Human-Derived Cortical Neurospheroids Coupled to Passive, High-Density and 3D MEAs: A Valid Platform for Functional Tests
Abstract
1. Introduction
2. Material and Methods
2.1. Human-Induced Pluripotent Stem Cells Generation and Maintenance
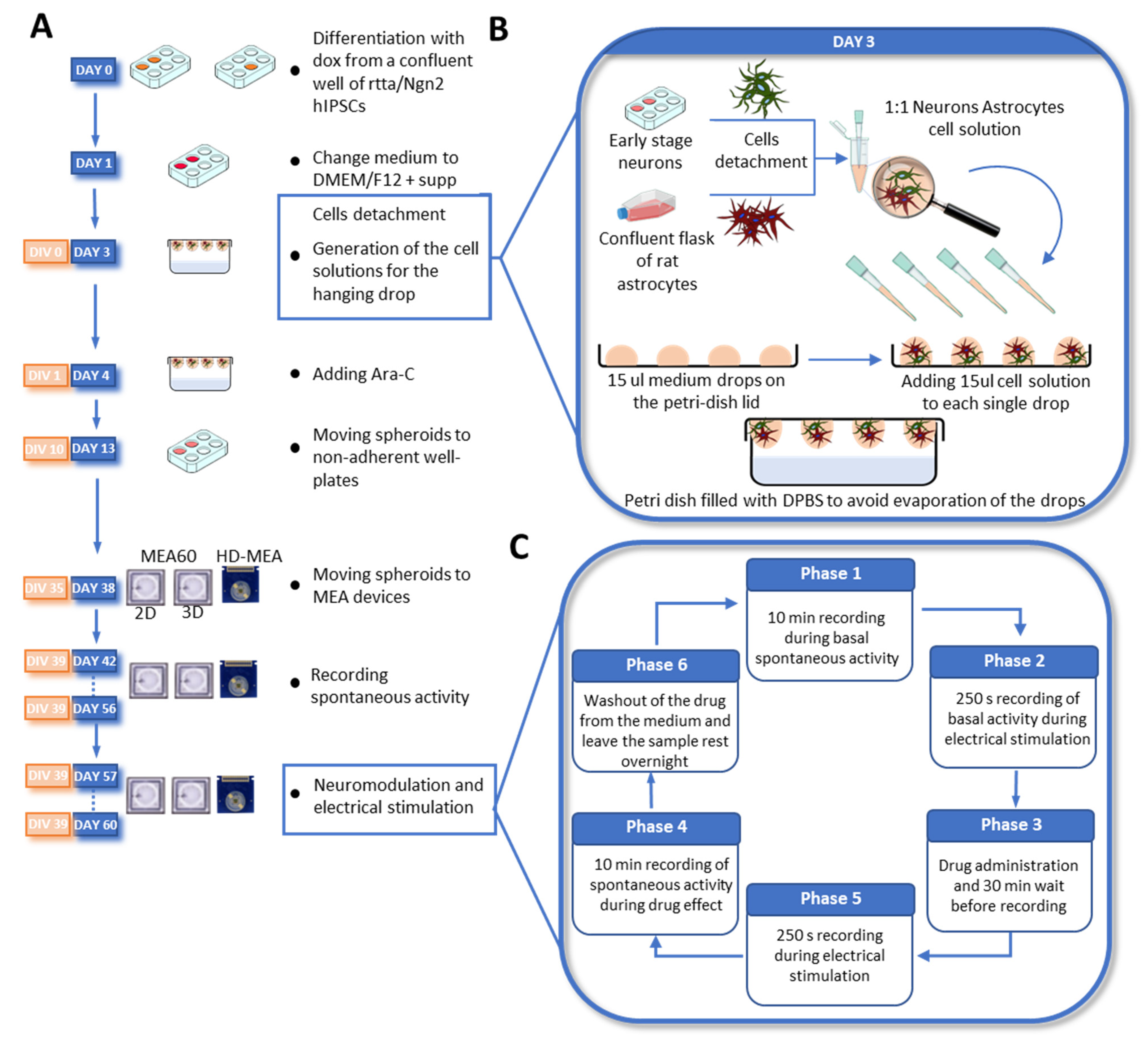
2.2. Neuronal Differentiation and Neurospheroids Generation
2.3. MEAs Devices
2.4. Data Acquisition and Analysis
2.5. Electrical Stimulation and Neuromodulation
2.6. Immunofluorescence
2.7. Statistical Analysis
3. Results
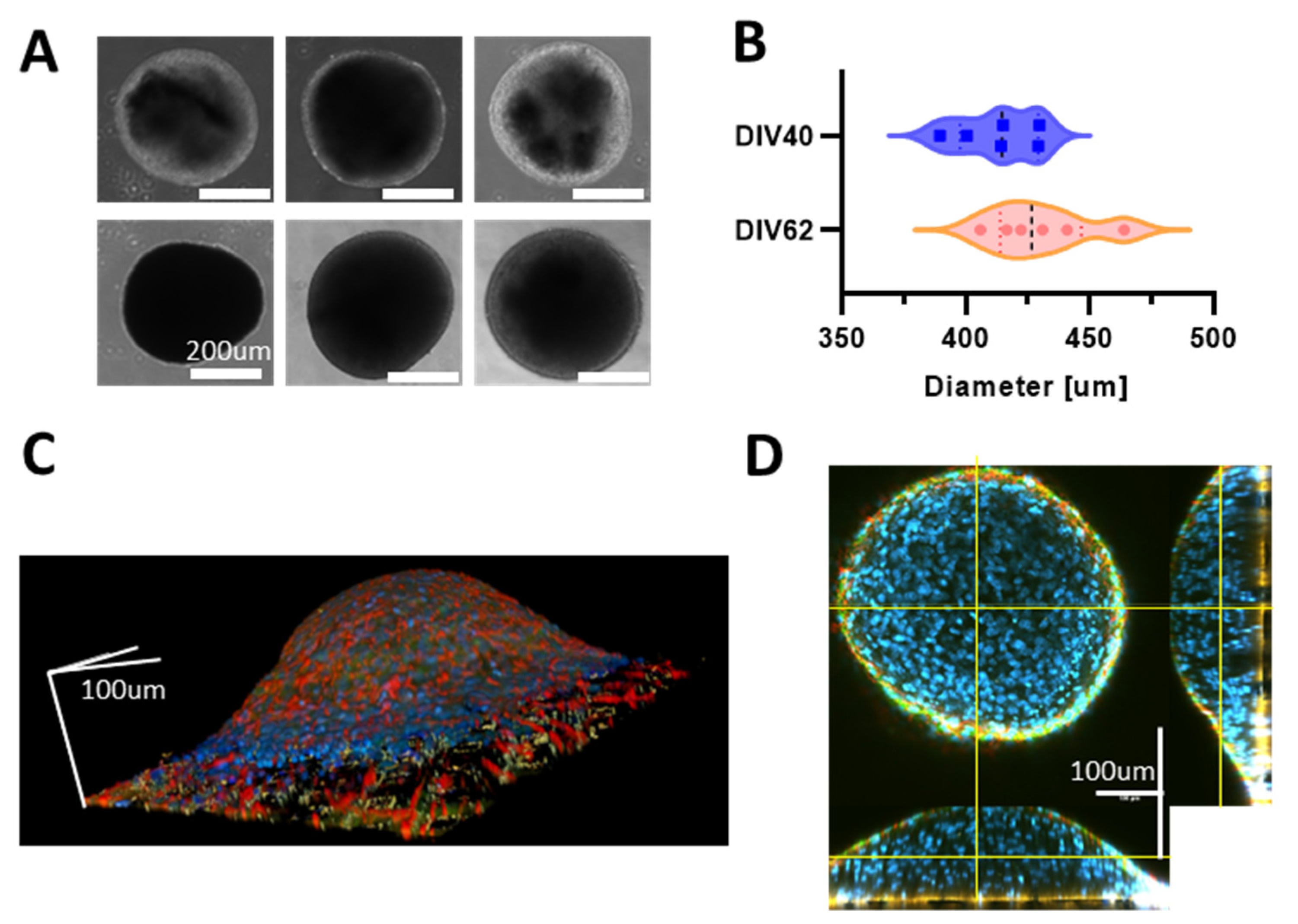
3.1. Human iPSCs-Derived Neurons Aggregate into Neurospheroids
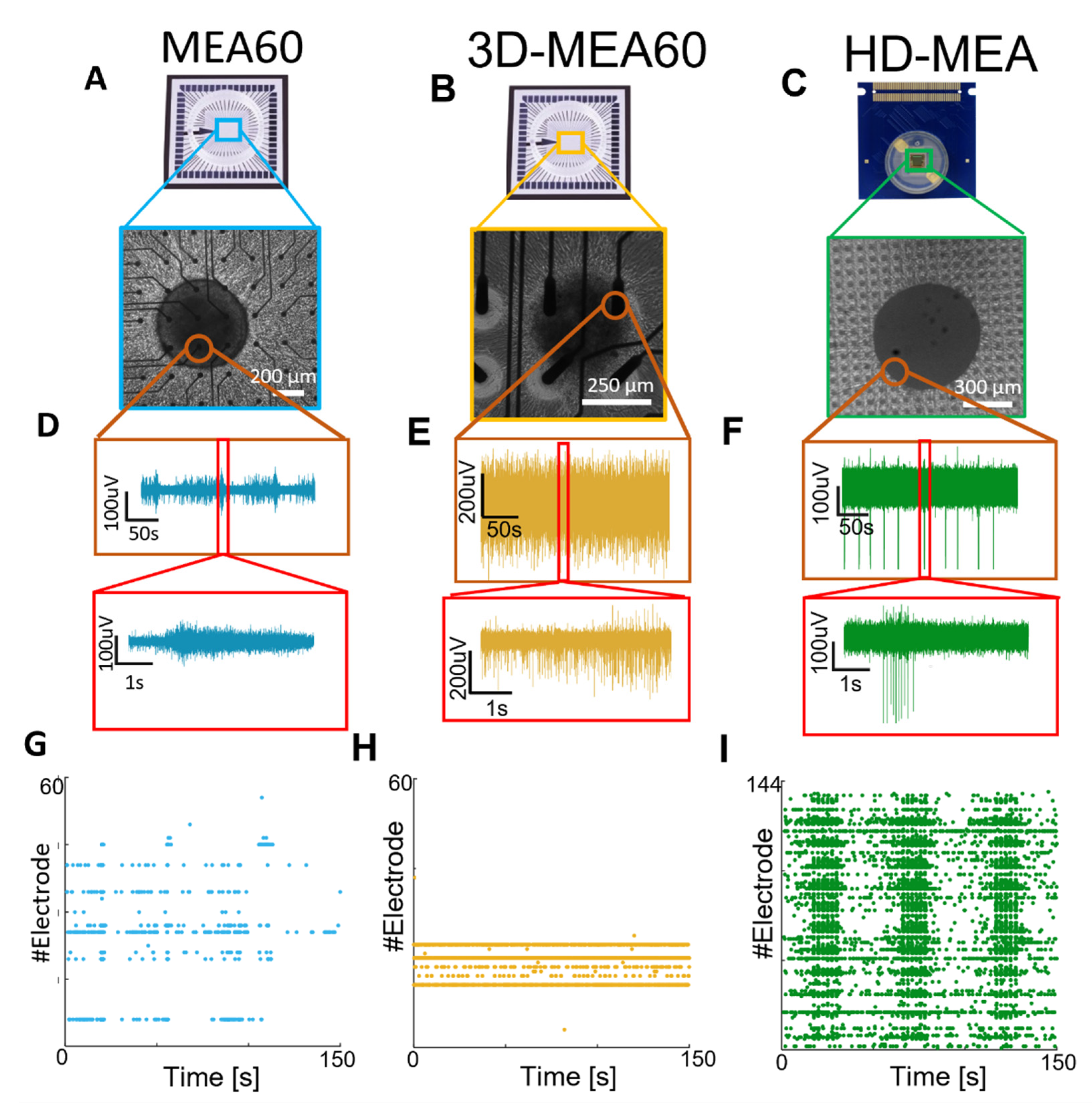
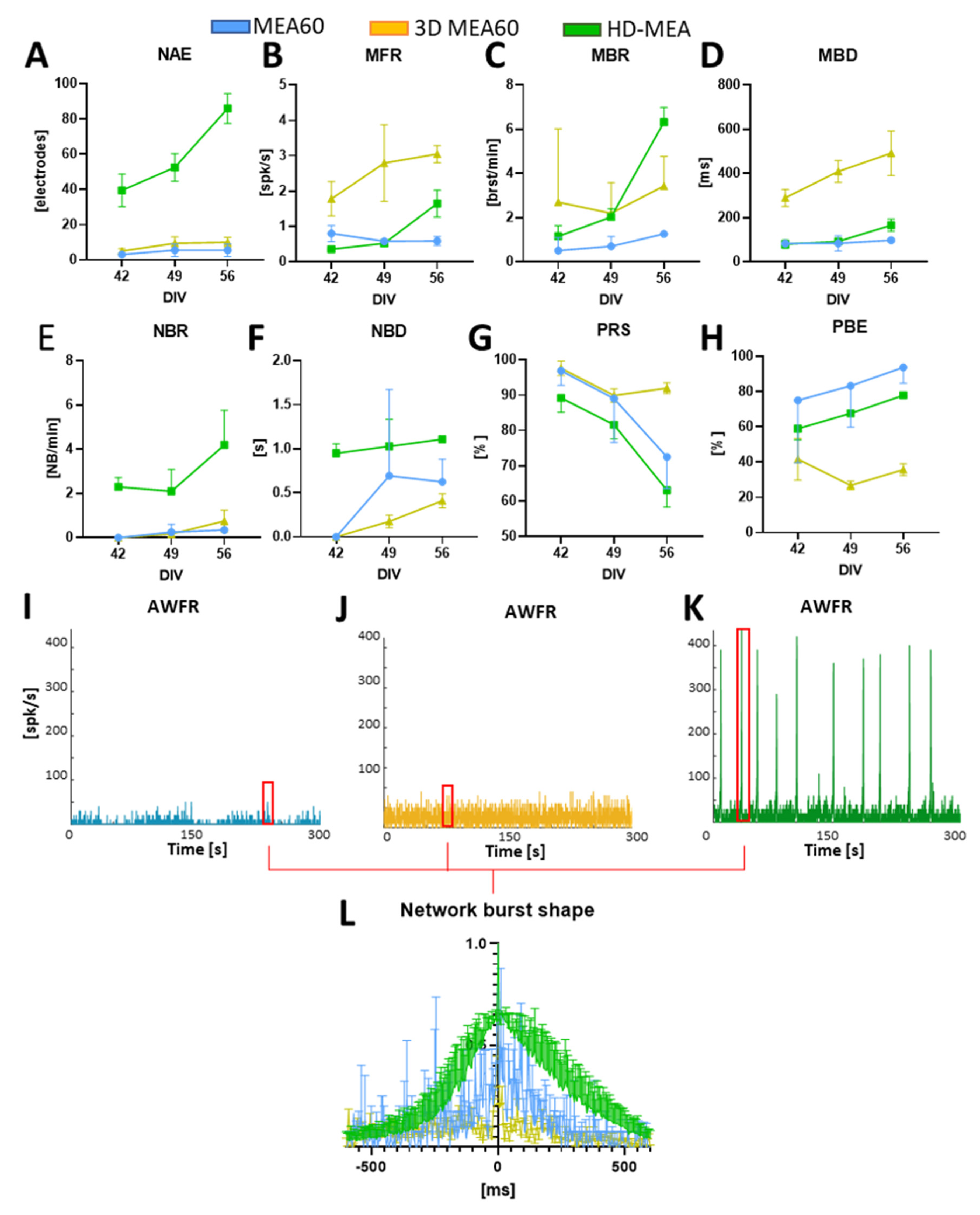
3.2. Neurospheroids Showed Electrophysiological Activity on MEAs
3.3. Neuromodulation Affects Spontaneous Activity and Electrical Induced Activity
3.4. Neuromodulation Affects Connectivity and Signal Propagation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vassallo, A.; Chiappalone, M.; Lopes, R.D.C.; Scelfo, B.; Novellino, A.; Defranchi, E.; Palosaari, T.; Weisschu, T.; Ramirez, T.; Martinoia, S. A multi-laboratory evaluation of microelectrode array-based measurements of neural network activity for acute neurotoxicity testing. Neurotoxicology 2017, 60, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Pelkonen, A.; Pistono, C.; Klecki, P.; Gómez-Budia, M.; Dougalis, A.; Konttinen, H.; Stanová, I.; Fagerlund, I.; Leinonen, V.; Korhonen, P. Functional characterization of human pluripotent stem cell-derived models of the brain with microelectrode arrays. Cells 2021, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- McCready, F.P.; Gordillo-Sampedro, S.; Pradeepan, K.; Martinez-Trujillo, J.; Ellis, J. Multielectrode arrays for functional phenotyping of neurons from induced pluripotent stem cell models of neurodevelopmental disorders. Biology 2022, 11, 316. [Google Scholar] [CrossRef] [PubMed]
- Chiappalone, M.; Bove, M.; Vato, A.; Tedesco, M.; Martinoia, S. Dissociated cortical networks show spontaneously correlated activity patterns during in vitro development. Brain Res. 2006, 1093, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Cotterill, E.; Hall, D.; Wallace, K.; Mundy, W.R.; Eglen, S.J.; Shafer, T.J. Characterization of early cortical neural network development in multiwell microelectrode array plates. J. Biomol. Screen. 2016, 21, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Van Pelt, J.; Corner, M.A.; Wolters, P.S.; Rutten, W.L.C.; Ramakers, G.J.A. Longterm stability and developmental changes in spontaneous network burst firing patterns in dissociated rat cerebral cortex cell cultures on multielectrode arrays. Neurosci. Lett. 2004, 361, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, D.A.; Pine, J.; Potter, S.M. An extremely rich repertoire of bursting patterns during the development of cortical cultures. BMC Neurosci. 2006, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Mossink, B.; Verboven, A.H.A.; van Hugte, E.J.H.; Klein Gunnewiek, T.M.; Parodi, G.; Linda, K.; Schoenmaker, C.; Kleefstra, T.; Kozicz, T.; van Bokhoven, H.; et al. Human neuronal networks on micro-electrode arrays are a highly robust tool to study disease-specific genotype-phenotype correlations in vitro. Stem Cell Rep. 2021, 16, 2182–2196. [Google Scholar] [CrossRef] [PubMed]
- Colombi, I.; Mahajani, S.; Frega, M.; Gasparini, L.; Chiappalone, M. Effects of antiepileptic drugs on hippocampal neurons coupled to micro-electrode arrays. Front. Neuroeng. 2013, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.M.; Frega, M. Past, present, and future of neuronal models in vitro. Adv. Neurobiol. 2019, 22, 3–17. [Google Scholar] [PubMed]
- Charkhkar, H.; Meyyappan, S.; Matveeva, E.; Moll, J.R.; McHail, D.G.; Peixoto, N.; Cliff, R.O.; Pancrazio, J.J. Amyloid beta modulation of neuronal network activity in vitro. Brain Res. 2015, 1629, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Martens, M.B.; Frega, M.; Classen, J.; Epping, L.; Bijvank, E.; Benevento, M.; Van Bokhoven, H.; Tiesinga, P.; Schubert, D.; Nadif Kasri, N. Euchromatin histone methyltransferase 1 regulates cortical neuronal network development. Sci. Rep. 2016, 6, 35756. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.A.; Luithardt, H.H.; Metea, M.R.; Strock, C.J. In vitro screening for seizure liability using microelectrode array technology. Toxicol. Sci. 2018, 163, 240–253. [Google Scholar] [CrossRef]
