A Single Active-Site Mutagenesis Confers Enhanced Activity and/or Changed Product Distribution to a Pentalenene Synthase from Streptomyces sp. PSKA01
Abstract
1. Introduction
2. Materials and Methods
2.1. General Materials and Culture Conditions
2.2. Plasmid Construction and Site-Directed Mutagenesis
2.3. PentS Mutant Library Construction and Screening
2.4. Shake-Flask Fermentation
2.5. Qualification and Quantification of Terpenoids
2.6. Protein Solubility Detection
2.7. Homology Modeling and Molecular Docking
2.8. Mutant Construction and Molecular Dynamics Simulations
3. Results
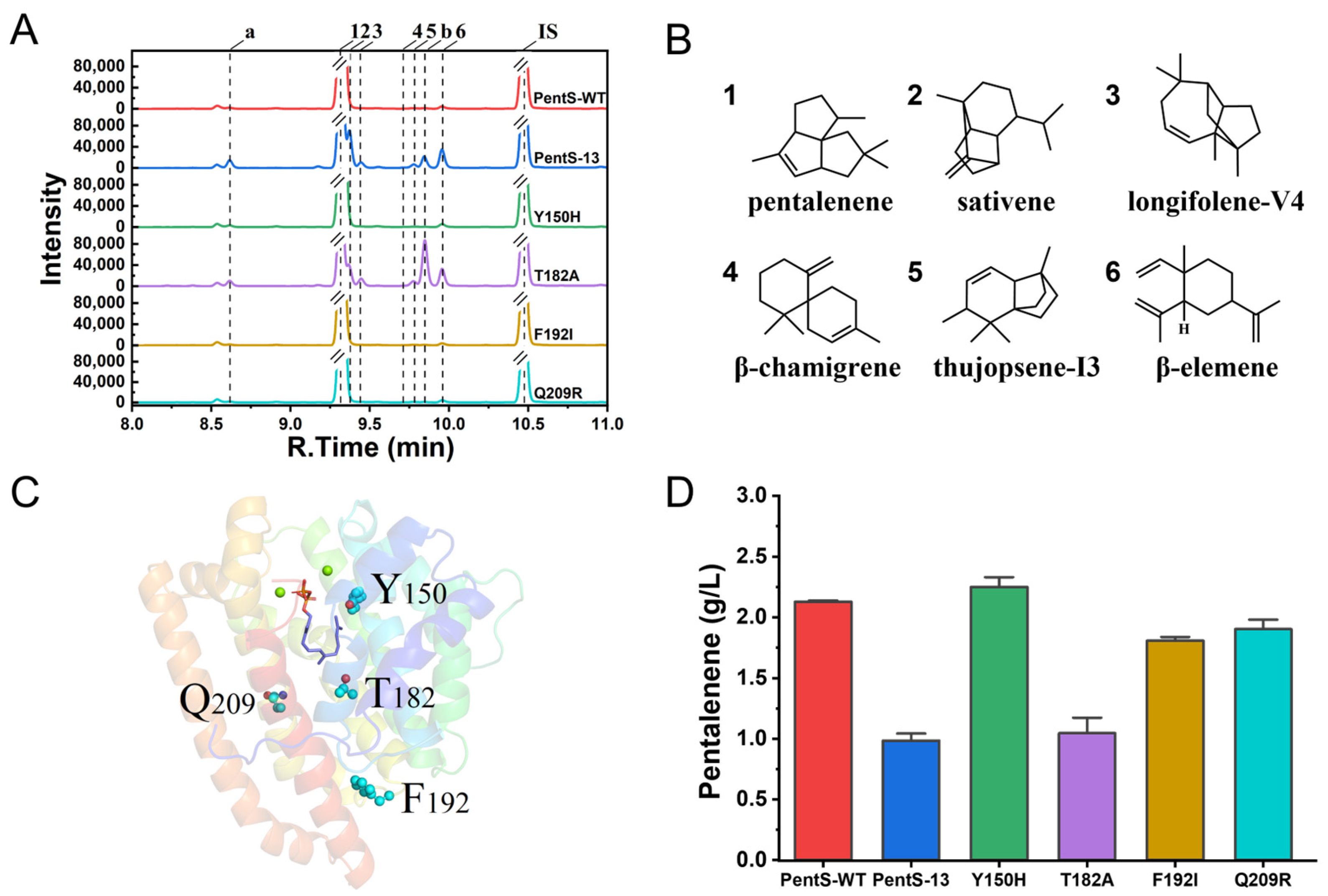
3.1. Screening of PentS Mutant Library and Mutation Identification
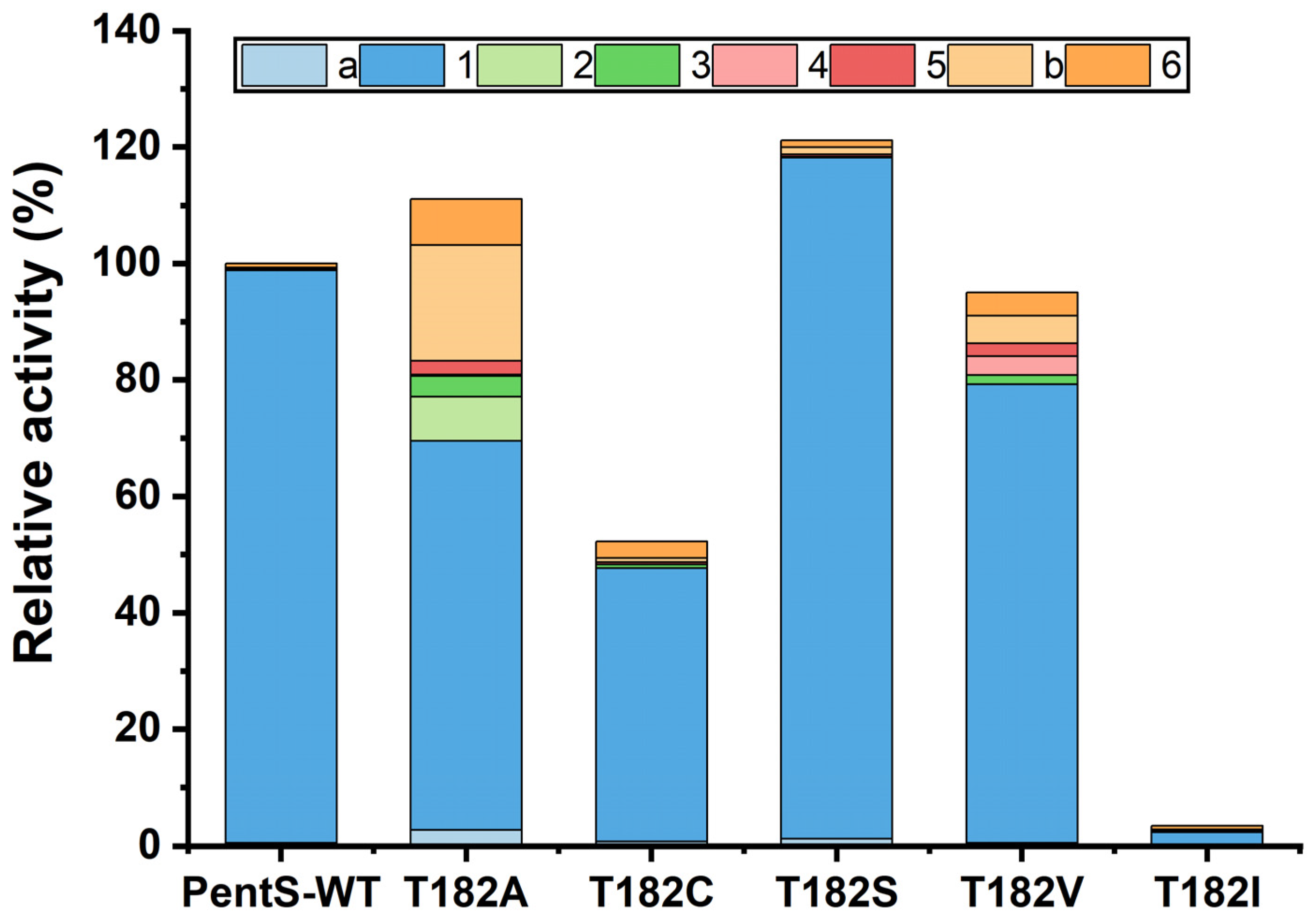
3.2. Site-Saturation Mutagenesis of Residue T182
3.3. Synthetic Pathways of Sesquiterpenes in Wild Type and Variants of PentS
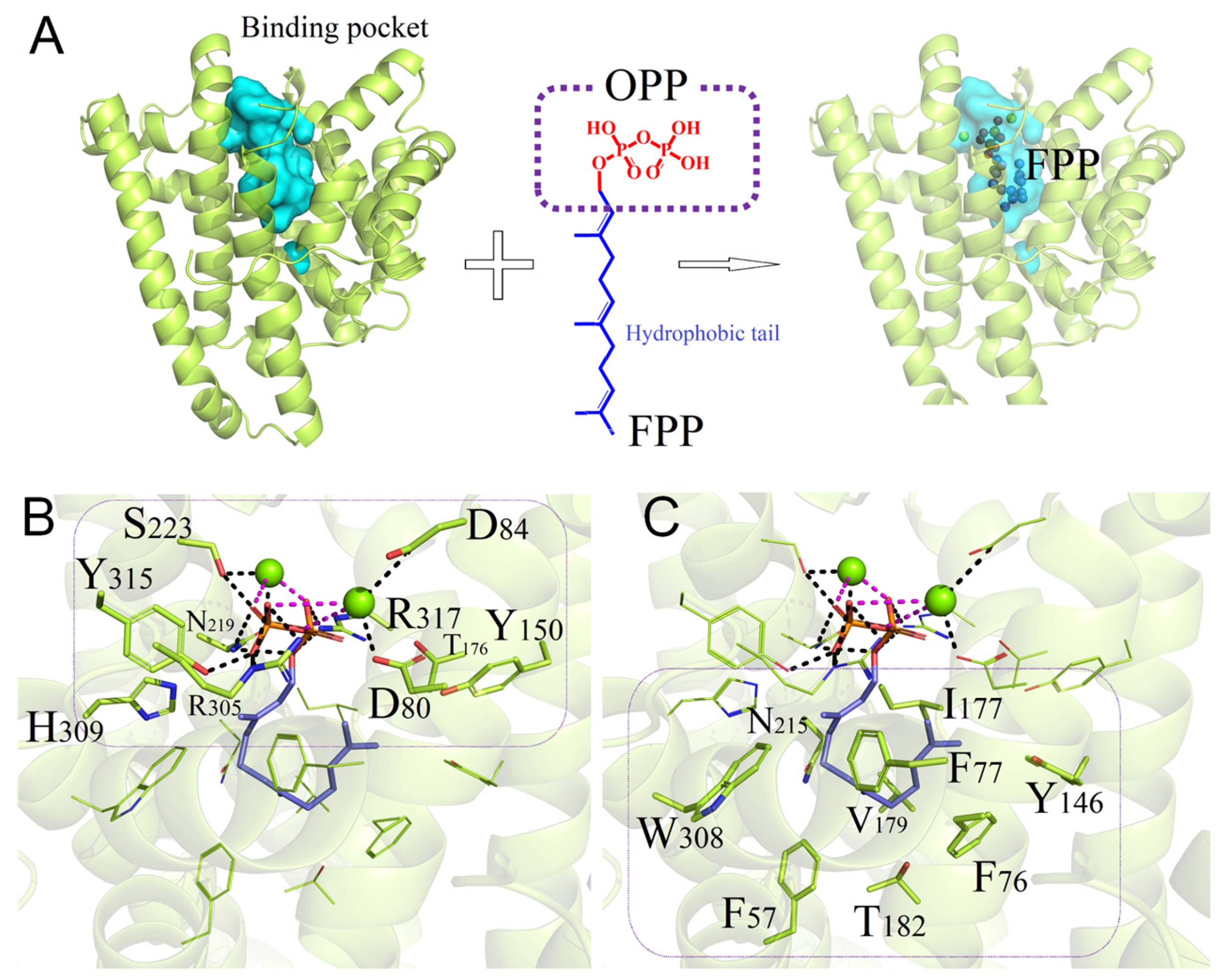
3.4. Binding Interactions between Wild-Type PentS and FPP
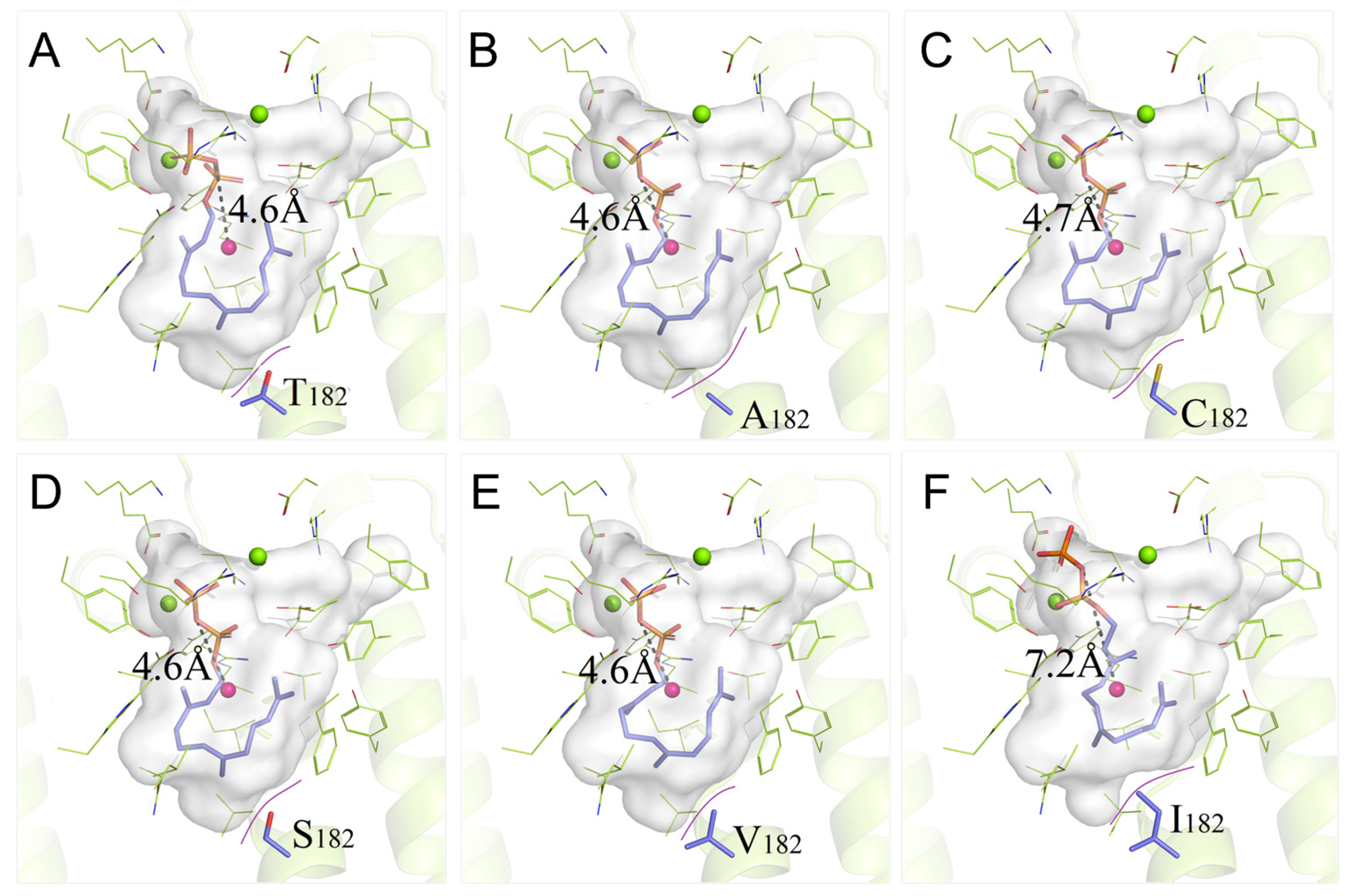
3.5. Binding Properties of PentS Variants with FPP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gong, D.Y.; Chen, X.Y.; Guo, S.X.; Wang, B.C.; Li, B. Recent advances and new insights in biosynthesis of dendrobine and sesquiterpenes. Appl. Microbiol. Biotechnol. 2021, 105, 6597–6606. [Google Scholar] [CrossRef] [PubMed]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Sarksian, R.; van der Donk, W.A. Divergent evolution of lanthipeptide stereochemistry. ACS Chem. Biol. 2022, 17, 2551–2558. [Google Scholar] [CrossRef]
