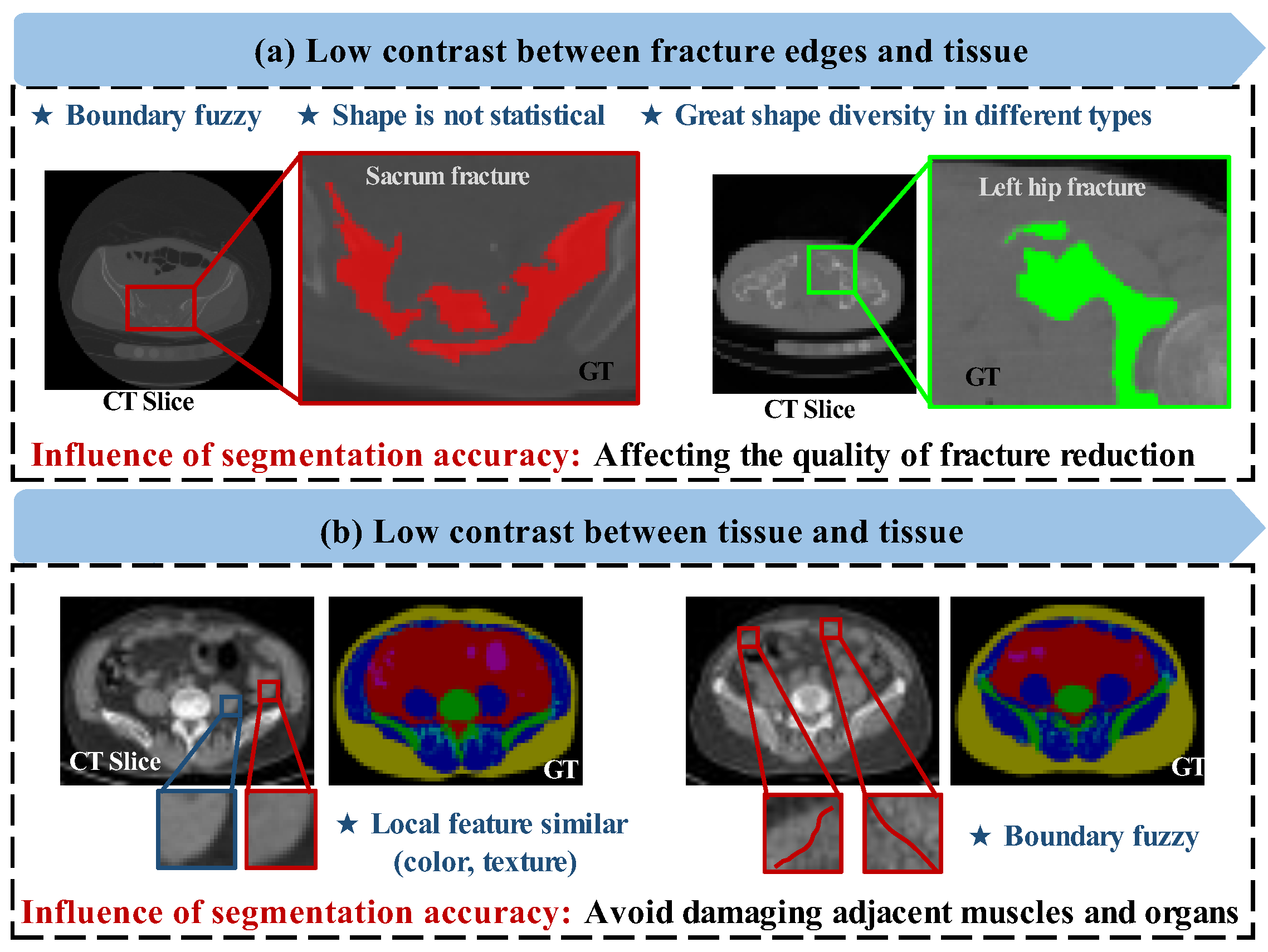
Figure 1.