- Ahtiainen, A.; Genocchi, B.; Tanskanen, J.; Barros, M.T.; Hyttinen, J.A.K.; Lenk, K. Astrocytes exhibit a protective role in neuronal firing patterns under chemically induced seizures in neuron–astrocyte co-cultures. Int. J. Mol. Sci. 2021, 22, 12770. [Google Scholar] [CrossRef] [PubMed]
- Prado, G.R.; Ross, J.D.; DeWeerth, S.P.; LaPlaca, M.C. Mechanical trauma induces immediate changes in neuronal network activity. J. Neural Eng. 2005, 2, 148. [Google Scholar] [CrossRef]
- MacLaren, E.J.; Charlesworth, P.; Coba, M.P.; Grant, S.G.N. Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro. Mol. Cell. Neurosci. 2011, 47, 93–99. [Google Scholar] [CrossRef]
- Gao, S.; Zhao, P.; Lin, C.; Sun, Y.; Wang, Y.; Zhou, Z.; Yang, D.; Wang, X.; Xu, H.; Zhou, F.; et al. Differentiation of human adipose-derived stem cells into neuron-like cells which are compatible with photocurable three-dimensional scaffolds. Tissue Eng. Part A 2014, 20, 1271–1284. [Google Scholar] [CrossRef]
- Hales, C.M.; Zeller-Townson, R.; Newman, J.P.; Shoemaker, J.T.; Killian, N.J.; Potter, S.M. Stimulus-evoked high frequency oscillations are present in neuronal networks on microelectrode arrays. Front. Neural Circuits 2012, 6, 29. [Google Scholar] [CrossRef]
- Bateup, H.S.; Johnson, C.A.; Denefrio, C.L.; Saulnier, J.L.; Kornacker, K.; Sabatini, B.L. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron 2013, 78, 510–522. [Google Scholar] [CrossRef]
- Vedunova, M.; Sakharnova, T.; Mitroshina, E.; Perminova, M.; Pimashkin, A.; Zakharov, Y.; Dityatev, A.; Mukhina, I. Seizure-like activity in hyaluronidase-treated dissociated hippocampal cultures. Front. Cell. Neurosci. 2013, 7, 149. [Google Scholar] [CrossRef]
- Jantzen, S.U.; Ferrea, S.; Wach, C.; Quasthoff, K.; Illes, S.; Scherfeld, D.; Hartung, H.-P.; Seitz, R.J.; Dihné, M. In vitro neuronal network activity in NMDA receptor encephalitis. BMC Neurosci. 2013, 14, 17. [Google Scholar] [CrossRef] [PubMed]
- Gullo, F.; Manfredi, I.; Lecchi, M.; Casari, G.; Wanke, E.; Becchetti, A. Multi-electrode array study of neuronal cultures expressing nicotinic β2-V287L subunits, linked to autosomal dominant nocturnal frontal lobe epilepsy. An in vitro model of spontaneous epilepsy. Front. Neural Circuits 2014, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Chen, Q.; Zhou, T.; Bozic, D.; Fu, Z.; Pan, J.Q.; Feng, G. Micro-electrode array recordings reveal reductions in both excitation and inhibition in cultured cortical neuron networks lacking Shank3. Mol. Psychiatry 2016, 21, 159–168. [Google Scholar] [CrossRef]
- McSweeney, K.M.; Gussow, A.B.; Bradrick, S.S.; Dugger, S.A.; Gelfman, S.; Wang, Q.; Petrovski, S.; Frankel, W.N.; Boland, M.J.; Goldstein, D.B. Inhibition of microRNA 128 promotes excitability of cultured cortical neuronal networks. Genome Res. 2016, 26, 1411–1416. [Google Scholar] [CrossRef] [PubMed]
- Theiss, S.; Maetzler, W.; Deuschle, C.; Lerche, H.; Koch, H.; Dihné, M. Dementia with Lewy bodies: Cerebrospinal fluid suppresses neuronal network activity. Neuroreport 2017, 28, 1061–1065. [Google Scholar] [CrossRef]
- Feng, X.; Bader, B.M.; Yang, F.; Segura, M.; Schultz, L.; Schröder, O.H.-U.; Rolfs, A.; Luo, J. Improvement of impaired electrical activity in NPC1 mutant cortical neurons upon DHPG stimulation detected by micro-electrode array. Brain Res. 2018, 1694, 87–93. [Google Scholar] [CrossRef]
- le Feber, J.; Dummer, A.; Hassink, G.C.; van Putten, M.J.A.M.; Hofmeijer, J. Evolution of Excitation-Inhibition Ratio in Cortical Cultures Exposed to Hypoxia. Front. Cell. Neurosci. 2018, 12, 183. [Google Scholar] [CrossRef]
- Muzzi, L.; Hassink, G.; Levers, M.; Jansman, M.; Frega, M.; Hofmeijer, J.; van Putten, M.; le Feber, J. Mild stimulation improves neuronal survival in an in vitro model of the ischemic penumbra. J. Neural Eng. 2019, 17, 16001. [Google Scholar] [CrossRef]
- Rogers, E.A.; Gross, G.W. Simultaneous electrophysiological and morphological assessment of functional damage to neural networks in vitro after 30–300 g impacts. Sci. Rep. 2019, 9, 14994. [Google Scholar] [CrossRef]
- Gao, F.; Gao, K.; He, C.; Liu, M.; Wan, H.; Wang, P. Multi-site dynamic recording for Aβ oligomers-induced Alzheimer’s disease in vitro based on neuronal network chip. Biosens. Bioelectron. 2019, 133, 183–191. [Google Scholar] [CrossRef]
- Moskalyuk, A.; Van De Vijver, S.; Verstraelen, P.; De Vos, W.H.; Kooy, R.F.; Giugliano, M. Single-cell and neuronal network alterations in an in vitro model of fragile X syndrome. Cereb. Cortex 2020, 30, 31–46. [Google Scholar] [CrossRef]