- Martin-Sanchezt, L.; Singh, K.S.; Avalos, M.; van Wezel, G.P.; Dickschat, J.S.; Garbeva, P. Phylogenomic analyses and distribution of terpene synthases among Streptomyces. Beilstein J. Org. Chem. 2019, 15, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Tian, T.; Alonso-Gutierrez, J.; Garabedian, B.; Wang, S.; Baidoo, E.E.K.; Benites, V.; Chen, Y.; Petzold, C.J.; Adams, P.D.; et al. Renewable production of high density jet fuel precursor sesquiterpenes from Escherichia coli. Biotechnol. Biofuels 2018, 11, 285. [Google Scholar] [CrossRef]
- Cane, D.E.; Oliver, J.S.; Harrison, P.H.M.; Abell, C.; Hubbard, B.R.; Kane, C.T.; Lattman, R. Biosynthesis of pentalenene and pentalenolactone. J. Am. Chem. Soc. 1990, 112, 4513–4524. [Google Scholar] [CrossRef]
- Tetzlaff, C.N.; You, Z.; Cane, D.E.; Takamatsu, S.; Omura, S.; Ikeda, H. A gene cluster for biosynthesis of the sesquiterpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry 2006, 45, 6179–6186. [Google Scholar] [CrossRef]
- Cane, D.E.; Pargellis, C. Partial purification and characterization of pentalenene synthase. Arch. Biochem. Biophys. 1987, 254, 421–429. [Google Scholar] [CrossRef]
- Cane, D.E.; Sohng, J.K.; Lamberson, C.R.; Rudnicki, S.M.; Wu, Z.; Lloyd, M.D.; Oliver, J.S.; Hubbard, B.R. Pentalenene synthase. Purification, molecular cloning, sequencing, and high-level expression in Escherichia coli of a terpenoid cyclase from Streptomyces UC5319. Biochemistry 1994, 33, 5846–5857. [Google Scholar] [CrossRef]
- Lesburg, C.A.; Zhai, G.Z.; Cane, D.E.; Christianson, D.W. Crystal structure of pentalenene synthase: Mechanistic insights on terpenoid cyclization reactions in biology. Science 1997, 277, 1820–1824. [Google Scholar] [CrossRef]
- Seemann, M.; Zhai, G.Z.; de Kraker, J.W.; Paschall, C.M.; Christianson, D.W.; Cane, D.E. Pentalenene synthase. Analysis of active site residues by site-directed mutagenesis. J. Am. Chem. Soc. 2002, 124, 7681–7689. [Google Scholar] [CrossRef]
