The Feasibility and Performance of Total Hip Replacement Prediction Deep Learning Algorithm with Real World Data
Abstract
1. Introduction
2. Materials and Methods
2.1. The Data Source and Label
2.2. Algorithm Design
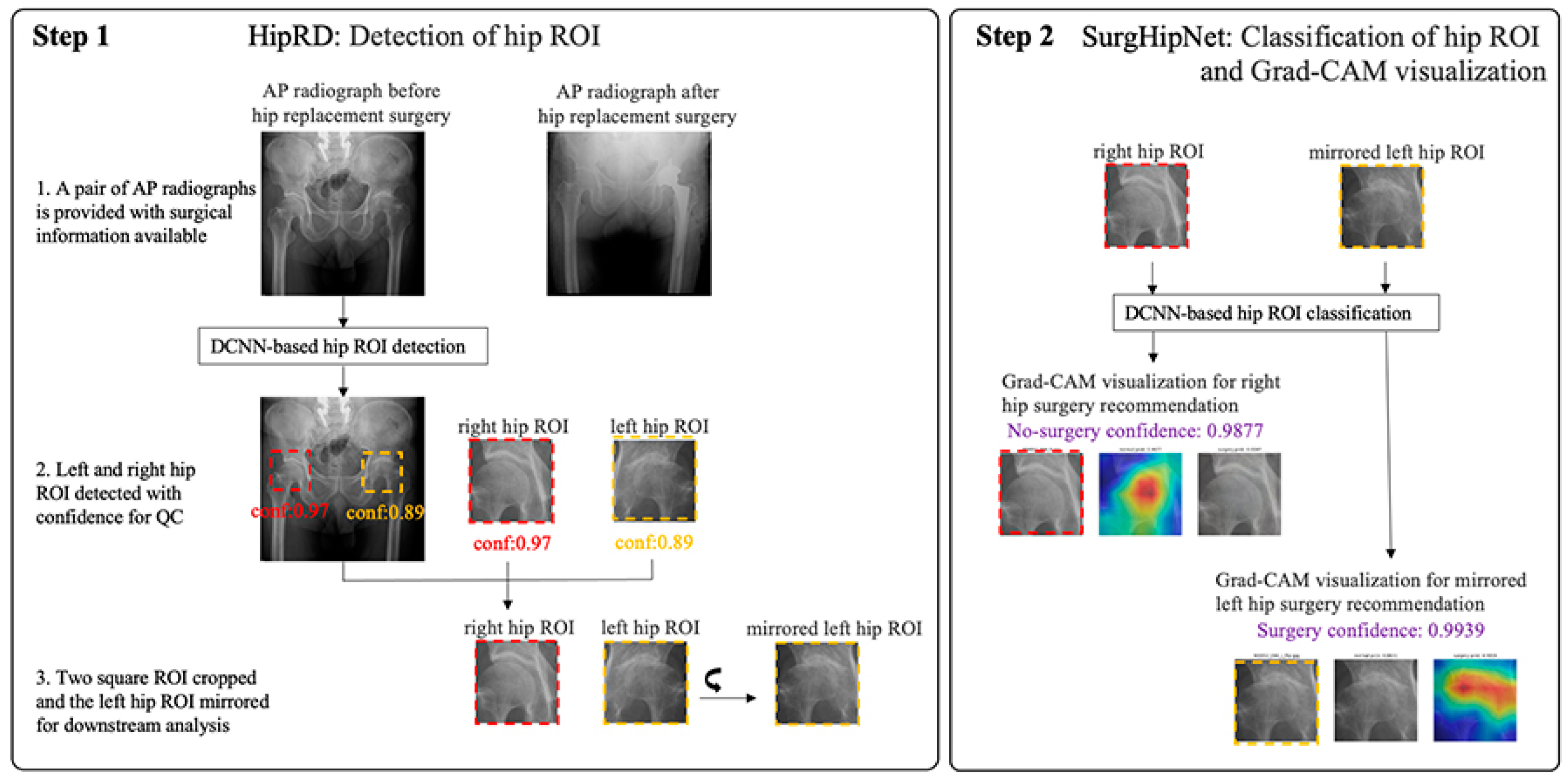
2.3. The Hip Localization
2.4. The Total Hip Replacement Classification and Visualization
2.5. The Real-World Data from 2018 to 2019
2.6. Statistical Analysis and Software
3. Results
3.1. The Patient’s Distribution of Training Dataset for SurgHipNet and the Performance of SurgHipNet in Testing Dataset
3.2. The 2018–2019 RWD Dataset Distribution
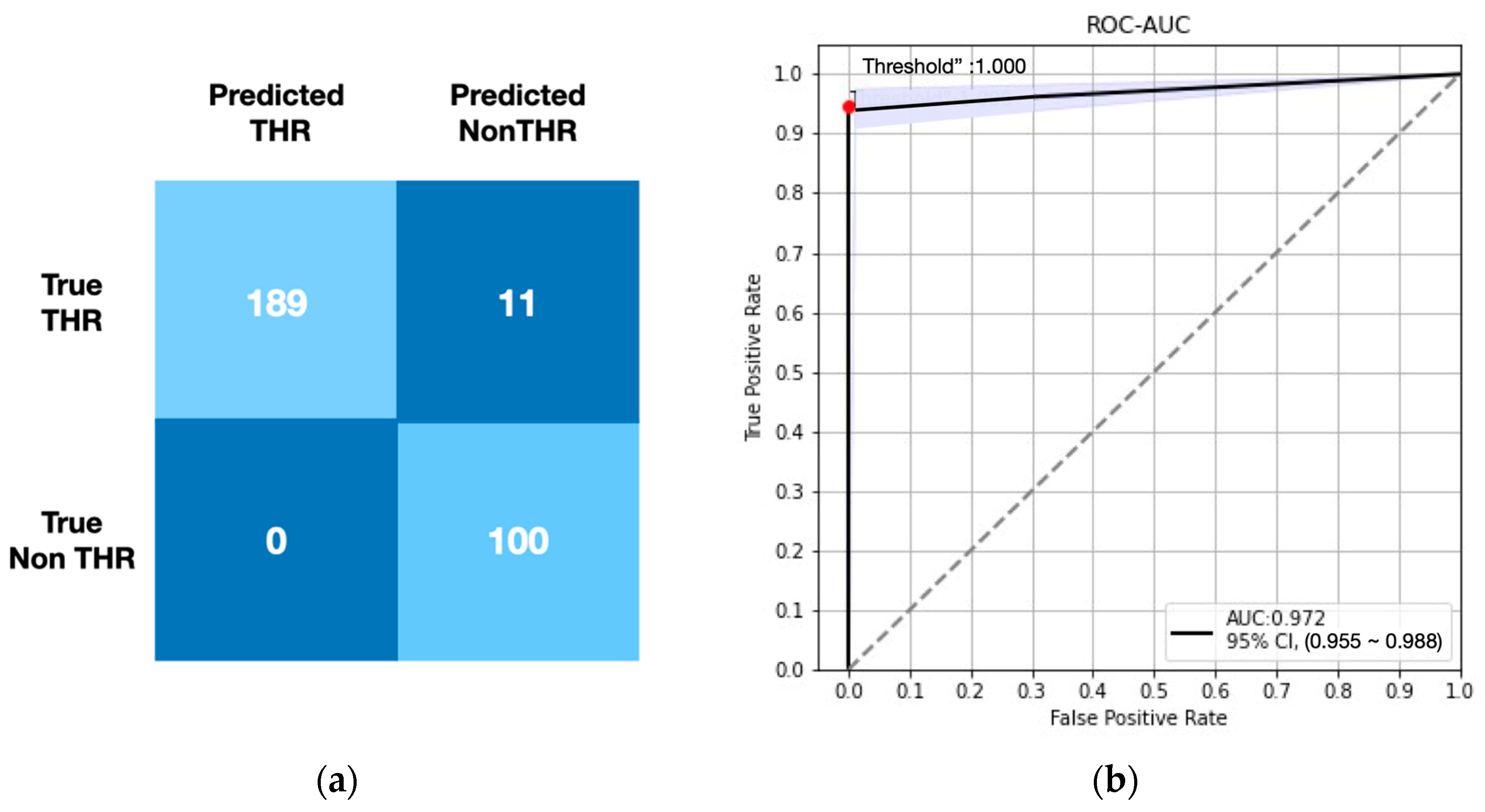
3.3. Performance of SurgHipNet on RWD Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Code Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kujala, U.M.; Kaprio, J.; Sarna, S. Osteoarthritis of Weight Bearing Joints of Lower Limbs in Former élite Male Athletes. BMJ 1994, 308, 231–234. [Google Scholar] [CrossRef]
- Spector, T.D.; Harris, P.A.; Hart, D.J.; Cicuttini, F.M.; Nandra, D.; Etherington, J.; Wolman, R.L.; Doyle, D.V. Risk of Osteoarthritis Associated with Long-Term Weight-Bearing Sports: A Radiologic Survey of the Hips and Knees in Female Ex-Athletes and Population Controls. Arthritis Rheum. 1996, 39, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Sathappan, S.S.; Strauss, E.J.; Ginat, D.; Upasani, V.; Di Cesare, P.E. Surgical Challenges in Complex Primary Total Hip Arthroplasty. Am. J. Orthop. 2007, 36, 534–541. [Google Scholar]
- Nadkarni, G.N.; Patel, A.A.; Ahuja, Y.; Annapureddy, N.; Agarwal, S.K.; Simoes, P.K.; Konstantinidis, I.; Kamat, S.; Archdeacon, M.; Thakar, C.V. Incidence, Risk Factors, and Outcome Trends of Acute Kidney Injury in Elective Total Hip and Knee Arthroplasty. Am. J. Orthop. 2016, 45, E12–E19. [Google Scholar] [PubMed]
