Blockchain-Federated and Deep-Learning-Based Ensembling of Capsule Network with Incremental Extreme Learning Machines for Classification of COVID-19 Using CT Scans
Abstract
:1. Introduction
2. Literature Review
3. Materials and Methods
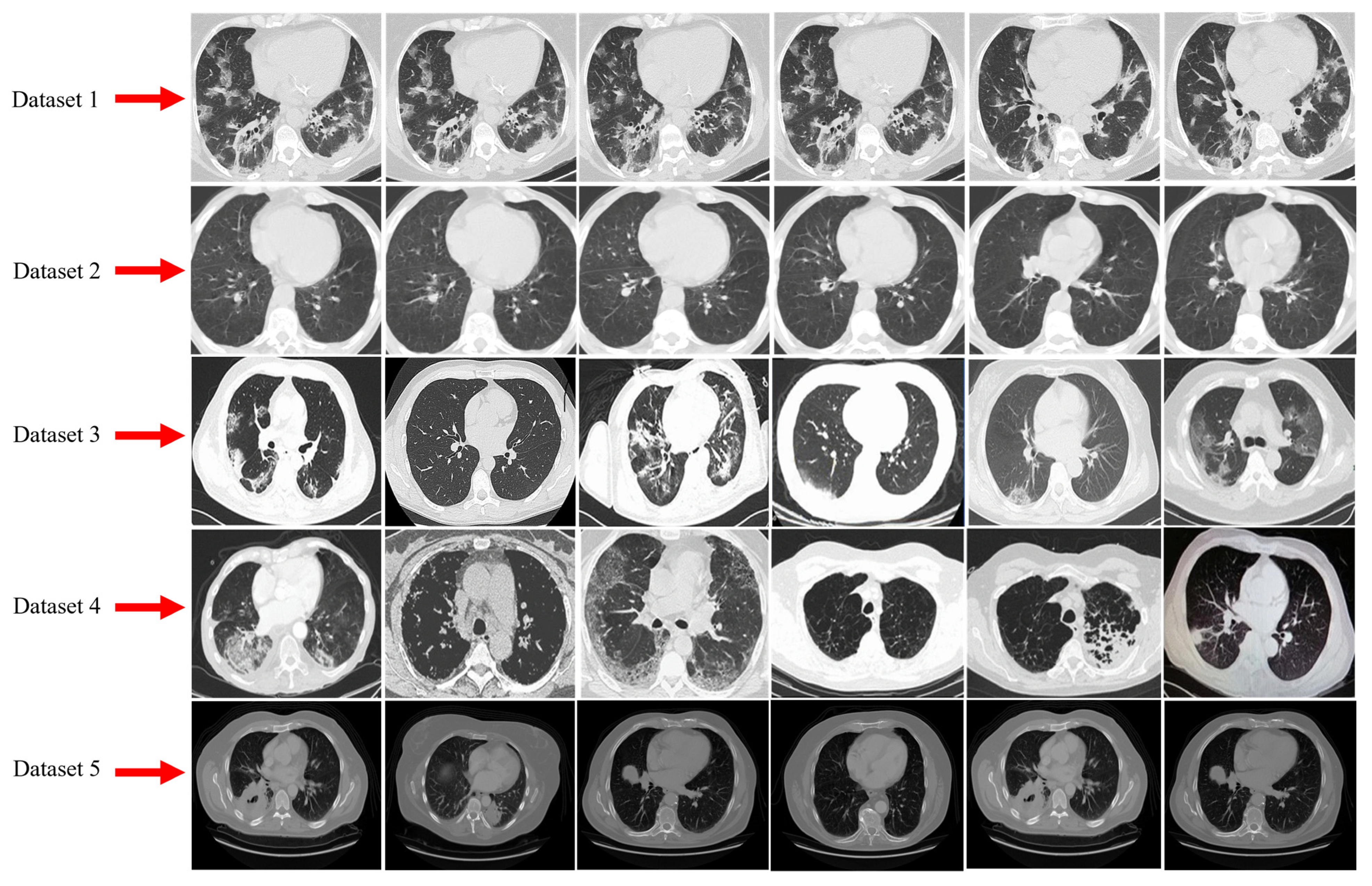
3.1. Dataset Description and Data Augmentation
3.1.1. Dataset-1
3.1.2. Dataset-2
3.1.3. Dataset-3
3.1.4. Dataset-4
3.1.5. Dataset-5
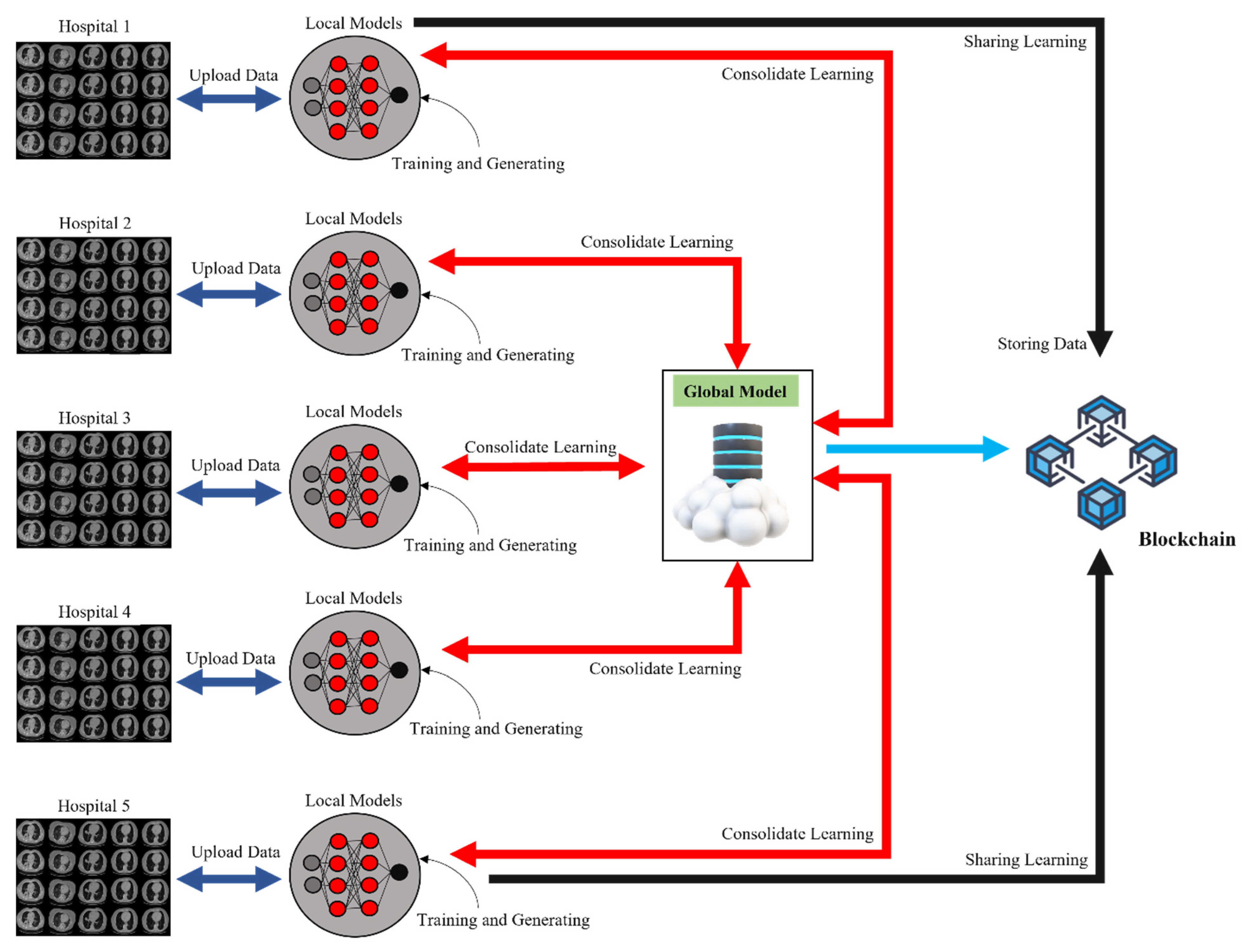