Some difficult samples of pelvic CT slices. (a) Low contrast between fracture edges and tissue. (b) Low contrast and local similarity between tissue and tissue. The boxes are the “interested region”, and the colors used are their corresponding GT marking color. The difficulties of segmentation are listed with asterisks.
Figure 1.
Some difficult samples of pelvic CT slices. (a) Low contrast between fracture edges and tissue. (b) Low contrast and local similarity between tissue and tissue. The boxes are the “interested region”, and the colors used are their corresponding GT marking color. The difficulties of segmentation are listed with asterisks.
Figure 2.
Schematic view of the proposed bi-directional constrained dual-task consistency method on CTPelvic1k dataset. The framework consists of two branches: supervised learning and semi-supervised learning. The below semi-supervised part is the combination of interpolation consistency regularization task and pseudo-label supervision task. The backbone models share the same architecture, and the weight of teacher model is the exponential moving average (EMA) of the student model. The “PDF” in the interpolation part represents the probability density function of beta distribution, “α” and “β” are the parameter of beta distribution, “μ” is the interpolation factor.
Figure 2.
Schematic view of the proposed bi-directional constrained dual-task consistency method on CTPelvic1k dataset. The framework consists of two branches: supervised learning and semi-supervised learning. The below semi-supervised part is the combination of interpolation consistency regularization task and pseudo-label supervision task. The backbone models share the same architecture, and the weight of teacher model is the exponential moving average (EMA) of the student model. The “PDF” in the interpolation part represents the probability density function of beta distribution, “α” and “β” are the parameter of beta distribution, “μ” is the interpolation factor.
Figure 3.
Illustration of interpolation consistency regularization, taking the Multi-tissue Pelvic dataset as an example. (a) describes the data augmentation process by pixel level interpolation, “μ” is the interpolation factor and follows the Beta distribution. The “PDF” represents the probability-density function of Beta distribution, and “α” and “β” are the parameters of beta distribution. (b) is the interpolation consistency regularization process.