- Weinstein, S.L. Natural History and Treatment Outcomes of Childhood Hip Disorders. Clin. Orthop. Relat. Res. 1997, 344, 227–242. [Google Scholar] [CrossRef]
- Schmidt, A.H.; Leighton, R.; Parvizi, J.; Sems, A.; Berry, D.J. Optimal Arthroplasty for Femoral Neck Fractures: Is Total Hip Arthroplasty the Answer? J. Orthop. Trauma 2009, 23, 428–433. [Google Scholar] [CrossRef]
- Wallis, J.A.; Taylor, N.F. Pre-Operative Interventions (non-Surgical and Non-Pharmacological) for Patients with Hip or Knee Osteoarthritis Awaiting Joint Replacement Surgery—A Systematic Review and Meta-Analysis. Osteoarthr. Cartil. 2011, 19, 1381–1395. [Google Scholar] [CrossRef]
- Lee, P.Y.F.; Rozewicz, S.; Othman, A.; Jury, C. Modern Non-Pharmacological and Non-Surgical Treatments for Hip Pain. J. Arthritis 2018, 7, 1–5. [Google Scholar]
- Hill, A.-M.; Ross-Adjie, G.; McPhail, S.M.; Monterosso, L.; Bulsara, M.; Etherton-Beer, C.; Powell, S.-J.; Hardisty, G. Incidence, Risk Factors and the Healthcare Cost of Falls Postdischarge after Elective Total Hip and Total Knee Replacement Surgery: Protocol for a Prospective Observational Cohort Study. BMJ Open 2016, 6, e011139. [Google Scholar] [CrossRef]
- MacLeod, A.M.; Huber, J.P.; Gollish, J.D. Functional Independence Training Program: An Example of a Sub-Acute Care Model for Patients Following Primary Joint Replacement. Healthc. Manag. Forum 1998, 11, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Zajc, J.; Fokter, S.K. Dual-Modular Stems for Primary Total Hip Arthroplasty. Encyclopedia 2022, 2, 893–911. [Google Scholar] [CrossRef]
- Quintana, J.M.; Aróstegui, I.; Azkarate, J.; Goenaga, J.I.; Elexpe, X.; Letona, J.; Arcelay, A. Evaluation of Explicit Criteria for Total Hip Joint Replacement. J. Clin. Epidemiol. 2000, 53, 1200–1208. [Google Scholar] [CrossRef]