3.2. Overview of FL and Blockchain
3.3. Local Client Model
3.3.1. Data Normalization
- Spatial Normalization of COVID-19 CT scans
- Signal Normalization of COVID-19 CT scans
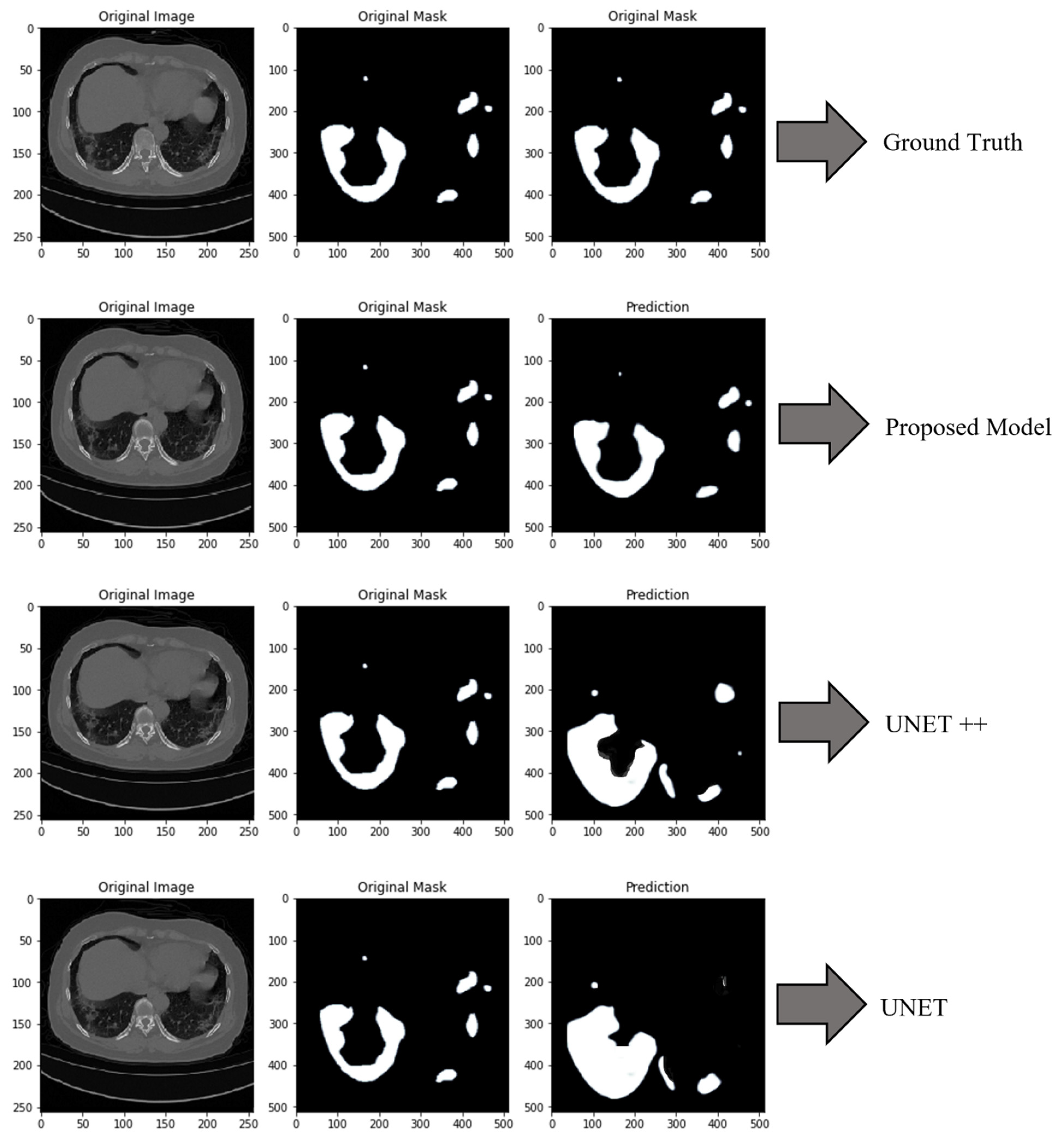
3.3.2. CT Scans Based Segmentation and Classification for COVID-19
3.3.3. Ensembling of CapsNet with IELMs
- CapsNet Architecture
| Algorithm 1: Routing Algorithm for Processing all Capsule |
| Input Parameters: Capsule = C; Layers = L; Weighted Sum = Ws |
| Output: Distributing the output of low-level to high-level capsule |
| 1. Foreach C in L: |
| 2. C (L + 1) do Ws = 0 |
| 3. While K = 1: |
| 4. C in L: |
| 5. do C(p,q) // see Equation (8) |
| 6. Foreach C in L + 1: |
| 7. do GK, SK // see Equation (7) |
| 8. Foreach C of j in L + 1: |
| 9. do E(a,b) // see Equation (5) |
| 10. Foreach C of K in L AND j in L + 1 |
| 11. do b(p,q) ← b(p,q) + |k |
| 12. Return b(p,q) |
- 2.