- Matos, J.O.; Kumar, R.P.; Ma, A.C.; Patterson, M.; Krauss, I.J.; Oprian, D.D. Mechanism underlying anti-Markovnikov addition in the reaction of pentalenene synthase. Biochemistry 2020, 59, 3271–3283. [Google Scholar] [CrossRef] [PubMed]
- Seemann, M.; Zhai, G.Z.; Umezawa, K.; Cane, D. Pentalenene synthase. Histidine-309 is not required for catalytic activity. J. Am. Chem. Soc. 1999, 121, 591–592. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Q.; Cao, Y.; Feng, X.; Zheng, Y.; Zou, H.; Liu, H.; Yang, J.; Xian, M. Microbial production of sabinene—A new terpene-based precursor of advanced biofuel. Microb. Cell Fact. 2014, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, Q.; Li, L.; Qin, W.; Yang, J.; Zhang, H.; Jiang, X.; Cheng, T.; Liu, W.; Xu, X.; et al. Metabolic engineering of Escherichia coli for high-specificity production of isoprenol and prenol as next generation of biofuels. Biotechnol. Biofuels 2013, 6, 57. [Google Scholar] [CrossRef]
- Mo, J.Y.; Maki, H.; Sekiguchi, M. Mutational specificity of the dnae173 mutator associated with a defect in the catalytic subunit of DNA polymerase III of Escherichia coli. J. Mol. Biol. 1991, 222, 925–936. [Google Scholar] [CrossRef]
- O’Maille, P.E.; Malone, A.; Dellas, N.; Hess, B.A.; Smentek, L.; Sheehan, I.; Greenhagen, B.T.; Chappell, J.; Manning, G.; Noel, J.P. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 2008, 4, 617–623. [Google Scholar] [CrossRef]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinf. 2006, 54, 5–6. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new endscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Sanner, M.F.; Olson, A.J.; Forli, S. The autodock suite at 30. Protein Sci. 2021, 30, 31–43. [Google Scholar] [CrossRef]
