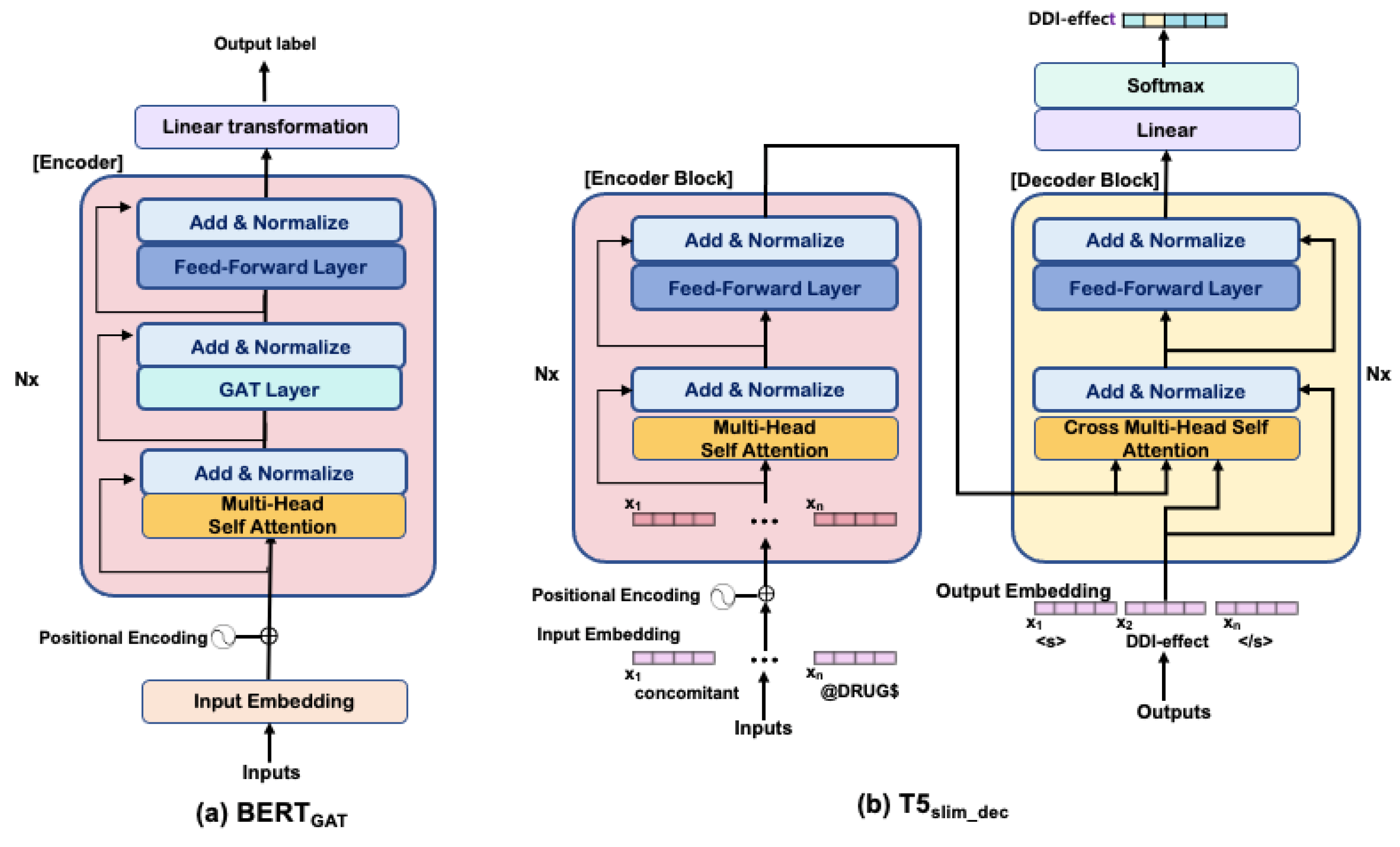
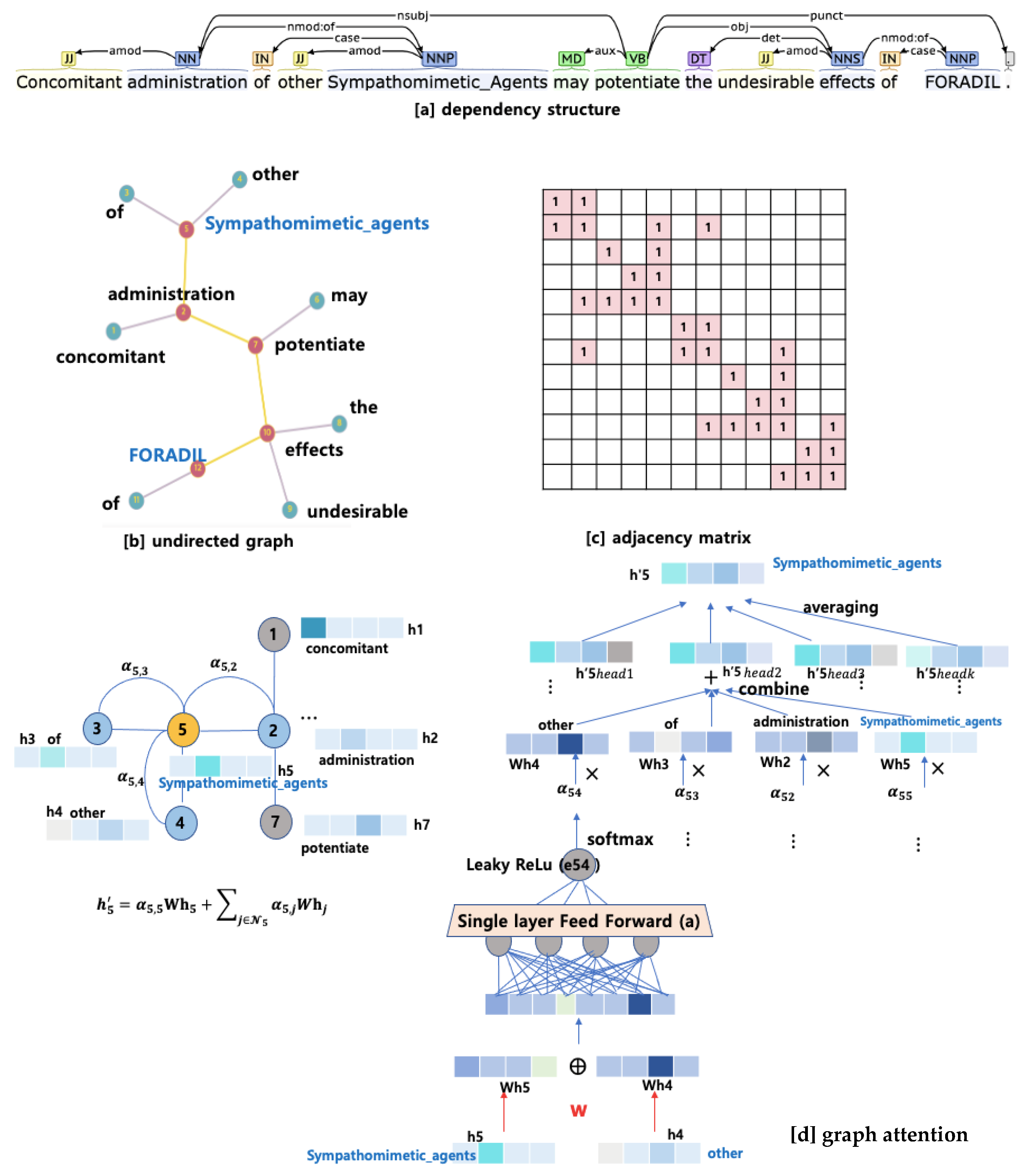
4.2. Self-Attention Using Dependency Graph: BERTGAT
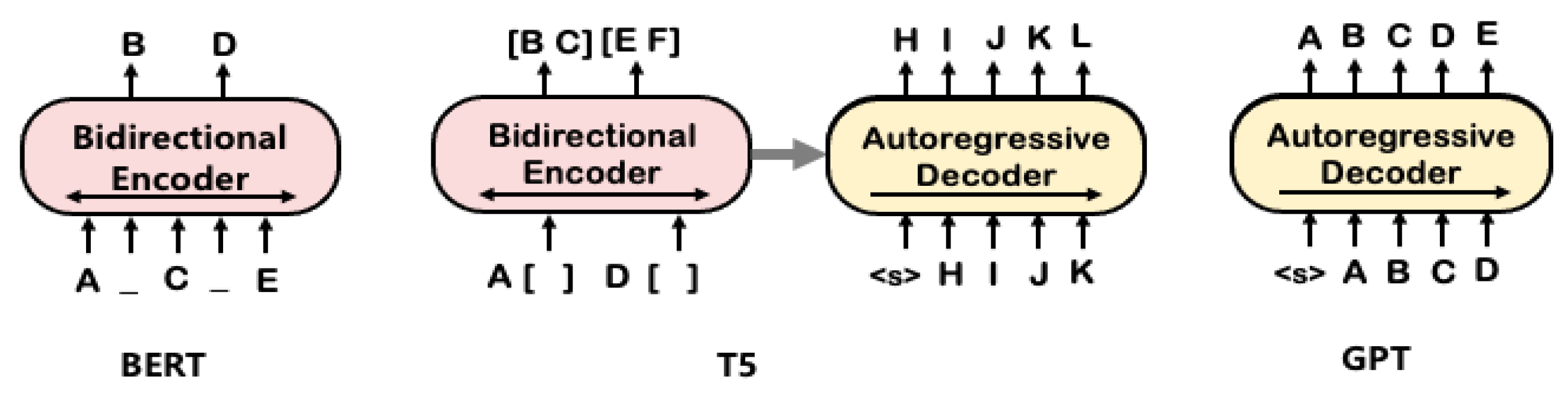
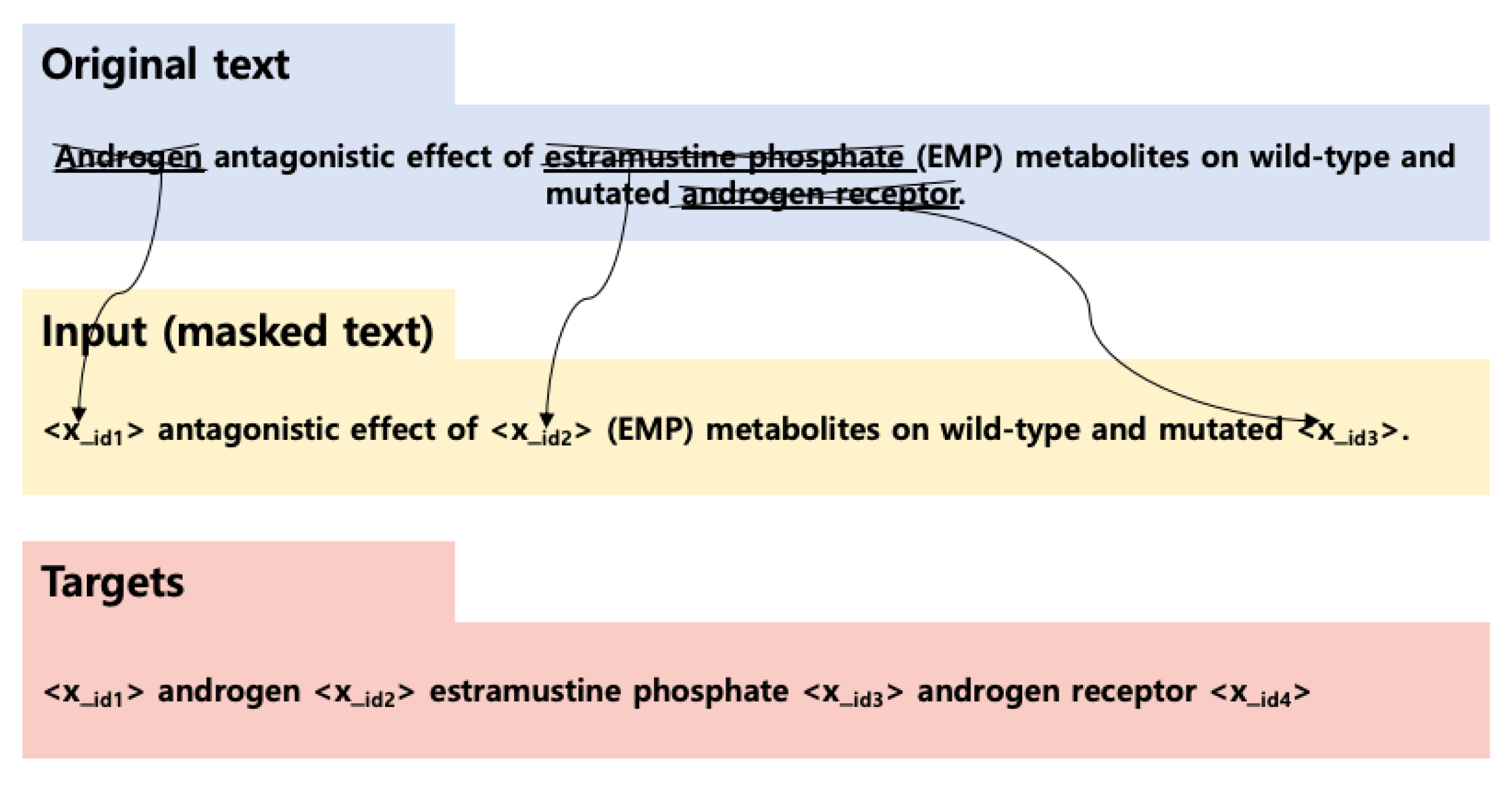
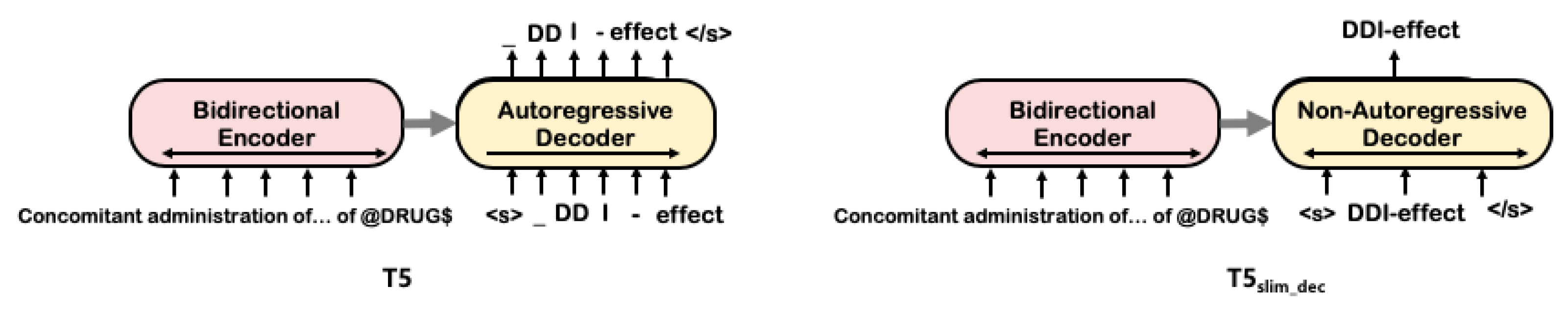
4.3. T5 with Non-Autoregressive Decoder: T5slim_dec
5. Results and Discussion
5.1. Experimental Setup
5.2. Baseline Models
5.3. Results of the Proposed Models
5.4. Comparisons with Other Systems
5.5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Q.; Allot, A.; Lu, Z. Keep up with the latest coronavirus research. Nature 2020, 579, 193. [Google Scholar] [CrossRef] [PubMed]
- PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov (accessed on 28 February 2023).
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 2000. Nucleic Acids Res. 1997, 25, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank. A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Wiegers, T.C.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2023. Nucleic Acids Res. 2023, 51, D1257–D1262. [Google Scholar] [CrossRef] [PubMed]
- Harmar, A.J.; Hills, R.A.; Rosser, E.M.; Jones, M.; Buneman, O.P.; Dunbar, D.R.; Greenhill, S.D.; Hale, V.A.; Sharman, J.L.; Bonner, T.I.; et al. IUPHAR-DB: The IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 2008, 37, D680–D685. [Google Scholar] [CrossRef] [PubMed]
- MEDLINE. Available online: https://www.nlm.nih.gov/medline/index.html (accessed on 28 February 2023).