- IELMs
| Algorithm 2: Pseudo Code of Proposed CapsNet with IELMs |
| Input Parameters: Input Image = Iinput; No. of iteration = Niterations; Capsule = C; Image Features = Fimage; Threshold = Tthreshold; IELMs = EIELM; Output Image = Ooutput; Disease = D. |
| Output: Classification of COVID-19-infected CT scans |
| 1. For n =1: |
| 2. Fimage = C (Iinput) |
| 3. Execute Algorithm 1 |
| 4. Ooutput = EIELM (Fimage) // see Equation (12) |
| 5. If Ooutput == Tthreshold |
| 6. D is True // COVID-19 detected |
| 7. Else |
| 8. D is False // Normal |
| 9. End |
3.4. DL Classifiers
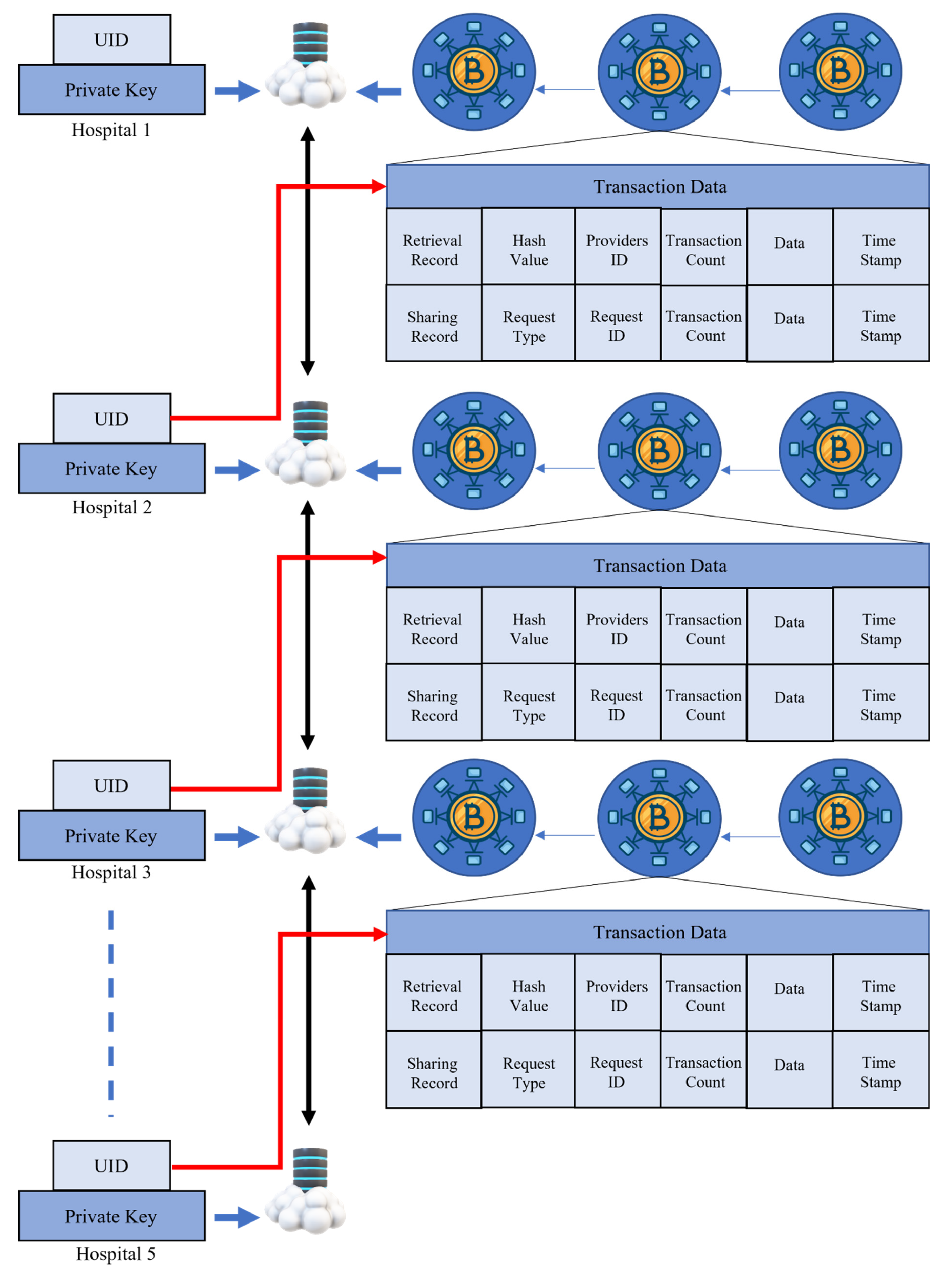
3.5. Blockchain-Based FL
- The local model Ti transaction is handed over from the Pi node to the Pj node.
- Local model Ti is sent up to the leader via node Pj.
- The leader sends out a broadcast to the Pi and Pj with the block node.
- Check that the Pi and Pj are correct, then wait for authorization.
- Finally, the blocks should be saved in the database of the retrieval blockchain.