- Gademan, M.G.J.; Hofstede, S.N.; Vliet Vlieland, T.P.M.; Nelissen, R.G.H.H.; Marang-van de Mheen, P.J. Indication Criteria for Total Hip or Knee Arthroplasty in Osteoarthritis: A State-of-the-Science Overview. BMC Musculoskelet. Disord. 2016, 17, 463. [Google Scholar] [CrossRef]
- Delaunay, S.; Dussault, R.G.; Kaplan, P.A.; Alford, B.A. Radiographic Measurements of Dysplastic Adult Hips. Skeletal Radiol. 1997, 26, 75–81. [Google Scholar] [CrossRef]
- Katz, J.N.; Arant, K.R.; Loeser, R.F. Diagnosis and Treatment of Hip and Knee Osteoarthritis: A Review. JAMA 2021, 325, 568–578. [Google Scholar] [CrossRef]
- Cheng, C.-T.; Wang, Y.; Chen, H.-W.; Hsiao, P.-M.; Yeh, C.-N.; Hsieh, C.-H.; Miao, S.; Xiao, J.; Liao, C.-H.; Lu, L. A Scalable Physician-Level Deep Learning Algorithm Detects Universal Trauma on Pelvic Radiographs. Nat. Commun. 2021, 12, 1066. [Google Scholar] [CrossRef]
- Asiri, N.; Hussain, M.; Al Adel, F.; Alzaidi, N. Deep Learning Based Computer-Aided Diagnosis Systems for Diabetic Retinopathy: A Survey. Artif. Intell. Med. 2019, 99, 101701. [Google Scholar] [CrossRef]
- Choi, J.; Hui, J.Z.; Spain, D.; Su, Y.-S.; Cheng, C.-T.; Liao, C.-H. Practical Computer Vision Application to Detect Hip Fractures on Pelvic X-Rays: A Bi-Institutional Study. Trauma Surg. Acute Care Open 2021, 6, e000705. [Google Scholar] [CrossRef]
- Gassenmaier, S.; Afat, S.; Nickel, D.; Mostapha, M.; Herrmann, J.; Othman, A.E. Deep Learning–accelerated T2-Weighted Imaging of the Prostate: Reduction of Acquisition Time and Improvement of Image Quality. Eur. J. Radiol. 2021, 137, 109600. [Google Scholar] [CrossRef]
- Ichikawa, S.; Sugimori, H.; Ichijiri, K.; Yoshimura, T.; Nagaki, A. Acquisition Time Reduction in Pediatric 99mTc-DMSA Planar Imaging Using Deep Learning. J. Appl. Clin. Med. Phys. 2023, e13978. [Google Scholar] [CrossRef]