Figure 3.
Illustration of interpolation consistency regularization, taking the Multi-tissue Pelvic dataset as an example. (a) describes the data augmentation process by pixel level interpolation, “μ” is the interpolation factor and follows the Beta distribution. The “PDF” represents the probability-density function of Beta distribution, and “α” and “β” are the parameters of beta distribution. (b) is the interpolation consistency regularization process.
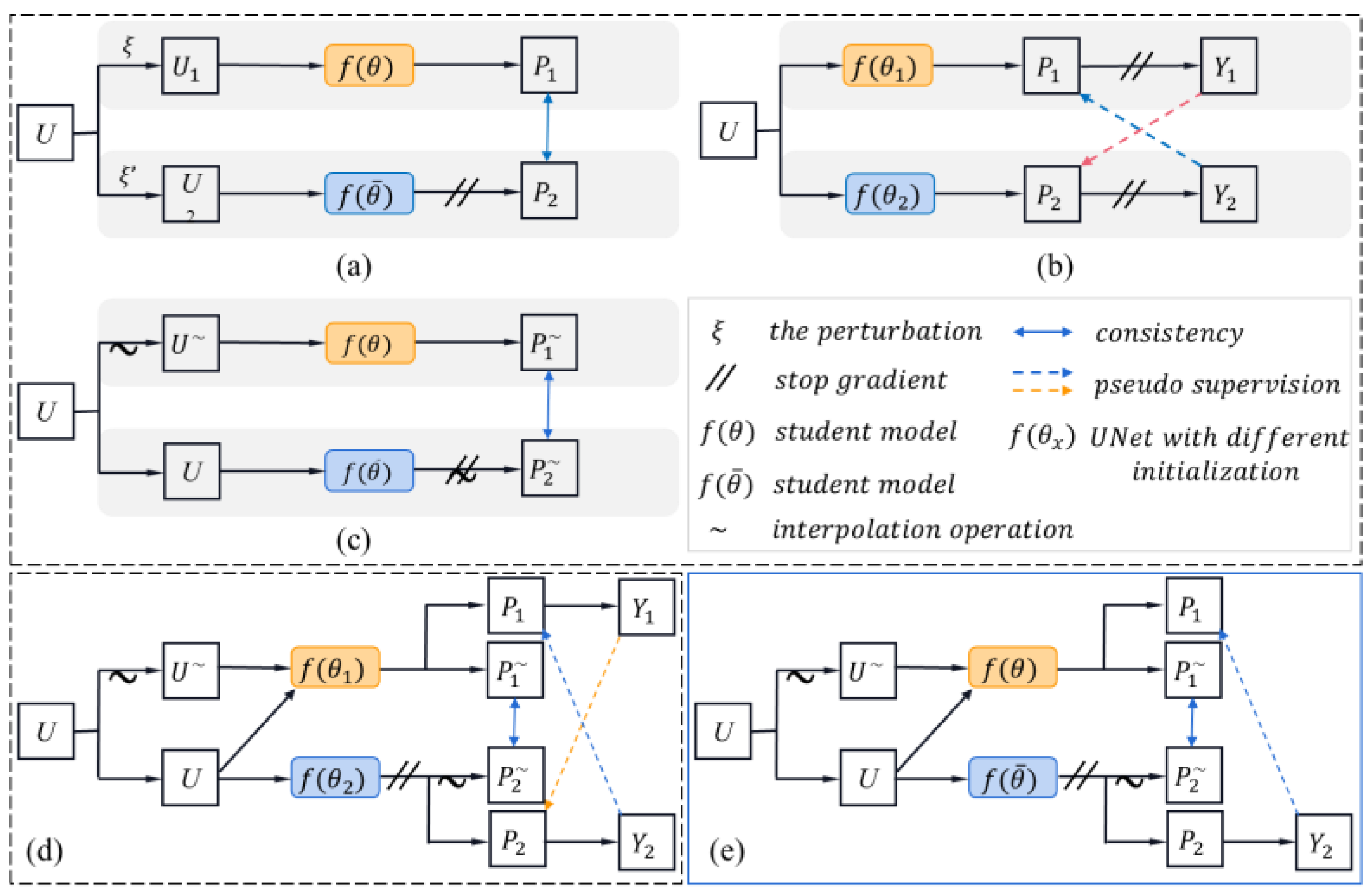
Figure 4.
Illustration of the diagrammatic architecture for related works; only the semi-supervised part is given here. (a) mean teacher, (b) cross-pseudo-supervision, (c) interpolation consistency training, (d) cross-pseudo-interpolation consistency learning, (e) pseudo-interpolation consistency training. (a–c) are the related advanced works, (d,e) are the proposed schemes. U denotes the unlabeled data, P is the prediction map, Y represents the pseudo-label, and represent network with different initialization. MT can be considered as the baseline for all work. Here, MT’ is used to represent two networks with different initialization. Therein, (b) can represented by MT’ + CP, (c) can be denoted as I + MT, (d) can be denoted as MT’ + CP + I, and (e) can be denoted as I + MT + P.
Figure 4.
Illustration of the diagrammatic architecture for related works; only the semi-supervised part is given here. (a) mean teacher, (b) cross-pseudo-supervision, (c) interpolation consistency training, (d) cross-pseudo-interpolation consistency learning, (e) pseudo-interpolation consistency training. (a–c) are the related advanced works, (d,e) are the proposed schemes. U denotes the unlabeled data, P is the prediction map, Y represents the pseudo-label, and represent network with different initialization. MT can be considered as the baseline for all work. Here, MT’ is used to represent two networks with different initialization. Therein, (b) can represented by MT’ + CP, (c) can be denoted as I + MT, (d) can be denoted as MT’ + CP + I, and (e) can be denoted as I + MT + P.