3.6. Performance Evaluation
4. Results and Discussions
4.1. Experimental Setup
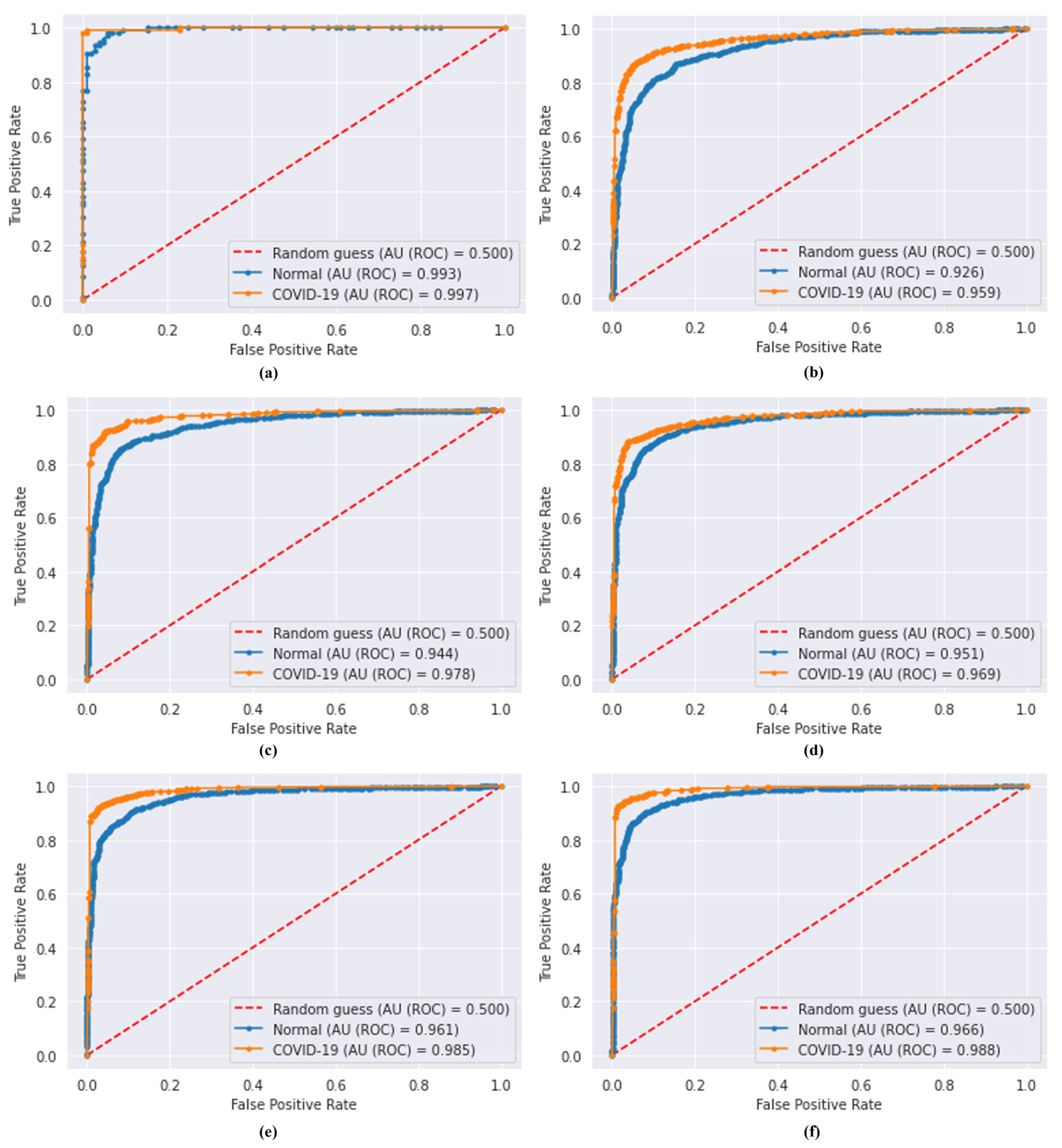
4.2. Comparison of the Proposed Model with DL Models
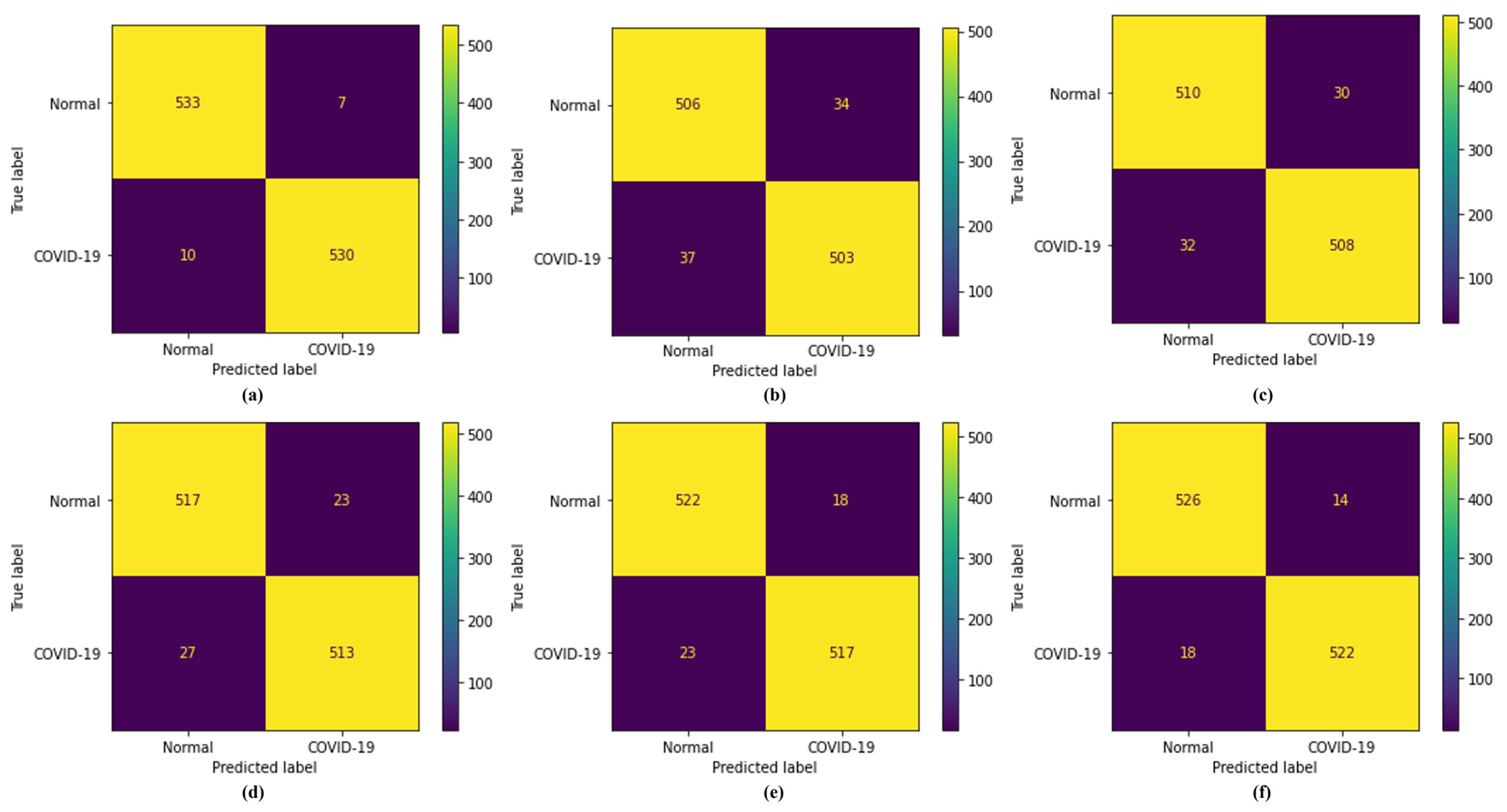
4.3. Analysis of FL Security
4.4. Comparison with State-of-the-Art Methods
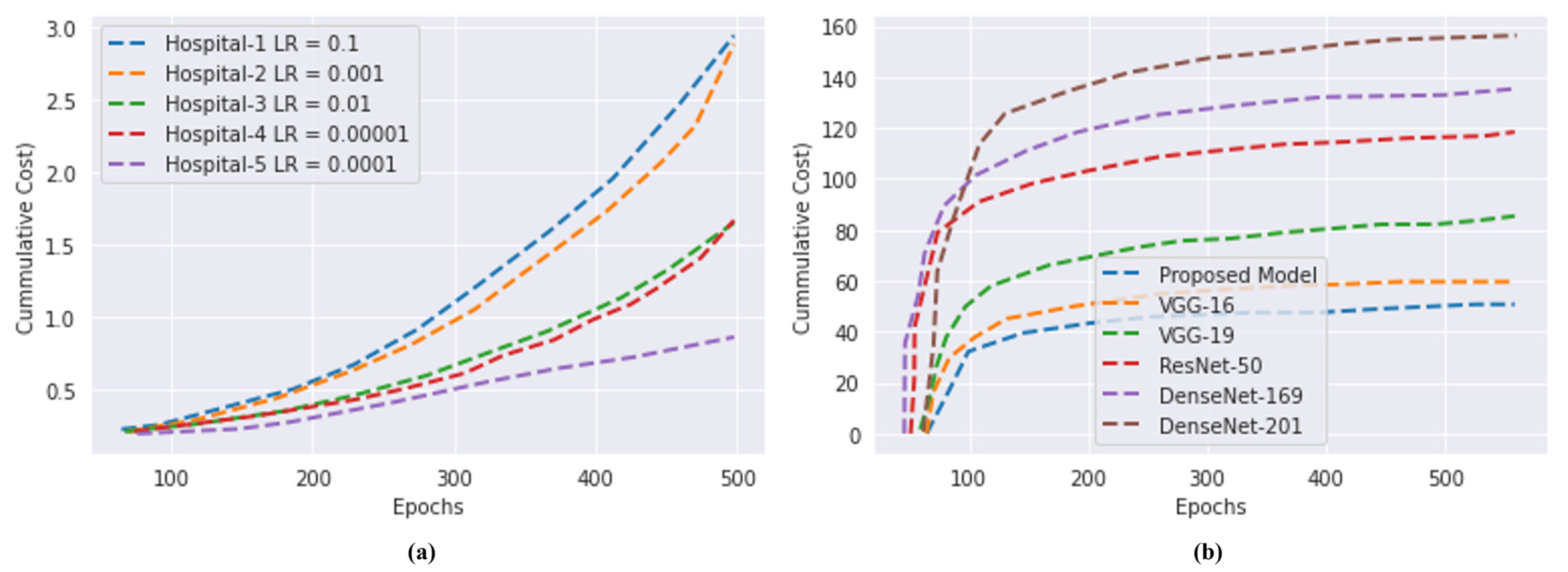
4.5. Computational Cost
4.6. Discussions
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, G.; Liu, X.; Li, C.; Xu, Z.; Ruan, J.; Zhu, H.; Meng, T.; Li, K.; Huang, N.; Zhang, S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging 2020, 39, 2653–2663. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Deng, X.; Fu, Q.; Zhou, Q.; Feng, J.; Ma, H.; Liu, W.; Zheng, C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging 2020, 39, 2615–2625. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Wei, B.; Hong, Y.; Li, T.; Cong, J.; Zhu, X.; Wei, H.; Zhang, W. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans. Med. Imaging 2020, 39, 2584–2594. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.; Huo, J.; Xia, L.; Shan, F.; Liu, J.; Mo, Z.; Yan, F.; Ding, Z.; Yang, Q.; Song, B.; et al. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging 2020, 39, 2595–2605. [Google Scholar] [CrossRef]
- Kang, H.; Xia, L.; Yan, F.; Wan, Z.; Shi, F.; Yuan, H.; Jiang, H.; Wu, D.; Sui, H.; Zhang, C.; et al. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans. Med. Imaging 2020, 39, 2606–2614. [Google Scholar] [CrossRef]
- Wang, J.; Bao, Y.; Wen, Y.; Lu, H.; Luo, H.; Xiang, Y.; Li, X.; Liu, C.; Qian, D. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imaging 2020, 39, 2572–2583. [Google Scholar] [CrossRef]
- Xu, G.; Li, H.; Liu, S.; Yang, K.; Lin, X. Verifynet: Secure and verifiable federated learning. IEEE Trans. Inf. Secur. 2020, 15, 911–926. [Google Scholar] [CrossRef]
- Lu, Y.; Huang, X.; Dai, Y.; Maharjan, S.; Zhang, Y. Blockchain and federated learning for privacy-preserved data sharing in industrial IoT. IEEE Trans. Ind. Inform. 2019, 16, 4177–4186. [Google Scholar] [CrossRef]