- Erata, E.; Gao, Y.; Purkey, A.M.; Soderblom, E.J.; McNamara, J.O.; Soderling, S.H. Cnksr2 loss in mice leads to increased neural activity and behavioral phenotypes of epilepsy-aphasia syndrome. J. Neurosci. 2021, 41, 9633–9649. [Google Scholar] [CrossRef] [PubMed]
- Mossink, B.; Van Rhijn, J.-R.; Wang, S.; Linda, K.; Vitale, M.R.; Zöller, J.E.M.; van Hugte, E.J.H.; Bak, J.; Verboven, A.H.A.; Selten, M. Cadherin-13 is a critical regulator of GABAergic modulation in human stem-cell-derived neuronal networks. Mol. Psychiatry 2022, 27, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Frega, M.; Linda, K.; Keller, J.M.; Gümüş-Akay, G.; Mossink, B.; van Rhijn, J.R.; Negwer, M.; Klein Gunnewiek, T.; Foreman, K.; Kompier, N.; et al. Neuronal network dysfunction in a model for Kleefstra syndrome mediated by enhanced NMDAR signaling. Nat. Commun. 2019, 10, 4928. [Google Scholar] [CrossRef] [PubMed]
- Gunnewiek, T.M.K.; Van Hugte, E.J.H.; Frega, M.; Guardia, G.S.; Foreman, K.; Panneman, D.; Mossink, B.; Linda, K.; Keller, J.M.; Schubert, D.; et al. m.3243A> G-induced mitochondrial dysfunction impairs human neuronal development and reduces neuronal network activity and synchronicity. Cell Rep. 2020, 31, 107538. [Google Scholar] [CrossRef]
- Linda, K.; Lewerissa, E.I.; Verboven, A.H.A.; Gabriele, M.; Frega, M.; Klein Gunnewiek, T.M.; Devilee, L.; Ulferts, E.; Hommersom, M.; Oudakker, A. Imbalanced autophagy causes synaptic deficits in a human model for neurodevelopmental disorders. Autophagy 2022, 18, 423–442. [Google Scholar] [CrossRef]
- Song, C.-G.; Zhang, Y.-Z.; Wu, H.-N.; Cao, X.-L.; Guo, C.-J.; Li, Y.-Q.; Zheng, M.-H.; Han, H. Stem cells: A promising candidate to treat neurological disorders. Neural Regen. Res. 2018, 13, 1294. [Google Scholar]
- Liu, W.; Deng, Y.; Liu, Y.; Gong, W.; Deng, W. Stem cell models for drug discovery and toxicology studies. J. Biochem. Mol. Toxicol. 2013, 27, 17–27. [Google Scholar] [CrossRef]
- Napoli, A.; Obeid, I. Comparative analysis of human and rodent brain primary neuronal culture spontaneous activity using micro-electrode array technology. J. Cell. Biochem. 2016, 117, 559–565. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Langer, R.; Vacanti, J.P.; Vacanti, C.A.; Atala, A.; Freed, L.E.; Vunjak-Novakovic, G. Tissue engineering: Biomedical applications. Tissue Eng. 1995, 1, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Wang, Y.; Podsiadlo, P.; Kotov, N.A. Biomedical applications of layer-by-layer assembly: From biomimetics to tissue engineering. Adv. Mater. 2006, 18, 3203–3224. [Google Scholar] [CrossRef]
- Chiaradia, I.; Lancaster, M.A. Brain organoids for the study of human neurobiology at the interface of in vitro and in vivo. Nat. Neurosci. 2020, 23, 1496–1508. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.; Haag, M.; Ugbode, C.; Tams, D.; Rattray, M.; Przyborski, S.; Bithell, A.; Whalley, B.J. Neuronal-glial populations form functional networks in a biocompatible 3D scaffold. Neurosci. Lett. 2015, 609, 198–202. [Google Scholar] [CrossRef]
- Shin, H.; Jeong, S.; Lee, J.H.; Sun, W.; Choi, N.; Cho, I.J. 3D high-density microelectrode array with optical stimulation and drug delivery for investigating neural circuit dynamics. Nat. Commun. 2021, 12, 492. [Google Scholar] [CrossRef]
- Lavik, E.; Teng, Y.D.; Snyder, E.; Langer, R. Seeding neural stem cells on scaffolds of PGA, PLA, and their copolymers. In Neural Stem Cells: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2002; pp. 89–96. [Google Scholar]
- Kunze, A.; Giugliano, M.; Valero, A.; Renaud, P. Micropatterning neural cell cultures in 3D with a multi-layered scaffold. Biomaterials 2011, 32, 2088–2098. [Google Scholar] [CrossRef]
- Fan, C.; Li, X.; Xiao, Z.; Zhao, Y.; Liang, H.; Wang, B.; Han, S.; Li, X.; Xu, B.; Wang, N. A modified collagen scaffold facilitates endogenous neurogenesis for acute spinal cord injury repair. Acta Biomater. 2017, 51, 304–316. [Google Scholar] [CrossRef]
- Alizadeh, R.; Zarrintaj, P.; Kamrava, S.K.; Bagher, Z.; Farhadi, M.; Heidari, F.; Komeili, A.; Gutiérrez, T.J.; Saeb, M.R. Conductive hydrogels based on agarose/alginate/chitosan for neural disorder therapy. Carbohydr. Polym. 2019, 224, 115161. [Google Scholar] [CrossRef]
- Tsai, E.C.; Dalton, P.D.; Shoichet, M.S.; Tator, C.H. Matrix inclusion within synthetic hydrogel guidance channels improves specific supraspinal and local axonal regeneration after complete spinal cord transection. Biomaterials 2006, 27, 519–533. [Google Scholar] [CrossRef]
- Willerth, S.M.; Sakiyama-Elbert, S.E. Approaches to neural tissue engineering using scaffolds for drug delivery. Adv. Drug Deliv. Rev. 2007, 59, 325–338. [Google Scholar] [CrossRef]