- Litjens, G.; Ciompi, F.; Wolterink, J.M.; de Vos, B.D.; Leiner, T.; Teuwen, J.; Išgum, I. State-of-the-Art Deep Learning in Cardiovascular Image Analysis. JACC Cardiovasc. Imaging 2019, 12, 1549–1565. [Google Scholar] [CrossRef] [PubMed]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes van Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.W.M.; The CAMELYON16 Consortium; Hermsen, M.; Manson, Q.F.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sha, L.; Lakin, J.R.; Bynum, J.; Bates, D.W.; Hong, P.; Zhou, L. Development and Validation of a Deep Learning Algorithm for Mortality Prediction in Selecting Patients With Dementia for Earlier Palliative Care Interventions. JAMA Netw. Open 2019, 2, e196972. [Google Scholar] [CrossRef]
- Kolossváry, M.; Raghu, V.K.; Nagurney, J.T.; Hoffmann, U.; Lu, M.T. Deep Learning Analysis of Chest Radiographs to Triage Patients with Acute Chest Pain Syndrome. Radiology 2023, 306, e221926. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, B.; Rouzrokh, P.; Faghani, S.; Moassefi, M.; Vahdati, S.; Mahmoudi, E.; Chalian, H.; Erickson, B.J. Machine Learning and Deep Learning in Cardiothoracic Imaging: A Scoping Review. Diagnostics 2022, 12, 2512. [Google Scholar] [CrossRef]
- Chea, P.; Mandell, J.C. Current Applications and Future Directions of Deep Learning in Musculoskeletal Radiology. Skelet. Radiol. 2020, 49, 183–197. [Google Scholar] [CrossRef]
- Jones, R.M.; Sharma, A.; Hotchkiss, R.; Sperling, J.W.; Hamburger, J.; Ledig, C.; O’Toole, R.; Gardner, M.; Venkatesh, S.; Roberts, M.M.; et al. Assessment of a Deep-Learning System for Fracture Detection in Musculoskeletal Radiographs. NPJ Digit. Med. 2020, 3, 144. [Google Scholar] [CrossRef]
- Harini, N.; Ramji, B.; Sriram, S.; Sowmya, V.; Soman, K.P. Musculoskeletal Radiographs Classification Using Deep Learning. In Deep Learning for Data Analytics; Elsevier: Amsterdam, The Netherlands, 2020; pp. 79–98. [Google Scholar]