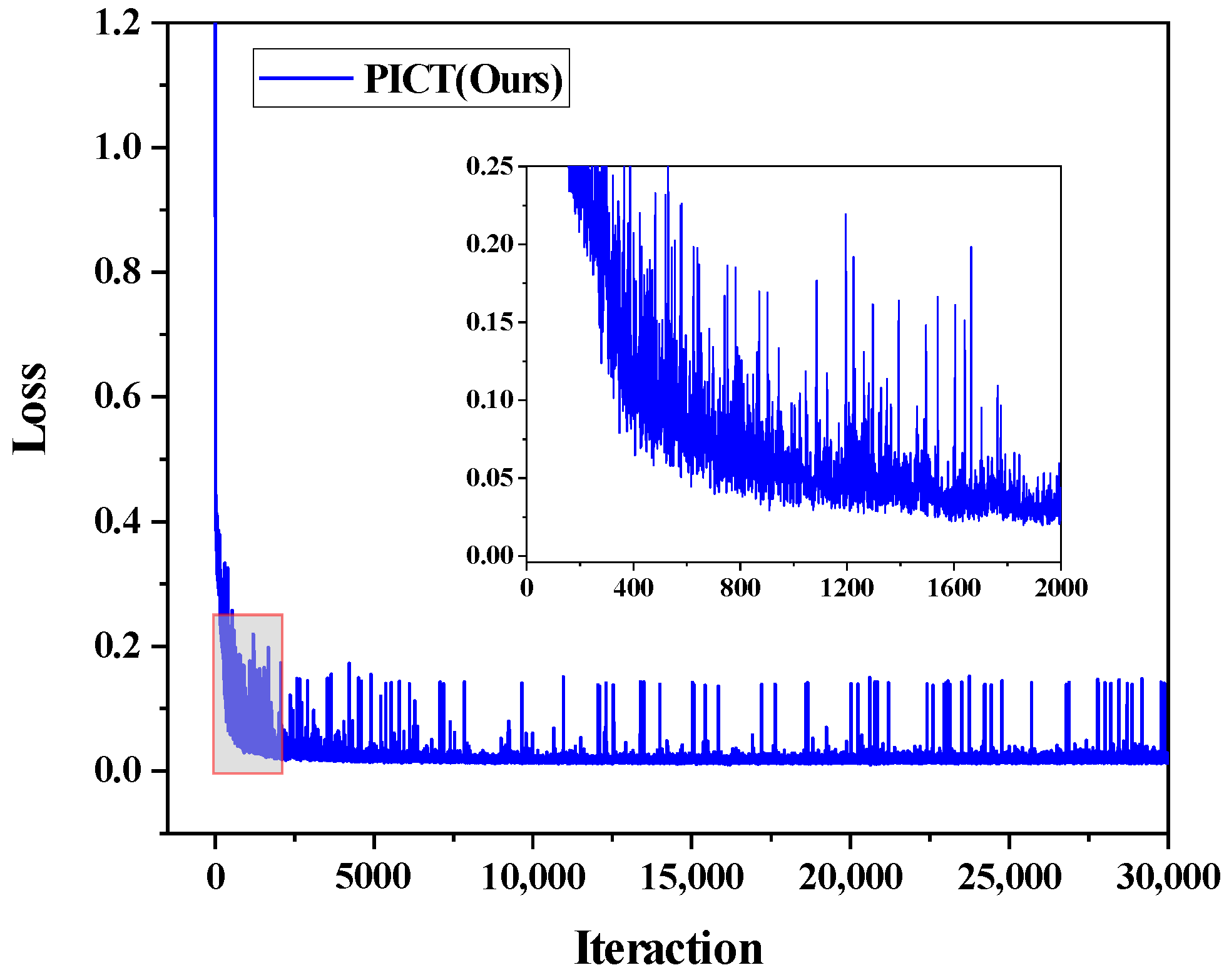
Figure 5.
The loss function of the proposed PICT on ACDC dataset with three labeled cases.
Figure 5.
The loss function of the proposed PICT on ACDC dataset with three labeled cases.
Figure 6.
Visual comparison on ACDC test images of the state-of-the-art methods. The top two rows are the results of using 3 labeled cases; the bottom two rows are the results of using 7 labeled cases. The yellow arrows indicate the misclassify situation of target; the yellow boxes indicate the misprediction in other area.