- Pautot, S.; Wyart, C.; Isacoff, E.Y. Colloid-guided assembly of oriented 3D neuronal networks. Nat. Methods 2008, 5, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Arnaldi, P.; Di Lisa, D.; Maddalena, L.; Carosio, F.; Fina, A.; Pastorino, L.; Monticelli, O. A facile approach for the development of high mechanical strength 3D neuronal network scaffold based on chitosan and graphite nanoplatelets. Carbohydr. Polym. 2021, 271, 118420. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, M.; Frega, M.; Martinoia, S.; Pesce, M.; Massobrio, P. Interfacing 3D Engineered Neuronal Cultures to Micro-Electrode Arrays: An Innovative In Vitro Experimental Model. J. Vis. Exp. 2015, e53080. [Google Scholar] [CrossRef]
- Muzzi, L.; Di Lisa, D.; Arnaldi, P.; Aprile, D.; Pastorino, L.; Martinoia, S.; Frega, M. Rapid generation of functional engineered 3D human neuronal assemblies: Network dynamics evaluated by micro-electrodes arrays. J. Neural Eng. 2021, 18, 66030. [Google Scholar] [CrossRef] [PubMed]
- Frega, M.; Tedesco, M.; Massobrio, P.; Pesce, M.; Martinoia, S. Network dynamics of 3D engineered neuronal cultures: A new experimental model for in-vitro electrophysiology. Sci. Rep. 2014, 4, 5489. [Google Scholar] [CrossRef]
- Jo, J.; Xiao, Y.; Sun, A.X.; Cukuroglu, E.; Tran, H.-D.; Göke, J.; Tan, Z.Y.; Saw, T.Y.; Tan, C.-P.; Lokman, H. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 2016, 19, 248–257. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Mariani, J.; Coppola, G.; Zhang, P.; Abyzov, A.; Provini, L.; Tomasini, L.; Amenduni, M.; Szekely, A.; Palejev, D.; Wilson, M. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 2015, 162, 375–390. [Google Scholar] [CrossRef]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Eur. J. Phys. Rehabil. Med. 2015, 44, 371–375. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef]
- Terrasso, A.P.; Pinto, C.; Serra, M.; Filipe, A.; Almeida, S.; Ferreira, A.L.; Pedroso, P.; Brito, C.; Alves, P.M. Novel scalable 3D cell based model for in vitro neurotoxicity testing: Combining human differentiated neurospheres with gene expression and functional endpoints. J. Biotechnol. 2015, 205, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Jian-Xiong, Y. 3-D spheroid culture of bone marrow mesenchymal stem cell of rhesus monkey with improved multi-differentiation potential to epithelial progenitors and neuron in vitro. Clin. Experiment. Ophthalmol. 2011, 39, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Fair, S.R.; Julian, D.; Hartlaub, A.M.; Pusuluri, S.T.; Malik, G.; Summerfied, T.L.; Zhao, G.; Hester, A.B.; Ackerman, W.E.; Hollingsworth, E.W.; et al. Electrophysiological Maturation of Cerebral Organoids Correlates with Dynamic Morphological and Cellular Development. Stem Cell Rep. 2020, 15, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesisin a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Jorfi, M.; D’Avanzo, C.; Tanzi, R.E.; Kim, D.Y.; Irimia, D. Human Neurospheroid Arrays for In Vitro Studies of Alzheimer’s Disease. Sci. Rep. 2018, 8, 2450. [Google Scholar] [CrossRef]
- Kato-Negishi, M.; Tsuda, Y.; Onoe, H.; Takeuchi, S. A neurospheroid network-stamping method for neural transplantation to the brain. Biomaterials 2010, 31, 8939–8945. [Google Scholar] [CrossRef]
- Boutin, M.E.; Kramer, L.L.; Livi, L.L.; Brown, T.; Moore, C.; Hoffman-Kim, D. A three-dimensional neural spheroid model for capillary-like network formation. J. Neurosci. Methods 2018, 299, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Nagamura-Inoue, T.; Shimazu, T.; Mori, Y.; Takahashi, A.; Tsunoda, H.; Yamaguchi, S.; Tojo, A. Neurosphere formation enhances the neurogenic differentiation potential and migratory ability of umbilical cord-mesenchymal stromal cells. Cytotherapy 2016, 18, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Durens, M.; Nestor, J.; Williams, M.; Herold, K.; Niescier, R.F.; Lunden, J.W.; Phillips, A.W.; Lin, Y.-C.; Dykxhoorn, D.M.; Nestor, M.W. High-throughput screening of human induced pluripotent stem cell-derived brain organoids. J. Neurosci. Methods 2020, 335, 108627. [Google Scholar] [CrossRef]
- Rybachuk, O.; Kopach, O.; Pivneva, T.; Kyryk, V. Isolation of Neural Stem Cells from the Embryonic Mouse Hippocampus for in vitro Growth or Engraftment into a Host Tissue. Bio-Protocol 2019, 9, e3165. [Google Scholar] [CrossRef]
