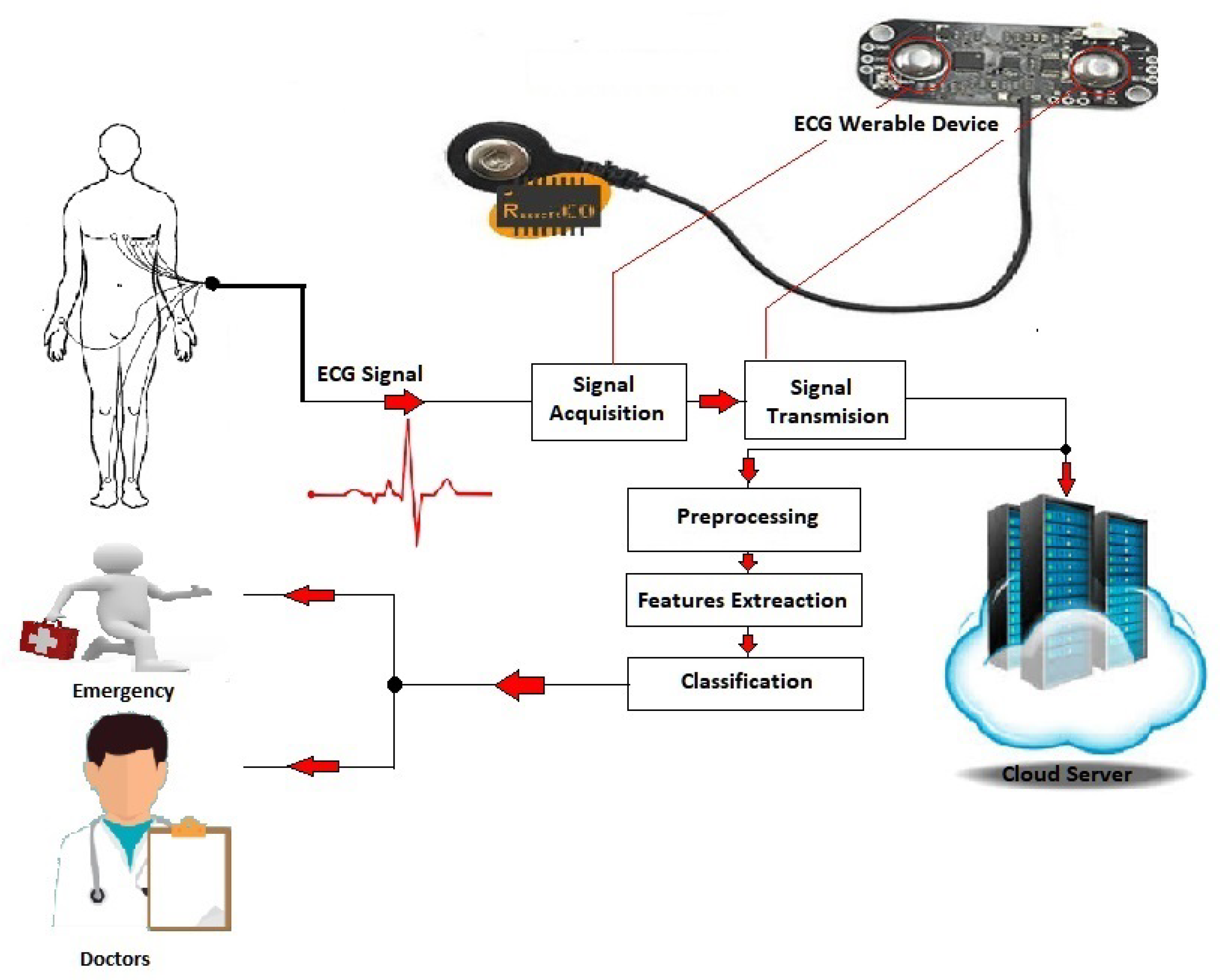
1. Introduction
The recent developments in biomedical sensors, the Internet of Medical Things (IoMT), and artificial intelligence (AI)-based techniques have increased interest in smart healthcare technologies [
1,
2]. Microelectronics, smart sensors, AI, 5G, and IoMT constitute the cornerstone of smart healthcare [
3,
4]. A smart healthcare system does not suffer fatigue; hence, it can process big data at a much higher speed than humans with greater accuracy [
5]. With smart healthcare systems, the diagnosis and treatment of diseases have become more intelligent. For instance, smart patient monitoring empowers the observation of a patient outside the traditional clinical settings, which offers a lower cost through reducing visits to physician offices and hospitalizations [
6].