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the NAACL-HLT 2019, Minneapolis, MN, USA, 2–7 June 2019; pp. 4171–4186. [Google Scholar]
- Brown, T.B.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. In Proceedings of the 34th Conference on Neural Information Processing Systems (NeurIPS 2020), Vancouver, Canada, 6–12 December 2020. [Google Scholar]
- Raffel, C.; Shazeer, N.; Roberts, A.; Lee, K.; Narang, S.; Matena, M.; Zhou, Y.; Li, W.; Liu, P.J. Exploring the Limits of Transfer Learning with a Unified Text-to-Text Transformer. J. Mach. Learn. Res. 2020, 21, 5485–5551. [Google Scholar]
- Krallinger, M. Overview of the Chemical-Protein relation extraction track. In Proceedings of the BioCreative VI Workshop, Bethesda, MD, USA, 20 October 2017. [Google Scholar]
- Segura-Bedmar, I.; Martínez, P.; Herrero-Zazo, M. SemEval-2013 Task 9: Extraction of Drug-Drug Interactions from Biomedical Texts (DDIExtraction 2013). In Proceedings of the Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Seventh International Workshop on Semantic Evaluation (SemEval 2013), Atlanta, GA, USA, 14–15 June 2013; pp. 341–350. [Google Scholar]
- Kanjirangat, V.; Rinaldi, F. Enhancing Biomedical Relation Extraction with Transformer Models using Shortest Dependency Path Features and Triplet Information. J. Biomed. Inform. 2021, 122, 103893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qi, P.; Manning, C.D. Graph Convolution over Pruned Dependency Trees Improves Relation Extraction. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 31 October–4 November 2018; pp. 2205–2215. [Google Scholar]
- Veličković, P.; Casanova, A.; Liò, P.; Cucurull, G.; Romero, A.; Bengio, Y. Graph attention networks. In Proceedings of the 6th International Conference on Learning Representations, Vancouver, Canada, 30 April 2018. [Google Scholar]
- Liu, S.; Tang, B.; Chen, Q.; Wang, X. Drug-Drug Interaction Extraction via Convolutional Neural Networks. Comput. Math. Methods Med. 2016, 2016, 6918381. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.K.; Anand, A. Drug-drug interaction extraction from biomedical texts using long short-term memory network. J. Biomed. Inform. 2018, 86, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. In Proceedings of the 20th China National Conference on Computational Linguistics, Hohhot, China, 13–15 August 2021; pp. 1218–1227. [Google Scholar]
- Song, K.; Tan, X.; Qin, T.; Lu, J.; Liu, T. MASS: Masked Sequence to Sequence Pre-training for Language Generation. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 5926–5936. [Google Scholar]
- Lewis, M.; Liu, Y.; Goyal, N.; Ghazvininejad, M.; Mohamed, A.; Levy, O.; Stoyanov, V.; Zet-tlemoyer, L. BART: Denoising Sequence-to-Sequence Pre-training for Natural Language Generation, Translation, and Comprehension. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 6–8 July 2020; pp. 7871–7880. [Google Scholar]
- Liu, X.; He, P.; Chen, W.; Gao, J. Multi-Task Deep Neural Networks for Natural Language Understanding. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, July 2019; pp. 4487–4496. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Advances in Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Beltagy, I.; Lo, K.; Cohan, A. SCIBERT: A Pretrained Language Model for Scientific Text. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong, China, 3–7 November 2019; pp. 3615–3620. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Yuxian, G.; Robert Tinn, R.; Hao Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. ACM Trans. Comput. Health 2021, 3, 1–23. [Google Scholar]
- Asada, M.; Miwa, M.; Sasaki, Y. Integrating heterogeneous knowledge graphs into drug–drug interaction extraction from the literature. Bioinformatics 2022, 39, btac754. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Liu, Y.; Tan, C.; Huang, S.; Huang, F. Improving Biomedical Pretrained Language Models with Knowledge. In Proceedings of the BioNLP 2021 Workshop, Online, 11 June 2021; pp. 180–190. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, L.; Lu, H.; Zhou, A.; Qin, X. Extracting drug-drug interactions from texts with BioBERT and multiple entity-aware attentions. J. Biomed. Inform. 2020, 106, 103451. [Google Scholar] [CrossRef] [PubMed]
- Yasunaga, M.; Jure Leskovec, J.; Liang, P. LinkBERT: Pretraining Language Models with Document Links. In Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics, Dublin, Ireland, 22–27 May 2022; pp. 8003–8016. [Google Scholar]
- Phan, L.N.; Anibal, J.T.; Tran, H.; Chanana, S.; Bahadıro, E.; Peltekian, A.; Altan-Bonnet, G. SciFive: A text-to-text transformer model for biomedical literature. arXiv 2021, arXiv:2106.03598. [Google Scholar]
- Sarrouti, M.; Tao, C.; Randriamihaja, Y.M. Comparing Encoder-Only and Encoder-Decoder Transformers for Relation Extraction from Biomedical Texts: An Empirical Study on Ten Benchmark Datasets. In Proceedings of the BioNLP 2022 Workshop, Dublin, Ireland, 26 May 2022; pp. 376–382. [Google Scholar] [CrossRef]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural Machine Translation by Jointly Learning to Align and Translate. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Fricke, S. Semantic Scholar. J. Med. Libr. Assoc. 2018, 106, 145–147. [Google Scholar] [CrossRef]
- Kudo, T.; Richardson, J. SentencePiece: A simple and language independent subword tokenizer and detokenizer for Neural Text Processing. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Brussels, Belgium, 2–4 November 2018; pp. 66–77. [Google Scholar]
- Dodge, J.; Sap, M.; Marasović, A.; Agnew, W.; Ilharco, G.; Groeneveld, D.; Mitchell, M.; Gardner, M. Documenting Large Webtext Corpora: A Case Study on the Colossal Clean Crawled Corpus. In Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, Online, 7–11 November 2021; pp. 1286–1305. [Google Scholar]
- Black, S.; Biderman, S.; Hallahan, E.; Anthony, Q.; Leo Gao, L.; Golding, L.; He, H.; Leahy, C.; McDonell, K.; Phang, J.; et al. GPT-NeoX-20B: An Open-Source Autoregressive Language Model. In Proceedings of the ACL Workshop on Challenges & Perspectives in Creating Large Language Models, Dublin, Ireland, 27 May 2022. [Google Scholar]
- Xu, Y.; GeLi, L.; Chen, Y.; Peng, H.; Jin, Z. Classifying relations via long short term memory networks along shortest dependency paths. In Proceedings of the EMNLP, Lisbon, Portugal, 17–21 September 2015. [Google Scholar]
- Marneffe, M.; Manning, C.D. Stanford Typed Dependencies Manual. 2016. Available online: https://downloads.cs.stanford.edu/nlp/software/dependencies_manual.pdf (accessed on 28 February 2023).