- Lecomte, A.; Giantomasi, L.; Rancati, S.; Boi, F.; Angotzi, G.N.; Berdondini, L. Surface-functionalized self-standing microdevices exhibit predictive localization and seamless integration in 3D neural spheroids. Adv. Biosyst. 2020, 4, 2000114. [Google Scholar] [CrossRef] [PubMed]
- Izsak, J.; Seth, H.; Andersson, M.; Vizlin-Hodzic, D.; Theiss, S.; Hanse, E.; Ågren, H.; Funa, K.; Illes, S. Robust generation of person-specific, synchronously active neuronal networks using purely isogenic human iPSC-3D neural aggregate cultures. Front. Neurosci. 2019, 13, 351. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kim, C.; Young Yang, J.; Lee, H.; Ahn, B.; Xu, L.; Yoon Kang, J.; Oh, K.W. Gravity-oriented microfluidic device for uniform and massive cell spheroid formation. Biomicrofluidics 2012, 6, 14114. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.-C.; Hsiao, A.Y.; Allen, S.G.; Torisawa, Y.; Ho, M.; Takayama, S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 2011, 136, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, A.; Mostafa, A.; Saavedra, C.; Kim, Y.; Le, P.; Faramarzi, V.; Feathers, R.W.; Berger, J.; Ramos-Cruz, K.P.; Adeniba, O. Three-dimensional microscale hanging drop arrays with geometric control for drug screening and live tissue imaging. Sci. Adv. 2021, 7, eabc1323. [Google Scholar] [CrossRef]
- Sakaguchi, H.; Kadoshima, T.; Soen, M.; Narii, N.; Ishida, Y.; Ohgushi, M.; Takahashi, J.; Eiraku, M.; Sasai, Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015, 6, 8896. [Google Scholar] [CrossRef]
- Yoon, S.-J.; Elahi, L.S.; Pașca, A.M.; Marton, R.M.; Gordon, A.; Revah, O.; Miura, Y.; Walczak, E.M.; Holdgate, G.M.; Fan, H.C. Reliability of human cortical organoid generation. Nat. Methods 2019, 16, 75–78. [Google Scholar] [CrossRef]
- Centeno, E.G.Z.; Cimarosti, H.; Bithell, A. 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling. Mol. Neurodegener. 2018, 13, 27. [Google Scholar] [CrossRef]
- Bose, R.; Banerjee, S.; Dunbar, G.L. Modeling neurological disorders in 3D organoids using human-derived pluripotent stem cells. Front. Cell Dev. Biol. 2021, 9, 640212. [Google Scholar] [CrossRef]
- Guy, B.; Zhang, J.S.; Duncan, L.H.; Johnston Jr, R.J. Human neural organoids: Models for developmental neurobiology and disease. Dev. Biol. 2021, 478, 102–121. [Google Scholar] [CrossRef]
- Trujillo, C.A.; Gao, R.; Negraes, P.D.; Gu, J.; Buchanan, J.; Preissl, S.; Wang, A.; Wu, W.; Haddad, G.G.; Chaim, I.A. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell 2019, 25, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Bourke, J.L.; Quigley, A.F.; Duchi, S.; O’Connell, C.D.; Crook, J.M.; Wallace, G.G.; Cook, M.J.; Kapsa, R.M.I. Three-dimensional neural cultures produce networks that mimic native brain activity. J. Tissue Eng. Regen. Med. 2018, 12, 490–493. [Google Scholar] [CrossRef] [PubMed]
- Rosas, M.; Porru, S.; Giugliano, V.; Antinori, S.; Scheggi, S.; Fadda, P.; Fratta, W.; Acquas, E.; Fattore, L. Sex-specific differences in cannabinoid-induced extracellular-signal-regulated kinase phosphorylation in the cingulate cortex, prefrontal cortex, and nucleus accumbens of Lister Hooded rats. Behav. Pharmacol. 2018, 29, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, C.A.; Adams, J.W.; Negraes, P.D.; Carromeu, C.; Tejwani, L.; Acab, A.; Tsuda, B.; Thomas, C.A.; Sodhi, N.; Fichter, K.M. Pharmacological reversal of synaptic and network pathology in human MECP2-KO neurons and cortical organoids. EMBO Mol. Med. 2021, 13, e12523. [Google Scholar] [CrossRef]
- Kathuria, A.; Lopez-Lengowski, K.; Vater, M.; McPhie, D.; Cohen, B.M.; Karmacharya, R. Transcriptome analysis and functional characterization of cerebral organoids in bipolar disorder. Genome Med. 2020, 12, 34. [Google Scholar] [CrossRef]
- Kathuria, A.; Lopez-Lengowski, K.; Jagtap, S.S.; McPhie, D.; Perlis, R.H.; Cohen, B.M.; Karmacharya, R. Transcriptomic landscape and functional characterization of induced pluripotent stem cell–derived cerebral organoids in schizophrenia. JAMA Psychiatry 2020, 77, 745–754. [Google Scholar] [CrossRef]
- Ghatak, S.; Dolatabadi, N.; Gao, R.; Wu, Y.; Scott, H.; Trudler, D.; Sultan, A.; Ambasudhan, R.; Nakamura, T.; Masliah, E. NitroSynapsin ameliorates hypersynchronous neural network activity in Alzheimer hiPSC models. Mol. Psychiatry 2021, 26, 5751–5765. [Google Scholar] [CrossRef]
- Szebényi, K.; Wenger, L.M.D.; Sun, Y.; Dunn, A.W.E.; Limegrover, C.A.; Gibbons, G.M.; Conci, E.; Paulsen, O.; Mierau, S.B.; Balmus, G. Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nat. Neurosci. 2021, 24, 1542–1554. [Google Scholar] [CrossRef]