The human body is known as a complex electromechanical system generating several types of biomedical signals, such as an electrocardiogram (ECG), which is a record of the dynamic changes of the human body that need to be monitored by smart healthcare systems. For instance, the EKG sensor measures cardiac electrical potential waveforms. It is used to create standard 3-lead electrocardiogram (EKG) tracings to record the electrical activity in the heart or to collect surface electromyography (sEMG) to study the contractions in the muscles of the arm, leg, or jaw. Simply, an ECG graphs heartbeats and rhythms. The classification of an ECG heartbeat plays a substantial role in smart healthcare systems [
7,
8], where the presence of multiple cardiovascular problems is generally indicated by an ECG. In the subsequent ECG waveform, diseases cause defects. However, early diagnosis via an ECG allows for the selection of suitable cardiac medication and is thus very important and helpful for reducing heart attacks [
9]. The method of detecting and classifying arrhythmia is not an easy task and may be very difficult even for professionals because sometimes it is important to examine multiple pulses of ECG data, obtained, for example, during hours, or even days, by a Holter clock. Furthermore, there is a possibility for errors by humans during the ECG recording study due to fatigue. Building a fully automatic arrhythmia detection or classification system is difficult. The difficulty comes from the large amount of data and the diversities in the ECG signals due to the nonlinearity, complexity, and low amplitude of ECG recordings, as well as the nonclinical conditions, such as noise [
10].
Despite all these difficulties, methods for ECG arrhythmia classification have been widely explored [
11,
12] but choosing the best technique for smart patient monitoring depends on the robustness and performance of these methods. Several convolutional neural network (CNN)-based approaches have been introduced for the task [
13,
14]. Bollepalli et al. [
10] proposed a CNN-based heartbeat detector to learn fused features from multiple ECG signals. It achieved an accuracy of 99.92% on the MITBIH database using two ECG channels. In [
15], a subject-adaptable ECG arrhythmia classification model was proposed and trained with unlabeled personal data. It achieved an average performance of 99.4% classification accuracy on the MIT-BIH database. In [
16], an end to-end deep multiscale fusion CNN model of multiple convolution kernels with different receptive fields was proposed, achieving an F1 score of 82.8% and 84.1% on two datasets. Chen et al. [
17] combined CNN with long short-term memory to classify six types of arrhythmia and achieved an average accuracy of 97.15% on the MIT-BIH database. A recent approach by Atal and Singh [
18] proposed using the bat-rider optimization to optimally tune a deep CNN to achieve an accuracy of 93.19% with a sensitivity of 93.9% on the MIT-BIH database. Unfortunately, most CNN-based methods are effective only for small numbers of arrhythmia classes, are computationally intensive, and need a very large amount of training data [
13]. This is a great challenge for using the CNN-based methods on real-time applications or wearable devices with limited hardware [
19].