Figure 6.
Visual comparison on ACDC test images of the state-of-the-art methods. The top two rows are the results of using 3 labeled cases; the bottom two rows are the results of using 7 labeled cases. The yellow arrows indicate the misclassify situation of target; the yellow boxes indicate the misprediction in other area.
Figure 7.
The mean score point-fold line of different methods on CTPelvic1k dataset with different ratio labeled cases.
Figure 7.
The mean score point-fold line of different methods on CTPelvic1k dataset with different ratio labeled cases.
Figure 8.
Representative cases of three methods on CTPelvic1k dataset. The green, blue, yellow, and red parts of the visual model represent the left hip bone, right hip bone, lumbar spine, and sacrum, respectively.
Figure 8.
Representative cases of three methods on CTPelvic1k dataset. The green, blue, yellow, and red parts of the visual model represent the left hip bone, right hip bone, lumbar spine, and sacrum, respectively.
Figure 9.
The mean score point-fold line of different methods on Multi-tissue Pelvic dataset with different ratio labeled cases.
Figure 9.
The mean score point-fold line of different methods on Multi-tissue Pelvic dataset with different ratio labeled cases.
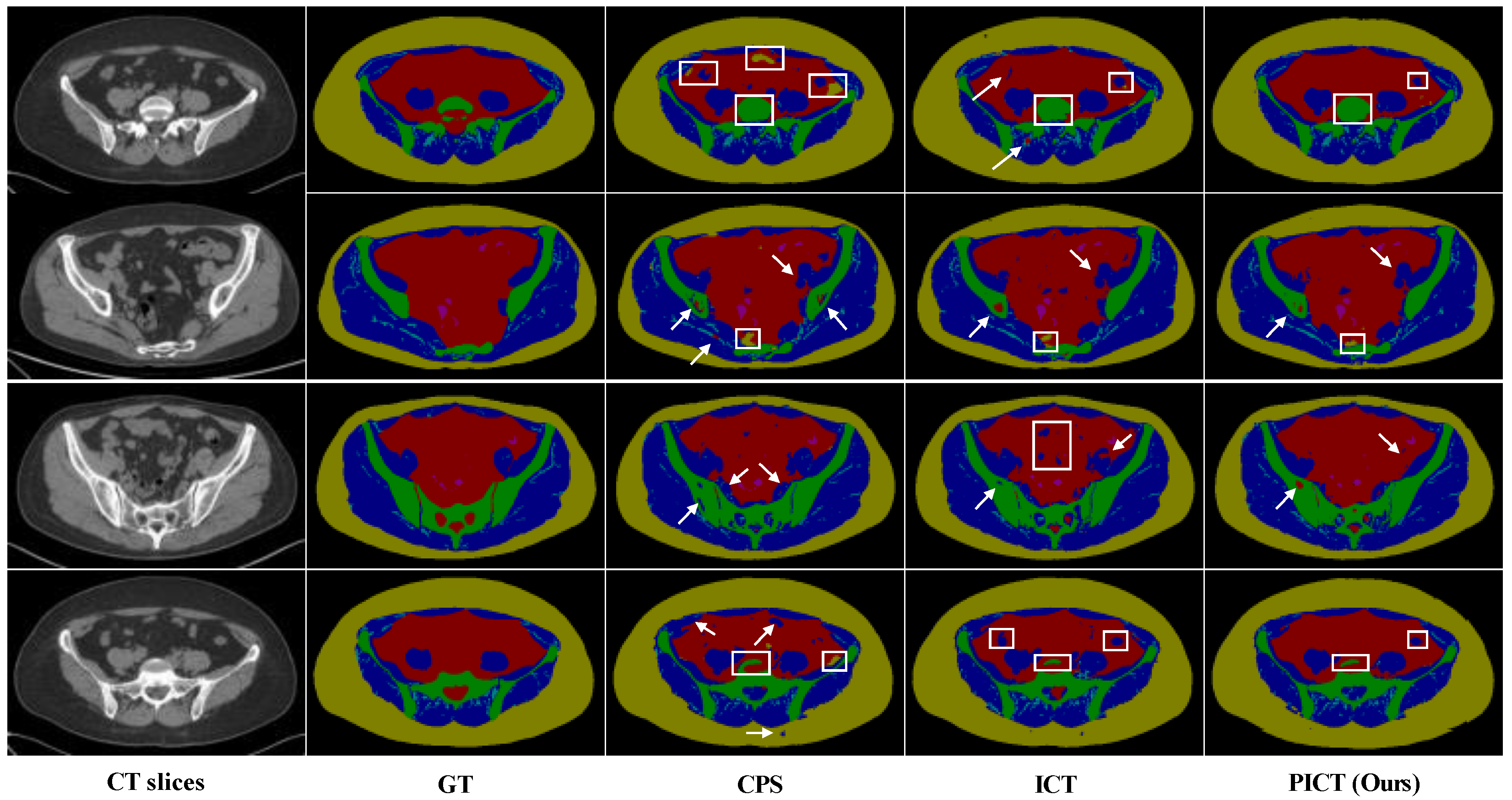
Figure 10.