- Rosignoli, S.; Paiardini, A. Boosting the full potential of pymol with structural biology plugins. Biomolecules 2022, 12, 1764. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Melillo, E.; Muntendam, R.; Quax, W.J.; Kayser, O. Heterologous expression of pentalenene synthase (PSS) from Streptomyces UC5319 in Xanthophyllomyces dendrorhous. J. Biotechnol. 2012, 161, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Faraldos, J.A.; Allemann, R.K. Inhibition of (+)-aristolochene synthase with iminium salts resembling eudesmane cation. Org. Lett. 2011, 13, 1202–1205. [Google Scholar] [CrossRef]
- Hong, Y.J.; Tantillo, D.J. Branching out from the bisabolyl cation. Unifying mechanistic pathways to barbatene, bazzanene, chamigrene, chamipinene, cumacrene, cuprenene, dunniene, isobazzanene, iso-γ-bisabolene, isochamigrene, laurene, microbiotene, sesquithujene, sesquisabinene, thujopsene, trichodiene, and widdradiene sesquiterpenes. J. Am. Chem. Soc. 2014, 136, 2450–2463. [Google Scholar] [CrossRef]
- Kumeta, Y.; Ito, M. Characterization of δ-guaiene synthases from cultured cells of Aquilaria, responsible for the formation of the sesquiterpenes in agarwood. Plant Physiol. 2010, 154, 1998–2007. [Google Scholar] [CrossRef]
- Salmon, M.; Laurendon, C.; Vardakou, M.; Cheema, J.; Defernez, M.; Green, S.; Faraldos, J.A.; O’Maille, P.E. Emergence of terpene cyclization in Artemisia annua. Nat. Commun. 2015, 6, 6143. [Google Scholar] [CrossRef]
- Harris, G.G.; Lombardi, P.M.; Pemberton, T.A.; Matsui, T.; Weiss, T.M.; Cole, K.E.; Koksal, M.; Murphy, F.V.; Vedula, L.S.; Chou, W.K.W.; et al. Structural studies of geosmin synthase, a bifunctional sesquiterpene synthase with αα domain architecture that catalyzes a unique cyclization-fragmentation reaction sequence. Biochemistry 2015, 54, 7142–7155. [Google Scholar] [CrossRef]