On the other hand, many research efforts have been devoted to ECG arrhythmia classification using ML classifiers, such as SVM, RF, kNN, linear discriminants, multilayered perceptron, and regression tree [
20,
21]. It is well known that the SVM classifier does not become trapped in the well-known local minima points, requires less training data, and is faster than CNN-based methods [
22]. In [
23], wavelet transform and ICA were used for the morphological features description of the segmented heartbeats. The features were fed into an SVM to classify an ECG into five classes. In [
24], least square twin SVM and kNN classifiers based on features’ sparse representation were used for cardiac arrhythmia recognition. The experiments were carried out on the MIT-BIH database in category and personalized schemes. A method based on improved fuzzy C-means clustering and Mahalanobis distance was introduced in [
25], while in [
26], abstract features from abductive interpretation of the ECG signals were utilized in heartbeat classification. Borui et al. [
27] proposed a deep learning model integrating a long short-term memory with SVM for ECG arrhythmia classification. Martis et al. [
28] evaluated the performance of several ML classifiers and concluded that the kNN and higher-order statistics features achieved an average accuracy of 97.65% and sensitivity of 98.16% on the MIT-BIH database. In [
29], the RF classifier was utilized with CNN and PQRST features for arrhythmia classification from imbalanced ECG data. The major drawback of ML classifiers (e.g., SVM) is their deficiency in interpreting the impact of ECG data features on different arrhythmia patterns for extracting the optimal features. Further, the performance of most ML classifiers is questionable because the interrelationship between the learning parameters is not well modeled, especially for data features with high dimensions.
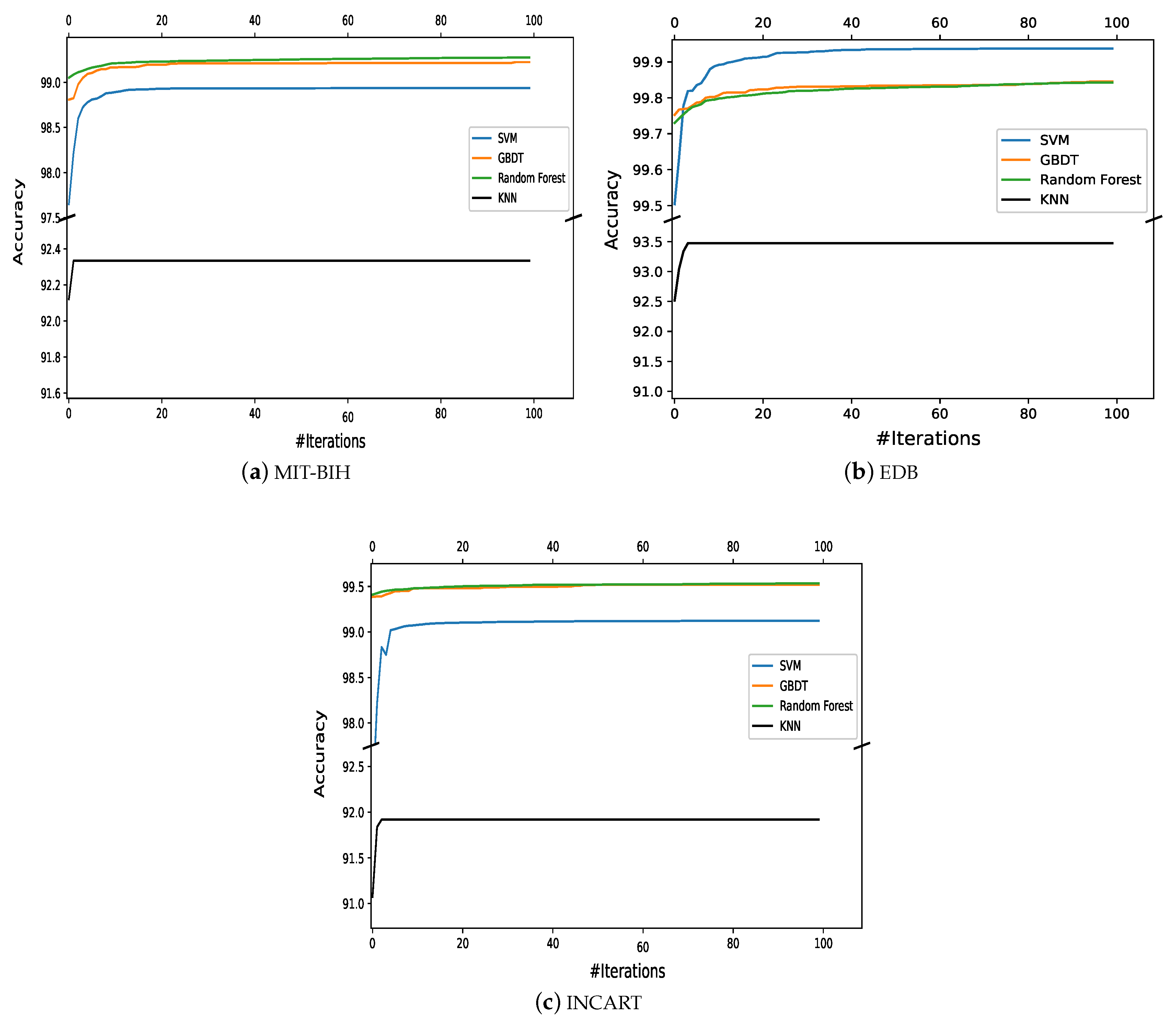
Despite the large amount of previous studies in the field, ECG arrhythmia classification has not been completely solved and remains a challenging problem. Consequently, there is room for improvement in several aspects, including classification, feature extraction, preprocessing, and ECG data segmentation. Most ML classifiers have some limitations; for example, SVM does not perform well with noisy data, while random forest (RF) suffers from interpretability issues and fails to determine the significance of variables. In addition, these ML classifiers have many parameters, and tuning such parameters has a crucial influence on the efficiency of the classification. Motivated by the advantages of the ML classifiers compared to the CNN-based methods, although they face a major challenge with a low classification accuracy, in this work, we focus on enhancing the classification