- Tanzi, L.; Vezzetti, E.; Moreno, R.; Moos, S. X-Ray Bone Fracture Classification Using Deep Learning: A Baseline for Designing a Reliable Approach. NATO Adv. Sci. Inst. Ser. E Appl. Sci. 2020, 10, 1507. [Google Scholar] [CrossRef]
- Wang, C.-W.; Khalil, M.-A.; Firdi, N.P. A Survey on Deep Learning for Precision Oncology. Diagnostics 2022, 12, 1489. [Google Scholar] [CrossRef]
- Alsentzer, E.; Li, M.M.; Kobren, S.N.; Kohane, I.S.; Zitnik, M. Undiagnosed Diseases Network Deep Learning for Diagnosing Patients with Rare Genetic Diseases. medRxiv 2022. [Google Scholar] [CrossRef]
- Hirschmann, A.; Cyriac, J.; Stieltjes, B.; Kober, T.; Richiardi, J.; Omoumi, P. Artificial Intelligence in Musculoskeletal Imaging: Review of Current Literature, Challenges, and Trends. Semin. Musculoskelet. Radiol. 2019, 23, 304–311. [Google Scholar] [CrossRef]
- Mahendraratnam, N.; Mercon, K.; Gill, M.; Benzing, L.; McClellan, M.B. Understanding Use of Real-World Data and Real-World Evidence to Support Regulatory Decisions on Medical Product Effectiveness. Clin. Pharmacol. Ther. 2022, 111, 150–154. [Google Scholar] [CrossRef]
- Arlett, P.; Kjaer, J.; Broich, K.; Cooke, E. Real-World Evidence in EU Medicines Regulation: Enabling Use and Establishing Value. Clin. Pharmacol. Ther. 2022, 111, 21–23. [Google Scholar] [CrossRef]
- Chen, D.; Liu, S.; Kingsbury, P.; Sohn, S.; Storlie, C.B.; Habermann, E.B.; Naessens, J.M.; Larson, D.W.; Liu, H. Deep Learning and Alternative Learning Strategies for Retrospective Real-World Clinical Data. NPJ Digit. Med. 2019, 2, 43. [Google Scholar] [CrossRef]
- Food and Drug Administration. In Proceedings of the Others Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD), Washington, DC, USA, 3 June 2019. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device (accessed on 6 February 2023).