Visual comparison of the state-of-the-art methods on Multi-tissues Pelvic test images. The top two rows are the results of using 35 labeled cases; the bottom two rows are the results of using 40 labeled cases. The white arrows and boxes indicate the misclassified pixels.
Figure 10.
Visual comparison of the state-of-the-art methods on Multi-tissues Pelvic test images. The top two rows are the results of using 35 labeled cases; the bottom two rows are the results of using 40 labeled cases. The white arrows and boxes indicate the misclassified pixels.
Table 1.
Dataset statistics of CTPelvic1k, Multi-tissue Pelvic, and ACDC.
Table 1.
Dataset statistics of CTPelvic1k, Multi-tissue Pelvic, and ACDC.
| Parameters | Multi-Tissue Pelvic | CTPelvic1k | ACDC |
|---|
| Mean size | 512 × 512 | 512 × 512 × 345 | 256 × 256 × 10 |
| Data number | 100 | 70 | 200 |
| Tra/Val/Ts | 80/5/15 | 50/10/10 | 140/20/40 |
| Category | 7 | 5 | 4 |
| Type | Individual | Public | Public |
| Pre-process | Center crop | Center crop | None |
| Patch size | 256 × 256 | 112 × 112 × 112 | 256 × 256 |
Table 2.
Ablation analysis results on ACDC dataset using 3 labeled cases. The bold fonts indicate the best results of the comparing variants.
Table 2.
Ablation analysis results on ACDC dataset using 3 labeled cases. The bold fonts indicate the best results of the comparing variants.
| Method | RV | Myo | LV | Mean |
|---|
|
|
|
|
|
|
|
|
|---|
| MT [33] | 56.77 | 37.83 | 67.76 | 16.61 | 76.46 | 24.37 | 67.00 | 26.27 |
| CPS (MT’ + CP) [35] | 61.46 | 18.59 | 70.90 | 12.54 | 78.67 | 18.73 | 70.34 | 16.62 |
| ICT (I + MT) [16] | 57.49 | 20.28 | 71.17 | 14.93 | 79.69 | 25.09 | 69.45 | 20.10 |
| CPICT (CP + I + MT) | 58.22 | 16.91 | 74.44 | 11.00 | 81.34 | 24.67 | 71.33 | 17.53 |
| PICT (P + I + MT) | 63.27 | 21.40 | 77.90 | 10.63 | 77.09 | 34.97 | 72.76 | 22.33 |
| PICT (CE) | 63.27 | 21.40 | 77.90 | 10.63 | 77.09 | 34.97 | 72.76 | 22.33 |
| PICT (CE + Dice) | 69.08 | 21.99 | 76.77 | 21.54 | 76.44 | 40.59 | 74.10 | 28.04 |
| PICT (Dice, Ours) | 80.06 | 15.25 | 82.42 | 7.07 | 83.39 | 25.96 | 81.95 | 16.09 |
Table 3.