- Liu, F.; Huang, T.; Lyu, S.; Shakeri, S.; Yu, H.; Li, J. EncT5: A Framework for Fine-tuning T5 as Non-autoregressive Models. arXiv 2021, arXiv:2110.08426. [Google Scholar]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. HuggingFace’s Transformers: State-of-the-Art Natural Language Processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Online, October 2020; pp. 38–45. [Google Scholar]
- Sun, X.; Dong, K.; Ma, L.; Sutcliffe, R.; He, F.; Chen, S.; Feng, J. Drug-Drug Interaction Extraction via Recurrent Hybrid Convolutional Neural Networks with an Improved Focal Loss. Entropy 2019, 21, 37. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.C.; Zhang, Y.; Bakhturina, E.; Puri, R.; Patwary, M.; Shoeybi, M.; Mani, R. BioMegatron: Larger Biomedical Domain Language Model. In Proceedings of the EMNLP, Online, 16–20 November 2020. [Google Scholar]
- Alrowili, S.; Vijay-Shanker, K. BioM-Transformers: Building Large Biomedical Language Models with BERT, ALBERT and ELECTRA. In Proceedings of the BioNLP 2021 Workshop, Online, 11 June 2021; pp. 221–227. [Google Scholar]

| Class Group | ChemProt Relations | Semantic Meaning |
|---|---|---|
| CPR:0 | UNDEFINED | |
| CPR:1 | PART-OF | Part-of |
| CPR:2 | DIRECT-REGULATOR, INDIRECT-REGULATOR, REGULATOR | Regulator |
| CPR:3 | ACTIVATOR, INDIRECT-UPREGULATOR, UPREGULATOR | Upregulator or activator |
| CPR:4 | DOWNREGULATOR, INDIRECT-DOWNREGULATOR, INHIBITOR | Downregulator or inhibitor |
| CPR:5 | AGONIST, AGONIST-ACTIVATOR, AGONIST-INHIBITOR | Agonist |
| CPR:6 | ANTAGONIST | Antagonist |
| CPR:7 | MODULATOR, MODULATOR-ACTIVATOR, MODULATOR-INHIBITOR | Modulator |
| CPR:8 | COFACTOR | Cofactor |
| CPR:9 | SUBSTRATE, SUBSTRATE_PRODUCT-OF, PRODUCT-OF | Substrate or product-of |
| CPR:10 | NOT | Not |
| Relation Class | Semantic Meaning |
|---|---|
| DDI-Mechanism | a pharmacokinetic interaction mechanism is described in a sentence |
| DDI-Effect | the effect of an interaction is described in a sentence |