- Voulodimos, A.; Protopapadakis, E.; Katsamenis, I.; Doulamis, A.; Doulamis, N. Deep learning models for COVID-19 infected area segmentation in CT images. In Proceedings of the 14th PErvasive Technologies Related to Assistive Environments Conference, Corfu, Greece, 29 June–2 July 2021; pp. 404–411. [Google Scholar]
- Loddo, A.; Pili, F.; Di Ruberto, C. Deep learning for COVID-19 diagnosis from ct images. Appl. Sci. 2021, 11, 8227. [Google Scholar] [CrossRef]
- Nair, R.; Adi Alhudhaif, A.; Koundal, D.; Doewes, R.I.; Sharma, P. Deep learning-based COVID-19 detection system using pulmonary CT scans. Turk. J. Electr. Eng. Comput. Sci. 2021, 29, 2716–2727. [Google Scholar] [CrossRef]
- Fusco, R.; Grassi, R.; Granata, V.; Setola, S.V.; Grassi, F.; Cozzi, D.; Pecori, B.; Izzo, F.; Petrillo, A. Artificial intelligence and COVID-19 using chest CT scan and chest X-ray images: Machine learning and deep learning approaches for diagnosis and treatment. J. Pers. Med. 2021, 11, 993. [Google Scholar] [CrossRef]
- Majid, A.; Khan, M.A.; Nam, Y.; Tariq, U.; Roy, S.; Mostafa, R.R.; Sakr, R.H. COVID19 classification using CT images via ensembles of deep learning models. Comput. Mater. Contin. 2021, 69, 319–337. [Google Scholar] [CrossRef]
- Fouladi, S.; Ebadi, M.J.; Safaei, A.A.; Bajuri, M.Y.; Ahmadian, A. Efficient deep neural networks for classification of COVID-19 based on CT images: Virtualization via software defined radio. Comput. Commun. 2021, 176, 234–248. [Google Scholar] [CrossRef]
- Saood, A.; Hatem, I. COVID-19 lung CT image segmentation using deep learning methods: U-Net versus SegNet. BMC Med. Imaging 2021, 21, 19. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, S.; Chen, Y.; Luo, F.; Kang, Z.; Cai, S.; Zhao, W.; Liu, J.; Zhao, D.; Li, Y. A deep-learning-based framework for severity assessment of COVID-19 with CT images. Expert Syst. Appl. 2021, 185, 115616. [Google Scholar] [CrossRef]