- Nadadhur, A.G.; Alsaqati, M.; Gasparotto, L.; Cornelissen-Steijger, P.; van Hugte, E.; Dooves, S.; Harwood, A.J.; Heine, V.M. Neuron-glia interactions increase neuronal phenotypes in tuberous sclerosis complex patient iPSC-derived models. Stem Cell Rep. 2019, 12, 42–56. [Google Scholar] [CrossRef]
- Winden, K.D.; Sundberg, M.; Yang, C.; Wafa, S.M.A.; Dwyer, S.; Chen, P.-F.; Buttermore, E.D.; Sahin, M. Biallelic mutations in TSC2 lead to abnormalities associated with cortical tubers in human iPSC-derived neurons. J. Neurosci. 2019, 39, 9294–9305. [Google Scholar] [CrossRef]
- Deneault, E.; Faheem, M.; White, S.H.; Rodrigues, D.C.; Sun, S.; Wei, W.; Piekna, A.; Thompson, T.; Howe, J.L.; Chalil, L. CNTN5-/+ or EHMT2-/+ human iPSC-derived neurons from individuals with autism develop hyperactive neuronal networks. eLife 2019, 8, e40092. [Google Scholar] [CrossRef] [PubMed]
- DeRosa, B.A.; El Hokayem, J.; Artimovich, E.; Garcia-Serje, C.; Phillips, A.W.; Van Booven, D.; Nestor, J.E.; Wang, L.; Cuccaro, M.L.; Vance, J.M. Convergent pathways in idiopathic autism revealed by time course transcriptomic analysis of patient-derived neurons. Sci. Rep. 2018, 8, 8423. [Google Scholar] [CrossRef] [PubMed]
- Alsaqati, M.; Heine, V.M.; Harwood, A.J. Pharmacological intervention to restore connectivity deficits of neuronal networks derived from ASD patient iPSC with a TSC2 mutation. Mol. Autism 2020, 11, 80. [Google Scholar] [CrossRef]
- Baldassari, S.; Musante, I.; Iacomino, M.; Zara, F.; Salpietro, V.; Scudieri, P. Brain organoids as model systems for genetic neurodevelopmental disorders. Front. Cell Dev. Biol. 2020, 8, 590119. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Song, H.; Ming, G. Modeling neurological disorders using brain organoids. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 111, pp. 4–14. [Google Scholar]
- Steinberg, D.J.; Repudi, S.; Saleem, A.; Kustanovich, I.; Viukov, S.; Abudiab, B.; Banne, E.; Mahajnah, M.; Hanna, J.H.; Stern, S. Modeling genetic epileptic encephalopathies using brain organoids. EMBO Mol. Med. 2021, 13, e13610. [Google Scholar] [CrossRef]
- Choi, S.H.; Kim, Y.H.; Hebisch, M.; Sliwinski, C.; Lee, S.; D’Avanzo, C.; Chen, H.; Hooli, B.; Asselin, C.; Muffat, J.; et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 2014, 515, 274–278. [Google Scholar] [CrossRef]
- Sun, A.X.; Ng, H.; Tan, E. Translational potential of human brain organoids. Ann. Clin. Transl. Neurol. 2018, 5, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pak, C.H.; Han, Y.; Ahlenius, H.; Zhang, Z.; Chanda, S.; Marro, S.; Patzke, C.; Acuna, C.; Covy, J.; et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013, 78, 785–798. [Google Scholar] [CrossRef]
- Frega, M.; van Gestel, S.H.C.; Linda, K.; van der Raadt, J.; Keller, J.; Van Rhijn, J.-R.; Schubert, D.; Albers, C.A.; Nadif Kasri, N. Rapid Neuronal Differentiation of Induced Pluripotent Stem Cells for Measuring Network Activity on Micro-electrode Arrays. J. Vis. Exp. 2017, volume, 54900. [Google Scholar] [CrossRef]
- Ichise, E.; Chiyonobu, T.; Ishikawa, M.; Tanaka, Y.; Shibata, M.; Tozawa, T.; Taura, Y.; Yamashita, S.; Yoshida, M.; Morimoto, M. Impaired neuronal activity and differential gene expression in STXBP1 encephalopathy patient iPSC-derived GABAergic neurons. Hum. Mol. Genet. 2021, 30, 1337–1348. [Google Scholar] [CrossRef]
- Yang, N.; Chanda, S.; Marro, S.; Ng, Y.-H.; Janas, J.A.; Haag, D.; Ang, C.E.; Tang, Y.; Flores, Q.; Mall, M. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods 2017, 14, 621–628. [Google Scholar] [CrossRef]
- Sundberg, M.; Bogetofte, H.; Lawson, T.; Jansson, J.; Smith, G.; Astradsson, A.; Moore, M.; Osborn, T.; Cooper, O.; Spealman, R. Improved cell therapy protocols for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells 2013, 31, 1548–1562. [Google Scholar] [CrossRef]
- Swistowski, A.; Peng, J.; Liu, Q.; Mali, P.; Rao, M.S.; Cheng, L.; Zeng, X. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells 2010, 28, 1893–1904. [Google Scholar] [CrossRef]
- Dimos, J.T.; Rodolfa, K.T.; Niakan, K.K.; Weisenthal, L.M.; Mitsumoto, H.; Chung, W.; Croft, G.F.; Saphier, G.; Leibel, R.; Goland, R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 2008, 321, 1218–1221. [Google Scholar] [CrossRef]
- Juopperi, T.A.; Kim, W.R.; Chiang, C.-H.; Yu, H.; Margolis, R.L.; Ross, C.A.; Ming, G.; Song, H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol. Brain 2012, 5, 17. [Google Scholar] [CrossRef]