| DDI-Advice | a recommendation or advice regarding the concomitant use of two drugs is described in an input sentence |
| DDI-Int | the sentence mentions that interaction occurs and does not provide any detailed information about the interaction |
| DDI-False | non-interacting entities |
| Dataset | CPR:0 | CPR:1 | CPR:2 | CPR:3 | CPR:4 | CPR:5 | CPR:6 | CPR:7 | CPR:8 | CPR:9 | CPR:10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| train | 0 | 550 | 1656 | 784 | 2278 | 173 | 235 | 29 | 34 | 727 | 242 |
| dev | 1 | 328 | 780 | 552 | 1103 | 116 | 199 | 19 | 2 | 457 | 175 |
| test | 2 | 482 | 1743 | 667 | 1667 | 198 | 293 | 25 | 25 | 644 | 267 |
| Corpus | Advice | Effect | Mechanism | Int | False |
|---|---|---|---|---|---|
| train | 826 | 1687 | 1319 | 188 | 15842 |
| test | 218 | 356 | 302 | 96 | 4782 |
| Dataset | Entity Type | Number of Entities |
|---|---|---|
| ChemProt | Protein–Chemical | 10,031 |
| DDI | Drug–Drug | 4920 |
| Method | ChemProt Accuracy (Micro F1-Score) | DDI Accuracy (Micro F1-Score) | |||||
|---|---|---|---|---|---|---|---|
| Entity Masking | Actual Relation Type | Class Group (CPR) | Entity Masking | 4Classes | 5Classes -False | ||
| SCIBERT | 0.8169 | 0.8844 | |||||
| SCIBERT | O | 0.7852 | 0.8764 | O | 0.8703 | 0.9292 | |
| BERTGAT | O | 0.8089 | 0.8812 | ||||
| GPT-Neo125M | 0.7647 | 0.8483 | |||||
| GPT-Neo1.3b | 0.8204 | 0.9010 | 0.8950 | 0.9261 | 0.6711 | ||
| GPT-Neo1.3b | O | 0.8282 | 0.9013 | O | 0.8978 | 0.9314 | 0.7263 |
| T5sciFive | 0.8408 | 0.9100 | 0.8808 | 0.9413 | 0.7268 | ||
| T5sciFive | O | 0.8223 | 0.9022 | O | 0.9031 | 0.9412 | 0.7324 |
| T5slim_dec | 0.8746 | 0.9429 | O | 0.9193 | 0.9533 | 0.7998 | |
| Relation Type | 4Classes | 5Classes | ||||||
|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1-Score | Support | Precision | Recall | F1-Score | Support | |
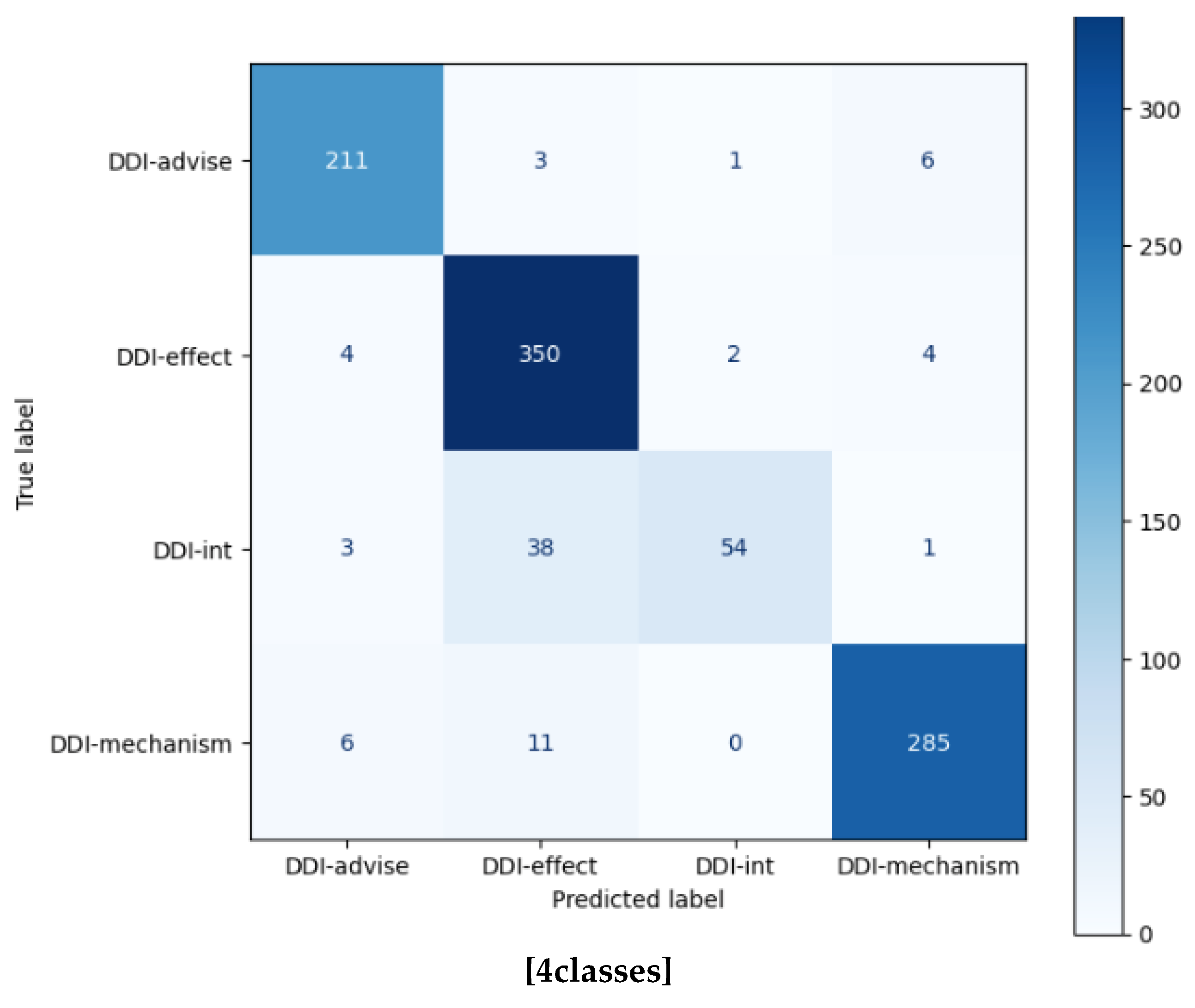
| DDI-advise | 0.9420 | 0.9548 | 0.9483 | 221 | 0.9019 | 0.8733 | 0.8874 | 221 |
| DDI-effect | 0.8706 | 0.9722 | 0.9186 | 360 | 0.7928 | 0.8611 | 0.8256 | 360 |
| DDI-false | 0.9767 | 0.9820 | 0.9794 | 4782 | ||||