The DSC score (%) comparison of CPS, ICT, and PICT on ACDC dataset under different basic models with using 7 labeled cases. The bold fonts indicate the best results of the comparing variants.
Table 3.
The DSC score (%) comparison of CPS, ICT, and PICT on ACDC dataset under different basic models with using 7 labeled cases. The bold fonts indicate the best results of the comparing variants.
| Type of Basic Models | LS | CPS | ICT | PICT (Ours) |
|---|
| E-Net [43] | 67.71 | 75.54 | 74.83 | 76.66 |
| P-Net [44] | 80.71 | 83.47 | 82.83 | 83.76 |
| U-Net [40] | 81.92 | 86.35 | 86.64 | 87.18 |
Table 4.
Quantitative comparison results of mean DSC score and of the state-of-art method on ACDC under 3 labeled cases and 7 labeled cases. The bold fonts indicate the best results of the comparing variants.
Table 4.
Quantitative comparison results of mean DSC score and of the state-of-art method on ACDC under 3 labeled cases and 7 labeled cases. The bold fonts indicate the best results of the comparing variants.
| Labled Number | Method | RV | Myo | LV | Mean |
|---|
|
|
|
|
|
|
|
|
|---|
| 3 cases | LS (baseline) | 48.11 | 45.76 | 62.76 | 23.27 | 72.06 | 24.87 | 60.98 | 31.30 |
| FS | 91.15 | 1.23 | 88.62 | 5.95 | 93.58 | 5.62 | 91.12 | 4.26 |
| MT [33] | 56.77 | 37.83 | 67.78 | 16.61 | 76.46 | 24.37 | 67.00 | 26.27 |
| UAMT [21] | 57.86 | 32.10 | 67.32 | 14.57 | 76.00 | 20.81 | 67.06 | 22.49 |
| URPC [45] | 63.73 | 33.13 | 69.59 | 15.95 | 79.19 | 18.61 | 70.89 | 22.56 |
| CPS [35] | 61.46 | 18.59 | 70.90 | 12.54 | 78.67 | 18.73 | 70.34 | 16.62 |
| ICT [16] | 57.48 | 20.28 | 71.17 | 14.83 | 79.69 | 25.09 | 69.45 | 20.06 |
| CNN-Trans [39] | 57.70 | 21.70 | 62.80 | 11.50 | 76.30 | 15.70 | 65.60 | 16.20 |
| PICT (Ours) | 80.06 | 15.25 | 82.41 | 7.06 | 83.39 | 25.96 | 81.95 | 16.09 |
| 7 cases | LS (baseline) | 79.42 | 8.39 | 79.61 | 3.06 | 86.75 | 18.97 | 81.93 | 13.39 |
| FS | 91.15 | 1.23 | 88.62 | 5.95 | 93.58 | 5.62 | 91.12 | 4.26 |
| MT [33] | 86.31 | 4.73 | 83.39 | 8.81 | 88.32 | 17.11 | 86.01 | 10.22 |
| UAMT [21] | 84.96 | 4.98 | 83.46 | 9.16 | 89.20 | 14.89 | 85.87 | 9.68 |
| URPC [45] | 85.77 | 4.65 | 83.79 | 7.44 | 89.08 | 7.44 | 86.21 | 6.85 |
| CPS [35] | 86.09 | 3.64 | 84.31 | 9.66 | 88.63 | 13.16 | 86.35 | 8.82 |
| ICT [16] | 86.49 | 4.48 | 84.12 | 9.27 | 89.33 | 11.38 | 86.64 | 8.37 |
| CNN-Trans [39] | 84.80 | 7.80 | 84.40 | 6.90 | 90.10 | 11.20 | 86.40 | 8.6 |
| PICT (Ours) | 86.74 | 3.22 | 85.16 | 3.24 | 89.66 | 9.93 | 87.18 | 5.46 |
Table 5.
Comparison of DSC score (%) of CPS, ICT, and PICT on CTPelvic1k with 10 labeled images as examples. The bold fonts indicate the best results of the comparing variants.