- Shokri, R.; Shmatikov, V. Privacy-preserving deep learning. In Proceedings of the 22nd ACM SIGSAC Conference On Computer and Communications Security, Denver, CO, USA, 12–16 October 2015; pp. 1310–1321. [Google Scholar]
- Song, F.; Qin, Z.; Xue, L.; Zhang, J.; Lin, X.; Shen, X. Privacy-preserving keyword similarity search over encrypted spatial data in cloud computing. IEEE Internet Things J. 2021, 9, 6184–6198. [Google Scholar] [CrossRef]
- Kong, L.; Liu, X.-Y.; Sheng, H.; Zeng, P.; Chen, G. Federated tensor mining for secure industrial internet of things. IEEE Trans. Ind. Inform. 2019, 16, 2144–2153. [Google Scholar] [CrossRef]
- Bonawitz, K.; Ivanov, V.; Kreuter, B.; Marcedone, A.; McMahan, H.B.; Patel, S.; Ramage, D.; Segal, A.; Seth, K. Practical secure aggregation for privacy-preserving machine learning. In Proceedings of the 2017 ACM SIGSAC Conference on Computer and Communications Security, Dallas, TX, USA, 30 October–3 November 2017; pp. 1175–1191. [Google Scholar]
- Zhang, X.; Ji, S.; Wang, H.; Wang, T. Private, yet practical, multiparty deep learning. In Proceedings of the 2017 IEEE 37th International Conference on Distributed Computing Systems (ICDCS), Atlanta, GA, USA, 5–8 June 2017; pp. 1442–1452. [Google Scholar]
- Lu, Y.; Huang, X.; Zhang, K.; Maharjan, S.; Zhang, Y. Blockchain empowered asynchronous federated learning for secure data sharing in internet of vehicles. IEEE Trans. Veh. Technol. 2020, 69, 4298–4311. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, J.J.Q.; Kang, J.; Niyato, D.; Zhang, S. Privacy-preserving traffic flow prediction: A federated learning approach. IEEE Internet Things J. 2020, 7, 7751–7763. [Google Scholar] [CrossRef]
- Qu, Y.; Gao, L.; Luan, T.H.; Xiang, Y.; Yu, S.; Li, B.; Zheng, G. Decentralized privacy using blockchain-enabled federated learning in fog computing. IEEE Internet Things J. 2020, 7, 5171–5183. [Google Scholar] [CrossRef]
- LaLonde, R.; Bagci, U. Capsules for object segmentation. arXiv 2018, arXiv:1804.04241. [Google Scholar]