- Gutta, P.; Tantillo, D.J. Theoretical studies on farnesyl cation cyclization: Pathways to pentalenene. J. Am. Chem. Soc. 2006, 128, 6172–6179. [Google Scholar] [CrossRef]
- Zu, L.S.; Xu, M.M.; Lodewyk, M.W.; Cane, D.E.; Peters, R.J.; Tantillo, D.J. Effect of isotopically sensitive branching on product distribution for pentalenene synthase: Support for a mechanism predicted by quantum chemistry. J. Am. Chem. Soc. 2012, 134, 11369–11371. [Google Scholar] [CrossRef]
- Ma, L.T.; Lee, Y.R.; Liu, P.L.; Cheng, Y.T.; Shiu, T.F.; Tsao, N.W.; Wang, S.Y.; Chu, F.H. Phylogenetically distant group of terpene synthases participates in cadinene and cedrane-type sesquiterpenes accumulation in Taiwania cryptomerioides. Plant Sci. 2019, 289, 110277. [Google Scholar] [CrossRef] [PubMed]
- Shitole, H.R.; Vyas, P.; Nayak, U.R. Alloisolongifolene, a unique acid-catalyzed isomer of longifolene. Tetrahedron Lett. 1984, 23, 702–706. [Google Scholar] [CrossRef]
- Berson, J.A.; Hammons, J.H.; McRowe, A.W.; Bergman, R.G.; Remanick, A.; Houston, D. Chemistry of methylnorbornyl cations. VI. The stereochemistry of vicinal hydride shift. Evidence for the nonclassical structure of 3-methyl-2-norbornyl cations. J. Am. Chem. Soc. 1967, 89, 2590–2600. [Google Scholar] [CrossRef]
- Lodewyk, M.W.; Gutta, P.; Tantillo, D.J. Computational studies on biosynthetic carbocation rearrangements leading to sativene, cyclosativene, α-ylangene, and β-ylangene. J. Org. Chem. 2008, 73, 6570–6579. [Google Scholar] [CrossRef]
- Dickschat, J.S.; Brock, N.L.; Citron, C.A.; Tudzynski, B. Biosynthesis of sesquiterpenes by the fungus Fusarium verticillioides. ChemBioChem 2011, 12, 2088–2095. [Google Scholar] [CrossRef]