Table 5.
Comparison of DSC score (%) of CPS, ICT, and PICT on CTPelvic1k with 10 labeled images as examples. The bold fonts indicate the best results of the comparing variants.
| Categories | Sacrum | LH | RH | LS |
|---|
| CPS [35] | 95.59 | 95.75 | 96.19 | 95.39 |
| ICT [16] | 95.98 | 96.05 | 95.82 | 95.81 |
| PICT (Ours) | 95.92 | 96.09 | 96.44 | 96.03 |
Table 6.
The mIoU (%) and mAcc for CPS, ICT, and PICT on Multi-tissue Pelvic with different ratio labeled cases. The bold fonts indicate the best results of the comparing variants.
Table 6.
The mIoU (%) and mAcc for CPS, ICT, and PICT on Multi-tissue Pelvic with different ratio labeled cases. The bold fonts indicate the best results of the comparing variants.
| Labeled Case | 5 | 10 | 15 | 20 | 25 | 30 | 35 | 40 |
|---|
| mIoU (%) | CPS [35] | 61.25 | 62.38 | 63.43 | 64.49 | 66.86 | 67.67 | 68.36 | 70.18 |
| ICT [16] | 59.36 | 60.80 | 63.51 | 64.62 | 67.63 | 68.32 | 69.29 | 69.05 |
| PICT (Ours) | 61.83 | 61.79 | 64.22 | 65.56 | 67.46 | 68.49 | 70.60 | 71.07 |
| mAcc (%) | CPS [35] | 73.85 | 74.73 | 75.15 | 77.15 | 78.56 | 79.62 | 79.11 | 80.97 |
| ICT [16] | 72.77 | 74.13 | 75.65 | 76.65 | 79.33 | 79.71 | 80.31 | 80.53 |
| PICT (Ours) | 73.84 | 74.30 | 76.60 | 77.92 | 78.91 | 79.97 | 80.60 | 81.75 |
Table 7.
Comparison of score (%), (%), (%) indexes of the 6 categories with 40 labeled images as an example on Multi-tissue Pelvic. The bold fonts indicate the best results of the comparing variants.
Table 7.
Comparison of score (%), (%), (%) indexes of the 6 categories with 40 labeled images as an example on Multi-tissue Pelvic. The bold fonts indicate the best results of the comparing variants.
| Categories | MIPC | Bone | Muscle | SAT | IMAT | IPG |
|---|
| DSC (%) | CPS [35] | 80.76 | 93.44 | 86.03 | 92.16 | 52.36 | 67.44 |
| ICT [16] | 78.99 | 93.58 | 86.25 | 90.97 | 52.55 | 63.67 |
| PICT (Ours) | 83.22 | 93.84 | 86.92 | 92.71 | 53.88 | 65.89 |
| mIoU (%) | CPS [35] | 69.21 | 87.93 | 75.78 | 85.83 | 36.09 | 66.24 |
| ICT [16] | 67.22 | 88.18 | 76.12 | 84.18 | 36.53 | 62.11 |
| PICT (Ours) | 72.54 | 88.62 | 77.17 | 86.78 | 37.51 | 64.46 |
| mAcc (%) | CPS [35] | 73.78 | 93.33 | 91.39 | 92.60 | 51.87 | 64.15 |
| ICT [16] | 70.35 | 93.40 | 92.80 | 91.93 | 55.20 | 60.36 |
| PICT (Ours) | 77.29 | 94.25 | 91.63 | 93.12 | 50.89 | 64.06 |
Table 8.
The time cost of ACDC. CTPelvic1k, Multi-tissue dataset; the unit is minutes.
Table 8.
The time cost of ACDC. CTPelvic1k, Multi-tissue dataset; the unit is minutes.
| Dataset | CPS | ICT | PICT (Ours) |
|---|
| ACDC | 132 | 100 | 105 |
| CTPelvic1k | 753 | 500 | 586 |
| Multi-tissue Pelvic | 31 | 24 | 26 |