- Brown, N.A.; Carey, C.H.; Gerry, E.I. FDA Releases Action Plan for Artificial Intelligence/Machine Learning-Enabled Software as a Medical Device. J. Robot. Artif. Intell. Law 2021, 4, 255–260. [Google Scholar]
- Bibault, J.-E.; Burgun, A.; Fournier, L.; Dekker, A.; Lambin, P. Chapter 18—Artificial Intelligence in Oncology. In Artificial Intelligence in Medicine; Xing, L., Giger, M.L., Min, J.K., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 361–381. ISBN 9780128212592. [Google Scholar]
- Liu, F.; Demosthenes, P. Real-World Data: A Brief Review of the Methods, Applications, Challenges and Opportunities. BMC Med. Res. Methodol. 2022, 22, 287. [Google Scholar] [CrossRef]
- Chodankar, D. Introduction to Real-World Evidence Studies. Perspect. Clin. Res. 2021, 12, 171–174. [Google Scholar] [CrossRef]
- von Schacky, C.E.; Sohn, J.H.; Liu, F.; Ozhinsky, E.; Jungmann, P.M.; Nardo, L.; Posadzy, M.; Foreman, S.C.; Nevitt, M.C.; Link, T.M.; et al. Development and Validation of a Multitask Deep Learning Model for Severity Grading of Hip Osteoarthritis Features on Radiographs. Radiology 2020, 295, 136–145. [Google Scholar] [CrossRef]
- Xue, Y.; Zhang, R.; Deng, Y.; Chen, K.; Jiang, T. A Preliminary Examination of the Diagnostic Value of Deep Learning in Hip Osteoarthritis. PLoS ONE 2017, 12, e0178992. [Google Scholar] [CrossRef]
- Tiulpin, A.; Thevenot, J.; Rahtu, E.; Lehenkari, P.; Saarakkala, S. Automatic Knee Osteoarthritis Diagnosis from Plain Radiographs: A Deep Learning-Based Approach. Sci. Rep. 2018, 8, 1727. [Google Scholar] [CrossRef]
- Üreten, K.; Arslan, T.; Gültekin, K.E.; Demir, A.N.D.; Özer, H.F.; Bilgili, Y. Detection of Hip Osteoarthritis by Using Plain Pelvic Radiographs with Deep Learning Methods. Skelet. Radiol. 2020, 49, 1369–1374. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.; Zhang, B.; Tan, J.; Shen, Y.; Geras, K.J.; Babb, J.S.; Cho, K.; Chang, G.; Deniz, C.M. Prediction of Total Knee Replacement and Diagnosis of Osteoarthritis by Using Deep Learning on Knee Radiographs: Data from the Osteoarthritis Initiative. Radiology 2020, 296, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Borjali, A.; Chen, A.; Muratoglu, O.; Varadarajan, K.M. Detecting Mechanical Loosening of Total Hip Arthroplasty Using Deep Convolutional Neural Network. In Orthopaedic Proceedings; The British Editorial Society of Bone & Joint Surgery: London, UK, 2020; Volume 102, p. 133. [Google Scholar]
- Liu, F.-Y.; Chen, C.-C.; Cheng, C.-T.; Wu, C.-T.; Hsu, C.-P.; Fu, C.-Y.; Chen, S.-C.; Liao, C.-H.; Lee, M.S. Automatic Hip Detection in Anteroposterior Pelvic Radiographs—A Labelless Practical Framework. J. Personalized Med. 2021, 11, 522. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Wu, C.-T.; Chung, C.-Y.; Chen, S.-C.; Lee, M.S.; Cheng, C.-T.; Liao, C.-H. Prediction of Total Hip Replacement by Using Deep Learning Algorithm on Plain Pelvic Radiographs: A Diagnostic Study. JMIR Prepr. 2023; submitted. [Google Scholar]