- Yarovaya, O.I.; Polovinka, M.P.; Korchagina, D.V.; Gatilov, Y.V.; Bagryanskaya, I.Y.; Shcherbukhin, V.V.; Shal’ko, A.A.; Zenkovets, G.A.; Barkhash, V.A. Acid-catalyzed rearrangements of (-)-thujopsene. Russ. J. Org. Chem. 2001, 37, 362–374. [Google Scholar] [CrossRef]
- Kumari, I.; Ahmed, M.; Akhter, Y. Evolution of catalytic microenvironment governs substrate and product diversity in trichodiene synthase and other terpene fold enzymes. Biochimie 2018, 144, 9–20. [Google Scholar] [CrossRef]
- Yoshikuni, Y.; Martin, V.J.J.; Ferrin, T.E.; Keasling, J.D. Engineering cotton (+)-δ-cadinene synthase to an altered function: Germacrene D-4-ol synthase. Chem. Biol. 2006, 13, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Kersten, R.D.; Diedrich, J.K.; Yates, J.R., III; Noel, J.P. Mechanism-based post-translational modification and inactivation in terpene synthases. ACS Chem. Biol. 2015, 10, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.W.; Li, J.X.; Liu, Z.X.; Mitsuhashi, T.; Zhang, Y.T.; Liu, H.L.; Ma, Y.H.; He, J.; Shinada, T.; Sato, T.; et al. Molecular basis for sesterterpene diversity produced by plant terpene synthases. Plant Commun. 2020, 1, 100051. [Google Scholar] [CrossRef]
- Srivastava, P.L.; Escorcia, A.M.; Huynh, F.; Miller, D.J.; Allemann, R.K.; van der Kamp, M.W. Redesigning the molecular choreography to prevent hydroxylation in germacradien-11-ol synthase catalysis. ACS Catal. 2021, 11, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Shukal, S.; Chen, X.X.; Zhang, C.Q. Systematic engineering for high-yield production of viridiflorol and amorphadiene in auxotrophic Escherichia coli. Metab. Eng. 2019, 55, 170–178. [Google Scholar] [CrossRef]