accuracy of the ML classifiers. To this end and to develop an efficient classifier model, we propose to optimize the learning parameters of these classifiers using a naturally inspired metaheuristic algorithm called the marine predators optimization algorithm (MPA). The parameters of the classifier are gradually optimized using the MPA algorithm, which introduces an optimal classifier model that can classify the ECG features efficiently. Four different machine learning classifiers are considered, namely SVM, GBDT, RF, and kNN. The performance of these classifiers without learning parameter optimization and with optimization (i.e., MPA-SVM, MPA-GBDT, MPA-RF, and MPA-kNN) are compared. The experiments are validated on the three common benchmarking databases: the MIT-BIH, EDB, and INCART.
The remainder of this paper is organized as follows.
Section 2 presents the methodology proposed to classify the ECG arrhythmia based on the optimization of the parameters of the ML classifiers. The experimental results and analysis as well as a comparison with the state of the art are presented in
Section 3. Finally, the paper is concluded in
Section 4.
4. Conclusions
This paper proposed an automatic arrhythmia classification method based on a new AI metaheuristic optimization algorithm and four ML classifiers for IoT-assisted smart healthcare systems. Multiclassifier models including the MPA-SVM, MPA-GBDT, MPA-RF, and MPA-kNN were introduced for classification with parameter optimization. The average classification accuracies achieved by the MPA with the SVM classifier were 99.48% (MIT-BIH), 99.90% (EDB), and 99.47% (INCART). The accuracies achieved by the MPA with the GBDT were 99.61% (MIT-BIH), 99.91% (EDB), and 99.72% (INCART); meanwhile, the MPA with the RF achieved 99.67% (MIT-BIH), 99.92% (EDB), and 99.73% (INCART), while the MPA with the kNN achieved 96.44% (MIT-BIH), 97.07% (EDB), and 93.51% (INCART). It is clear that the RF showed the most accurate results of these methods. Hence, it can be concluded that incorporating the MPA scheme can effectively optimize the ML classifiers, even a lazy one, such as the kNN. The achieved performance by the optimization step was ranked among the highest reported to date.
In future works, to enhance the ability to predict heart problems, other optimization algorithms can be investigated. For efficient methods to extract features and perform classification, it is necessary to incorporate the real-time surveillance of cardiac patients. Using powerful classification models (e.g., deep learning) is the possible next step of this research. To have meaningful classification outcomes with greater accuracy, these powerful classification models can be combined with the MPA algorithm, as it performed very well and enhanced the accuracy of the classification process.