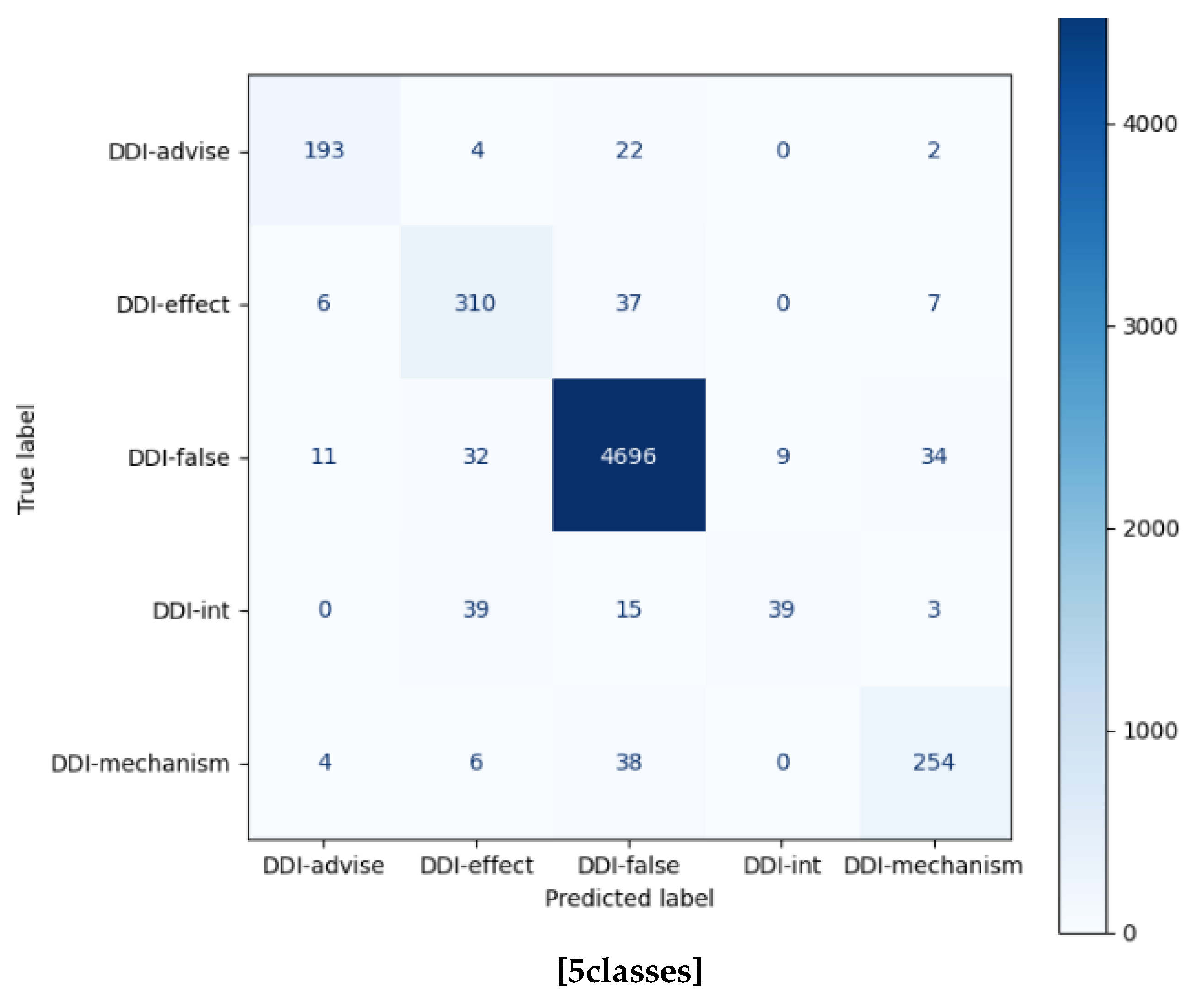
| DDI-int | 0.9474 | 0.5625 | 0.7059 | 96 | 0.8125 | 0.4062 | 0.5417 | 360 |
| DDI-mechanism | 0.9628 | 0.9437 | 0.9532 | 302 | 0.8467 | 0.8411 | 0.8439 | 302 |
| Accuracy | 0.9193 | 979 | 0.9533 | 5761 | ||||
| Macro avg. | 0.9307 | 0.8583 | 0.8815 | 979 | 0.8661 | 0.7927 | 0.8156 | 5761 |
| Weighted avg. | 0.9227 | 0.9193 | 0.9151 | 979 | 0.9528 | 0.9533 | 0.9518 | 5761 |
| Micro avg. | 0.9193 | 0.9193 | 0.9193 | 979 | 0.9533 | 0.9533 | 0.9533 | 5761 |
| Relation Type | Precision | Recall | F1-Score | Support |
|---|---|---|---|---|
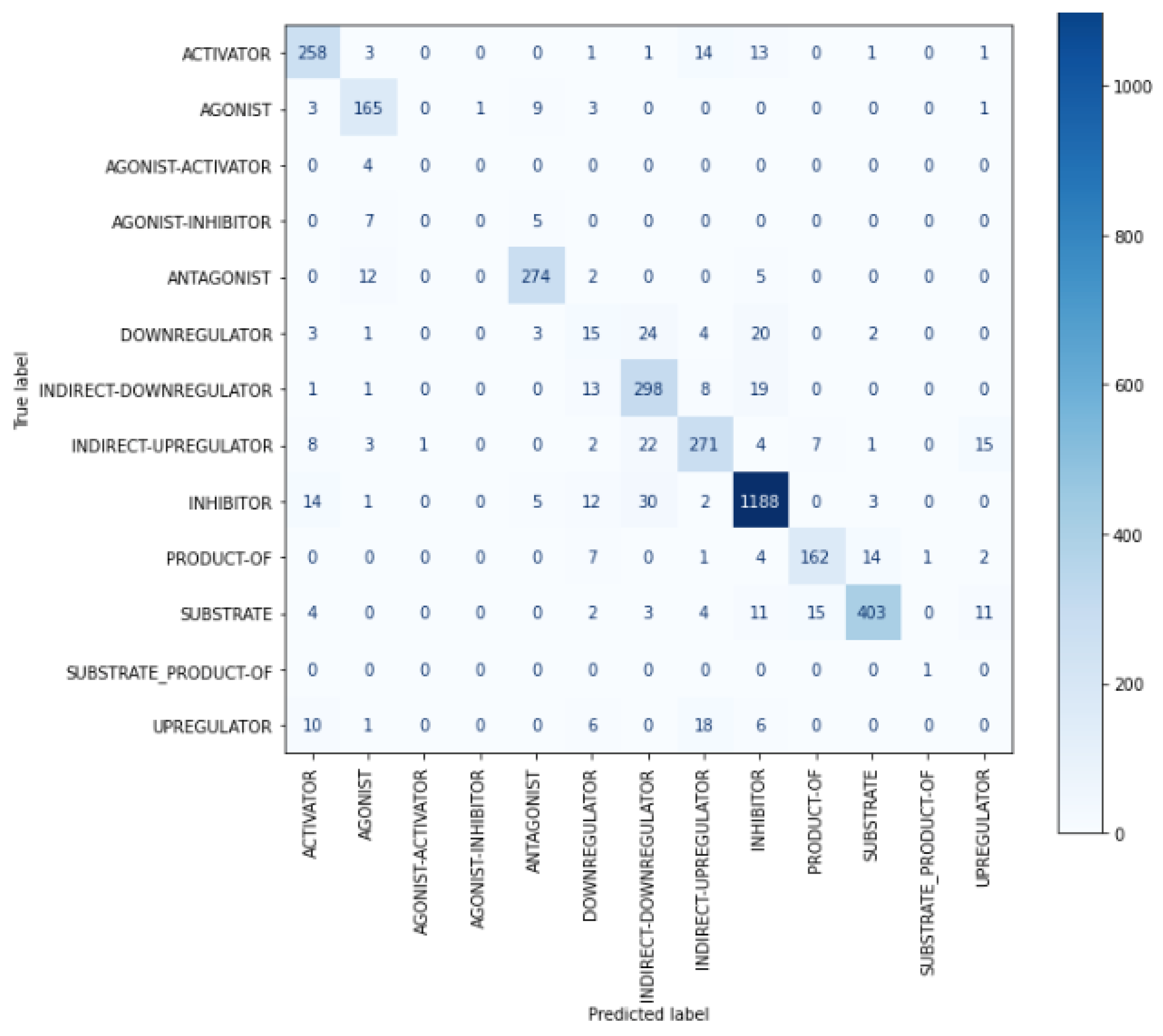
| ACTIVATOR | 0.8571 | 0.8836 | 0.8702 | 292 |
| AGONIST | 0.8333 | 0.9066 | 0.8684 | 182 |
| AGONIST-ACTIVATOR | 0 | 0 | 0 | 4 |
| AGONIST-INHIBITOR | 0 | 0 | 0 | 12 |
| ANTAGONIST | 0.9257 | 0.9352 | 0.9304 | 293 |
| DOWNREGULATOR | 0.2381 | 0.2083 | 0.2222 | 72 |
| INDIRECT-DOWNREGULATOR | 0.7884 | 0.8765 | 0.8301 | 340 |
| INDIRECT-UPREGULATOR | 0.8416 | 0.8114 | 0.8262 | 334 |
| INHIBITOR | 0.9354 | 0.9466 | 0.941 | 1255 |
| PRODUCT-OF | 0.8804 | 0.8482 | 0.864 | 191 |
| SUBSTRATE | 0.9505 | 0.8896 | 0.919 | 453 |
| SUBSTRATE_PRODUCT-OF | 0.5 | 1 | 0.6667 | 1 |
| UPREGULATOR | 0 | 0 | 0 | 41 |
| Accuracy | 0.8746 | 3470 | ||
| Macro avg. | 0.5961 | 0.6389 | 0.6106 | 3470 |
| Weighted avg. | 0.8682 | 0.8746 | 0.8709 | 3470 |
| Micro avg. | 0.8746 | 0.8746 | 0.8746 | 3470 |
| Interaction Type | T5slim_dec (4Classes) | T5slim_dec (5Classes) | Zhu et al. [28] | Sun et al. [41] |
|---|---|---|---|---|
| DDI-advise | 0.9483 | 0.8874 | 0.860 | 0.805 |
| DDI-effect | 0.9186 | 0.8256 | 0.801 | 0.734 |
| DDI-int | 0.7059 | 0.5417 | 0.566 | 0.589 |
| DDI-mechanism | 0.9532 | 0.8439 | 0.846 | 0.782 |
| Interaction Type | T5slim_dec F1-Score | Asada et al. F1-Score [26] |
|---|---|---|
| ACTIVATOR | 0.8702 | 0.771 |
| AGONIST | 0.8684 | 0.790 |
| AGONIST-ACTIVATOR | 0 | 0 |
| AGONIST-INHIBITOR | 0 | 0 |
| ANTAGONIST | 0.9304 | 0.919 |
| DOWNREGULATOR | 0.2222 | ? |
| INDIRECT-DOWNREGULATOR | 0.8301 | 0.779 |