- Kollner, T.G.; Schnee, C.; Gershenzon, J.; Degenhardt, J. The variability of sesquiterpenes emitted from two Zea mays cultivars is controlled by allelic variation of two terpene synthase genes encoding stereoselective multiple product enzymes. Plant Cell 2004, 16, 1115–1131. [Google Scholar] [CrossRef] [PubMed]
- Vattekkatte, A.; Boland, W. Biosynthetic origin of complex terpenoid mixtures by multiproduct enzymes, metal cofactors, and substrate isomers. Nat. Prod. Chem. Res. 2020, 8, 372. [Google Scholar] [CrossRef]
- Yang, J.; Xian, M.; Su, S.; Zhao, G.; Nie, Q.; Jiang, X.; Zheng, Y.; Liu, W. Enhancing production of bio-isoprene using hybrid MVA pathway and isoprene synthase in E. coli. PLoS ONE 2012, 7, e33509. [Google Scholar] [CrossRef]

| Name | Relevant Genotype | Sources |
|---|---|---|
| Plasmids | ||
| pACYCDuet-1 | P15A ori; CmR; PT7 | Novagen |
| pTrcHis2B | pBR322 ori; AmpR; Ptrc | Invitrogen |
| pET28a | pBR322 ori, f1 ori; KanR; PT7 | Novagen |
| pA | P15A ori; CmR; PT7:: mvaE-mvaS | [15] |
| pT | pBR322 ori; AmpR; Ptrc:: ERG12-ERG8-ERG19 | Lab stock |
| pE | pBR322 ori, f1 ori; KanR; PT7:: PentS (WT or mutants)-ispA-IDI | This work |
| Strains | ||
| E. coli Trans1-T1 | F-ψ80 (lacZ)ΔM15ΔlacX74 hsdR (rk−,mk+)ΔrecA1398endA1tonA | TransGen Biotech |
| E. coli BL21(DE3) | E. coli B dcm ompT hsdS(rB− mB−) gal | TaKaRa |
| EC1 | E. coli BL21(DE3) harboring pA | This work |
| EC2 | E. coli BL21(DE3) harboring pA, pT and pE | This work |
| Chassis | Name | Source | Titer (mg/L) | Reference |
|---|---|---|---|---|
| Saccharomyces cerevisiae | PentS | Streptomyces sp. UC5319 | 334.7 | [5] |
| Xanthophyllomyces dendrorhous | PPS | Streptomyces sp. UC5319 | 0.25–0.68 | [23] |
| E. coli | PentS | Streptomyces sp. UC5319 | 780.3 | [5] |
| E. coli | PentS | Streptomyces sp. PSKA01 | 2130 | This work |
| Variant | VVdW | Polarity | VCavity | ∆G | DO-C | Variant | VVdW | Polarity | VCavity | ∆G | DO-C |