- Buitinck, L.; Louppe, G.; Blondel, M.; Pedregosa, F.; Mueller, A.; Grisel, O.; Niculae, V.; Prettenhofer, P.; Gramfort, A.; Grobler, J.; et al. API Design for Machine Learning Software: Experiences from the Scikit-Learn Project. arXiv 2013, arXiv:1309.0238. [Google Scholar] [CrossRef]
- Croft, P.; Cooper, C.; Wickham, C.; Coggon, D. Defining Osteoarthritis of the Hip for Epidemiologic Studies. Am. J. Epidemiol. 1990, 132, 514–522. [Google Scholar] [CrossRef]
- Steinberg, M.E.; Hayken, G.D.; Steinberg, D.R. A Quantitative System for Staging Avascular Necrosis. J. Bone Jt. Surg. Br. 1995, 77, 34–41. [Google Scholar] [CrossRef]
- Cheng, C.-T.; Ho, T.-Y.; Lee, T.-Y.; Chang, C.-C.; Chou, C.-C.; Chen, C.-C.; Chung, I.-F.; Liao, C.-H. Application of a Deep Learning Algorithm for Detection and Visualization of Hip Fractures on Plain Pelvic Radiographs. Eur. Radiol. 2019, 29, 5469–5477. [Google Scholar] [CrossRef]
- Sherman, R.E.; Anderson, S.A.; Dal Pan, G.J.; Gray, G.W.; Gross, T.; Hunter, N.L.; LaVange, L.; Marinac-Dabic, D.; Marks, P.W.; Robb, M.A.; et al. Real-World Evidence—What Is It and What Can It Tell Us. N. Engl. J. Med. 2016, 375, 2293–2297. [Google Scholar] [CrossRef]
- Wu, J.; Wang, C.; Toh, S.; Pisa, F.E.; Bauer, L. Use of Real-World Evidence in Regulatory Decisions for Rare Diseases in the United States-Current Status and Future Directions. Pharmacoepidemiol. Drug Saf. 2020, 29, 1213–1218. [Google Scholar] [CrossRef]
- Anthimopoulos, M.; Christodoulidis, S.; Ebner, L.; Christe, A.; Mougiakakou, S. Lung Pattern Classification for Interstitial Lung Diseases Using a Deep Convolutional Neural Network. IEEE Trans. Med. Imaging 2016, 35, 1207–1216. [Google Scholar] [CrossRef]
- Kleesiek, J.; Urban, G.; Hubert, A.; Schwarz, D.; Maier-Hein, K.; Bendszus, M.; Biller, A. Deep MRI Brain Extraction: A 3D Convolutional Neural Network for Skull Stripping. Neuroimage 2016, 129, 460–469. [Google Scholar] [CrossRef]
- Johansson, F.D.; Collins, J.E.; Yau, V.; Guan, H.; Kim, S.C.; Losina, E.; Sontag, D.; Stratton, J.; Trinh, H.; Greenberg, J.; et al. Predicting Response to Tocilizumab Monotherapy in Rheumatoid Arthritis: A Real-World Data Analysis Using Machine Learning. J. Rheumatol. 2021, 48, 1364–1370. [Google Scholar] [CrossRef]
- Dreyer, N.A.; Garner, S. Registries for Robust Evidence. JAMA 2009, 302, 790–791. [Google Scholar] [CrossRef]
- Izmirly, P.M.; Parton, H.; Wang, L.; McCune, W.J.; Lim, S.S.; Drenkard, C.; Ferucci, E.D.; Dall’Era, M.; Gordon, C.; Helmick, C.G.; et al. Prevalence of Systemic Lupus Erythematosus in the United States: Estimates From a Meta-Analysis of the Centers for Disease Control and Prevention National Lupus Registries. Arthritis Rheumatol. 2021, 73, 991–996. [Google Scholar] [CrossRef]
- Lacaze, P.; Millis, N.; Fookes, M.; Zurynski, Y.; Jaffe, A.; Bellgard, M.; Winship, I.; McNeil, J.; Bittles, A.H. Rare Disease Registries: A Call to Action. Intern. Med. J. 2017, 47, 1075–1079. [Google Scholar] [CrossRef]