| INDIRECT-UPREGULATOR | 0.8262 | 0.752 |
| INHIBITOR | 0.941 | 0.853 |
| PRODUCT-OF | 0.864 | 0.669 |
| SUBSTRATE | 0.919 | 0.708 |
| SUBSTRATE_PRODUCT-OF | 0.6667 | 0 |
| UPREGULATOR | 0 | ? |
| Method | Accuracy (Micro F1-Score) DDI | Accuracy (Micro F1-Score) ChemProt | ||
|---|---|---|---|---|
| Our Experiment | Our Experiment | |||
| CNN (Liu et al., 2016) [16] | 0.6701 | |||
| BiLSTM (Sahu and Anand, 2018) [17] | 0.6939 | |||
| BioBERT (Lee et al., 2019) [24] | 0.7646 | |||
| SCIBERT (Beltagy et al., 2019) [23] | 0.8364 | 0.8169 | ||
| BioMegatron (Shin et al., 2020) [42] | 0.77 | |||
| KeBioLM (Yuan et al., 2021) [27] | 0.8190 | 0.775 | ||
| PubMedBERT(Gu et al., 2021) [25] | 0.8236 | 0.7724 | ||
| SciFive (Phan et al., 2021) [30] | 0.8367 | 0.90314classes | 0.8895 | 0.9100CPR |
| BioM-BERT (Alrowili et al., 2021) [43] | 0.80 | |||
| BioLinkBERT (Yasunaga et al., 2022) [29] | 0.8335 | 0.7998 | ||
| PubMedBERT+HKG (Asada et al., 2022) [26] | 0.8540 | |||
| BioBERT+multi entity-aware attention (Zhu et al.) [28] | 0.8090 | |||
| Our Method (T5slim_dec) | 0.95335classes 0.91154classes | 0.8746 0.9429CPR | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Yoon, J.; Kwon, O. Biomedical Relation Extraction Using Dependency Graph and Decoder-Enhanced Transformer Model. Bioengineering 2023, 10, 586. https://doi.org/10.3390/bioengineering10050586
Kim S, Yoon J, Kwon O. Biomedical Relation Extraction Using Dependency Graph and Decoder-Enhanced Transformer Model. Bioengineering. 2023; 10(5):586. https://doi.org/10.3390/bioengineering10050586
Chicago/Turabian StyleKim, Seonho, Juntae Yoon, and Ohyoung Kwon. 2023. "Biomedical Relation Extraction Using Dependency Graph and Decoder-Enhanced Transformer Model" Bioengineering 10, no. 5: 586. https://doi.org/10.3390/bioengineering10050586
APA StyleKim, S., Yoon, J., & Kwon, O. (2023). Biomedical Relation Extraction Using Dependency Graph and Decoder-Enhanced Transformer Model. Bioengineering, 10(5), 586. https://doi.org/10.3390/bioengineering10050586