|---|---|---|---|---|---|---|---|---|---|---|---|
| T182 | 93 | P | 797.82 | −9.19 | 4.6 | T182I | 124 | H | 779.96 | −8.67 | 7.2 |
| T182F | 135 | H | 760.11 | −8.31 | 10.5 | T182C | 86 | P | 801.36 | −9.06 | 4.7 |
| T182W | 163 | H | 730.21 | −7.30 | 9.5 | T182A | 67 | H | 824.06 | −9.10 | 4.6 |
| T182M | 124 | H | 768.73 | −7.56 | 9.2 | T182V | 105 | H | 790.29 | −8.93 | 4.6 |
| T182K | 135 | C+ | 756.24 | −8.37 | 9.0 | T182S | 73 | P | 815.81 | −9.16 | 4.6 |
| T182R | 148 | C+ | 745.56 | −6.53 | 8.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Fang, S.; Zhao, L.; Men, X.; Zhang, H. A Single Active-Site Mutagenesis Confers Enhanced Activity and/or Changed Product Distribution to a Pentalenene Synthase from Streptomyces sp. PSKA01. Bioengineering 2023, 10, 392. https://doi.org/10.3390/bioengineering10030392
Liu H, Fang S, Zhao L, Men X, Zhang H. A Single Active-Site Mutagenesis Confers Enhanced Activity and/or Changed Product Distribution to a Pentalenene Synthase from Streptomyces sp. PSKA01. Bioengineering. 2023; 10(3):392. https://doi.org/10.3390/bioengineering10030392
Chicago/Turabian StyleLiu, Hongshuang, Senbiao Fang, Lin Zhao, Xiao Men, and Haibo Zhang. 2023. "A Single Active-Site Mutagenesis Confers Enhanced Activity and/or Changed Product Distribution to a Pentalenene Synthase from Streptomyces sp. PSKA01" Bioengineering 10, no. 3: 392. https://doi.org/10.3390/bioengineering10030392
APA StyleLiu, H., Fang, S., Zhao, L., Men, X., & Zhang, H. (2023). A Single Active-Site Mutagenesis Confers Enhanced Activity and/or Changed Product Distribution to a Pentalenene Synthase from Streptomyces sp. PSKA01. Bioengineering, 10(3), 392. https://doi.org/10.3390/bioengineering10030392