- Suga, M.; Kondo, T.; Inoue, H. Modeling neurological disorders with human pluripotent stem cell-derived astrocytes. Int. J. Mol. Sci. 2019, 20, 3862. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; van Rhijn, J.-R.; Akkouh, I.; Kogo, N.; Maas, N.; Bleeck, A.; Ortiz, I.S.; Lewerissa, E.; Wu, K.M.; Schoenmaker, C. Loss-of-function variants in the schizophrenia risk gene SETD1A alter neuronal network activity in human neurons through the cAMP/PKA pathway. Cell Rep. 2022, 39, 110790. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Ishikawa, M.; Fujimori, K.; Maeda, T.; Kushima, I.; Arioka, Y.; Mori, D.; Nakatake, Y.; Yamagata, B.; Nio, S. In vitro modeling of the bipolar disorder and schizophrenia using patient-derived induced pluripotent stem cells with copy number variations of PCDH15 and RELN. eNeuro 2019, 6, ENEURO.0403-18.2019. [Google Scholar] [CrossRef] [PubMed]
- Maccione, A.; Gandolfo, M.; Massobrio, P.; Novellino, A.; Martinoia, S.; Chiappalone, M. A novel algorithm for precise identification of spikes in extracellularly recorded neuronal signals. J. Neurosci. Methods 2009, 177, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Eytan, D.; Minerbi, A.; Ziv, N.; Marom, S. Dopamine-induced dispersion of correlations between action potentials in networks of cortical neurons. J. Neurophysiol. 2004, 92, 1817–1824. [Google Scholar] [CrossRef]
- Qian, X.; Song, H.; Ming, G. Brain organoids: Advances, applications and challenges. Development 2019, 146, dev166074. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Corsini, N.S.; Wolfinger, S.; Gustafson, E.H.; Phillips, A.W.; Burkard, T.R.; Otani, T.; Livesey, F.J.; Knoblich, J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017, 35, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Sharf, T.; van der Molen, T.; Glasauer, S.M.K.; Guzman, E.; Buccino, A.P.; Luna, G.; Cheng, Z.; Audouard, M.; Ranasinghe, K.G.; Kudo, K. Functional neuronal circuitry and oscillatory dynamics in human brain organoids. Nat. Commun. 2022, 13, 4403. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Tang, W.C.; Chen, J.; Chew, K.; Seiler, M.J.; Browne, A. Retinal Organoids Cultured by Microfluidic Bioreactor Demonstrated Functionality Measured by a High-Density Microelectrode Array System. Invest. Ophthalmol. Vis. Sci. 2022, 63, 3784-F0205. [Google Scholar]
- Gong, W.; Senčar, J.; Bakkum, D.J.; Jäckel, D.; Obien, M.E.J.; Radivojevic, M.; Hierlemann, A.R. Multiple single-unit long-term tracking on organotypic hippocampal slices using high-density microelectrode arrays. Front. Neurosci. 2016, 10, 537. [Google Scholar] [CrossRef]
- Obien, M.E.J.; Andreas, H.; Frey, U. Accurate signal-source localization in brain slices by means of high-density microelectrode arrays. Sci. Rep. 2019, 9, 788. [Google Scholar] [CrossRef]
- Müller, J.; Ballini, M.; Livi, P.; Chen, Y.; Radivojevic, M.; Shadmani, A.; Viswam, V.; Jones, I.L.; Fiscella, M.; Diggelmann, R.; et al. High-resolution CMOS MEA platform to study neurons at subcellular, cellular, and network levels. Lab Chip 2015, 15, 2767–2780. [Google Scholar] [CrossRef]
- Bakkum, D.J.; Chao, Z.C.; Potter, S.M. Spatio-temporal electrical stimuli shape behavior of an embodied cortical network in a goal-directed learning task. J. Neural Eng. 2008, 5, 310. [Google Scholar] [CrossRef]
- Heikkilä, T.J.; Ylä-Outinen, L.; Tanskanen, J.M.A.; Lappalainen, R.S.; Skottman, H.; Suuronen, R.; Mikkonen, J.E.; Hyttinen, J.A.K.; Narkilahti, S. Human embryonic stem cell-derived neuronal cells form spontaneously active neuronal networks in vitro. Exp. Neurol. 2009, 218, 109–116. [Google Scholar] [CrossRef]
- Odawara, A.; Katoh, H.; Matsuda, N.; Suzuki, I. Physiological maturation and drug responses of human induced pluripotent stem cell-derived cortical neuronal networks in long-term culture. Sci. Rep. 2016, 6, 26181. [Google Scholar] [CrossRef]
- Suresh, J.; Radojicic, M.; Pesce, L.L.; Bhansali, A.; Wang, J.; Tryba, A.K.; Marks, J.D.; Van Drongelen, W. Network burst activity in hippocampal neuronal cultures: The role of synaptic and intrinsic currents. J. Neurophysiol. 2016, 115, 3073–3089. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Song, X.; Xiang, G.; Zhu, J.; Cheng, J. Effects of disinhibition on spatiotemporal pattern of neuronal first recruitment in neuronal networks. Prog. Nat. Sci. 2009, 19, 615–621. [Google Scholar] [CrossRef]
- Eytan, D.; Brenner, N.; Marom, S. Selective adaptation in networks of cortical neurons. J. Neurosci. 2003, 23, 9349–9356. [Google Scholar] [CrossRef] [PubMed]
- Yvon, C.; Rubli, R.; Streit, J. Patterns of spontaneous activity in unstructured and minimally structured spinal networks in culture. Exp. Brain Res. 2005, 165, 139–151. [Google Scholar] [CrossRef]