- Gerke, S.; Babic, B.; Evgeniou, T.; Cohen, I.G. The Need for a System View to Regulate Artificial Intelligence/machine Learning-Based Software as Medical Device. NPJ Digit. Med. 2020, 3, 53. [Google Scholar] [CrossRef]
- Suzuki, K. Overview of Deep Learning in Medical Imaging. Radiol. Phys. Technol. 2017, 10, 257–273. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Oh, Y.; Park, S.; Ye, J.C. Deep Learning COVID-19 Features on CXR Using Limited Training Data Sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of Deep Learning Technique to Manage COVID-19 in Routine Clinical Practice Using CT Images: Results of 10 Convolutional Neural Networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef] [PubMed]
- Panwar, H.; Gupta, P.K.; Siddiqui, M.K.; Morales-Menendez, R.; Bhardwaj, P.; Singh, V. A Deep Learning and Grad-CAM Based Color Visualization Approach for Fast Detection of COVID-19 Cases Using Chest X-Ray and CT-Scan Images. Chaos Solitons Fractals 2020, 140, 110190. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Zhang, L.; Shi, W.; Liu, J.; He, W.; Jiang, Z. A Visualization Method Based on the Grad-CAM for Medical Image Segmentation Model. In Proceedings of the 2021 International Conference on Electronic Information Engineering and Computer Science (EIECS), Changchun, China, 23–26 September 2021; pp. 242–247. [Google Scholar]
| TP | TN | FP | FN | ACC | Sn | Sp | NPV | F1 | AUC (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|---|
| 2008–2017 Test data | 92 | 372 | 3 | 8 | 0.977 | 0.920 | 0.992 | 0.979 | 0.944 | 0.994 (0.990–0.998) |
| 2018–2019 RWD data | 189 | 100 | 11 | 0 | 0.972 | 0.945 | 1.000 | 0.900 | 0.972 | 0.972 (0.955–0.988) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.-C.; Huang, J.-F.; Lin, W.-C.; Cheng, C.-T.; Chen, S.-C.; Fu, C.-Y.; Lee, M.S.; Liao, C.-H.; Chung, C.-Y. The Feasibility and Performance of Total Hip Replacement Prediction Deep Learning Algorithm with Real World Data. Bioengineering 2023, 10, 458. https://doi.org/10.3390/bioengineering10040458
Chen C-C, Huang J-F, Lin W-C, Cheng C-T, Chen S-C, Fu C-Y, Lee MS, Liao C-H, Chung C-Y. The Feasibility and Performance of Total Hip Replacement Prediction Deep Learning Algorithm with Real World Data. Bioengineering. 2023; 10(4):458. https://doi.org/10.3390/bioengineering10040458
Chicago/Turabian StyleChen, Chih-Chi, Jen-Fu Huang, Wei-Cheng Lin, Chi-Tung Cheng, Shann-Ching Chen, Chih-Yuan Fu, Mel S. Lee, Chien-Hung Liao, and Chia-Ying Chung. 2023. "The Feasibility and Performance of Total Hip Replacement Prediction Deep Learning Algorithm with Real World Data" Bioengineering 10, no. 4: 458. https://doi.org/10.3390/bioengineering10040458
APA StyleChen, C.-C., Huang, J.-F., Lin, W.-C., Cheng, C.-T., Chen, S.-C., Fu, C.-Y., Lee, M. S., Liao, C.-H., & Chung, C.-Y. (2023). The Feasibility and Performance of Total Hip Replacement Prediction Deep Learning Algorithm with Real World Data. Bioengineering, 10(4), 458. https://doi.org/10.3390/bioengineering10040458