- Rao, V.R.; Finkbeiner, S. NMDA and AMPA receptors: Old channels, new tricks. Trends Neurosci. 2007, 30, 284–291. [Google Scholar] [CrossRef]
- Sperk, G. Kainic acid seizures in the rat. Prog. Neurobiol. 1994, 42, 1–32. [Google Scholar] [CrossRef]
- Fisher, R.S.; Alger, B.E. Electrophysiological mechanisms of kainic acid-induced epileptiform activity in the rat hippocampal slice. J. Neurosci. 1984, 4, 1312–1323. [Google Scholar] [CrossRef]
- Jimbo, Y.; Robinson, H.P.C. Propagation of spontaneous synchronized activity in cortical slice cultures recorded by planar electrode arrays. Bioelectrochemistry 2000, 51, 107–115. [Google Scholar] [CrossRef]
- Mzezewa, R.; Lotila, J.; Kiiski, H.; Vinogradov, A.; Kapucu, F.E.; Peltola, J.; Hagman, S.; Narkilahti, S. A kainic acid-induced seizure model in human pluripotent stem cell-derived cortical neurons for studying the role of IL-6 in the functional activity. Stem Cell Res. 2022, 60, 102665. [Google Scholar] [CrossRef]
- Odawara, A.; Matsuda, N.; Ishibashi, Y.; Yokoi, R.; Suzuki, I. Toxicological evaluation of convulsant and anticonvulsant drugs in human induced pluripotent stem cell-derived cortical neuronal networks using an MEA system. Sci. Rep. 2018, 8, 10416. [Google Scholar] [CrossRef]
- Avoli, M. Mechanisms of epileptiform synchronization in cortical neuronal networks. Curr. Med. Chem. 2014, 21, 653–662. [Google Scholar] [CrossRef]
- Poli, D.; Pastore, V.P.; Massobrio, P. Functional connectivity in in vitro neuronal assemblies. Front. Neural Circuits 2015, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Chiappalone, M.; Massobrio, P.; Martinoia, S. Network plasticity in cortical assemblies. Eur. J. Neurosci. 2008, 28, 221–237. [Google Scholar] [CrossRef]
- Sporns, O. Graph theory methods: Applications in brain networks. Dialogues Clin. Neurosci. 2018, 20, 111–121. [Google Scholar] [CrossRef]
- Shahaf, G.; Marom, S. Learning in networks of cortical neurons. J. Neurosci. 2001, 21, 8782–8788. [Google Scholar] [CrossRef] [PubMed]
- Diering, G.H.; Huganir, R.L. The AMPA receptor code of synaptic plasticity. Neuron 2018, 100, 314–329. [Google Scholar] [CrossRef]
- Haselmann, H.; Mannara, F.; Werner, C.; Planagumà, J.; Miguez-Cabello, F.; Schmidl, L.; Grünewald, B.; Petit-Pedrol, M.; Kirmse, K.; Classen, J. Human autoantibodies against the AMPA receptor subunit GluA2 induce receptor reorganization and memory dysfunction. Neuron 2018, 100, 91–105. [Google Scholar] [CrossRef]
- Collingridge, G. The role of NMDA receptors in learning and memory. Nature 1987, 330, 604–605. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Tsien, J.Z. Memory and the NMDA receptors. N. Engl. J. Med. 2009, 361, 302. [Google Scholar] [CrossRef]
- Le Feber, J.; Stegenga, J.; Rutten, W.L.C. The effect of slow electrical stimuli to achieve learning in cultured networks of rat cortical neurons. PLoS ONE 2010, 5, e8871. [Google Scholar] [CrossRef]
- Jimbo, Y.; Robinson, H.P.C.; Kawana, A. Strengthening of synchronized activity by tetanic stimulation in cortical cultures: Application of planar electrode arrays. IEEE Trans. Biomed. Eng. 1998, 45, 1297–1304. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muzzi, L.; Di Lisa, D.; Falappa, M.; Pepe, S.; Maccione, A.; Pastorino, L.; Martinoia, S.; Frega, M. Human-Derived Cortical Neurospheroids Coupled to Passive, High-Density and 3D MEAs: A Valid Platform for Functional Tests. Bioengineering 2023, 10, 449. https://doi.org/10.3390/bioengineering10040449
Muzzi L, Di Lisa D, Falappa M, Pepe S, Maccione A, Pastorino L, Martinoia S, Frega M. Human-Derived Cortical Neurospheroids Coupled to Passive, High-Density and 3D MEAs: A Valid Platform for Functional Tests. Bioengineering. 2023; 10(4):449. https://doi.org/10.3390/bioengineering10040449
Chicago/Turabian StyleMuzzi, Lorenzo, Donatella Di Lisa, Matteo Falappa, Sara Pepe, Alessandro Maccione, Laura Pastorino, Sergio Martinoia, and Monica Frega. 2023. "Human-Derived Cortical Neurospheroids Coupled to Passive, High-Density and 3D MEAs: A Valid Platform for Functional Tests" Bioengineering 10, no. 4: 449. https://doi.org/10.3390/bioengineering10040449
APA StyleMuzzi, L., Di Lisa, D., Falappa, M., Pepe, S., Maccione, A., Pastorino, L., Martinoia, S., & Frega, M. (2023). Human-Derived Cortical Neurospheroids Coupled to Passive, High-Density and 3D MEAs: A Valid Platform for Functional Tests. Bioengineering, 10(4), 449. https://doi.org/10.3390/bioengineering10040449