- Sabour, S.; Frosst, N.; Hinton, G.E. Dynamic routing between capsules. In Proceedings of the Advances in Neural Information Processing Systems 30, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Chattu, V.K. A review of artificial intelligence, big data, and blockchain technology applications in medicine and global health. Big Data Cogn. Comput. 2021, 5, 41. [Google Scholar]
- Kim, H.; Park, J.; Bennis, M.; Kim, S.-L. Blockchained on-device federated learning. IEEE Commun. Lett. 2019, 24, 1279–1283. [Google Scholar] [CrossRef]
- Martinez, I.; Francis, S.; Hafid, A.S. Record and reward federated learning contributions with blockchain. In Proceedings of the 2019 International Conference on Cyber-Enabled Distributed Computing and Knowledge Discovery (CyberC), Guilin, China, 17–19 October 2019; pp. 50–57. [Google Scholar]
- He, C.; Mushtaq, E.; Ding, J.; Avestimehr, S. Fednas: Federated deep learning via neural architecture search. CVPR Workshop 2021, 3618. [Google Scholar]
- Bao, X.; Su, C.; Xiong, Y.; Huang, W.; Hu, Y. Flchain: A blockchain for auditable federated learning with trust and incentive. In Proceedings of the 2019 5th International Conference on Big Data Computing and Communications (BIGCOM), QingDao, China, 9–11 August 2019; pp. 151–159. [Google Scholar]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. Covid-caps: A capsule network-based framework for identification of COVID-19 cases from x-ray images. Pattern Recognit. Lett. 2020, 138, 638–643. [Google Scholar] [CrossRef]
- Umer, M.; Hong, C.S. FLchain: Federated learning via MEC-enabled blockchain network. In Proceedings of the 2019 20th Asia-Pacific Network Operations and Management Symposium (APNOMS), Matsue, Japan, 18–20 September 2019; pp. 1–4. [Google Scholar]
- Xuan, S.; Jin, M.; Li, X.; Yao, Z.; Yang, W.; Man, D. DAM-SE: A Blockchain-Based Optimized Solution for the Counterattacks in the Internet of Federated Learning Systems. Secur. Commun. Netw. 2021, 2021, 9965157. [Google Scholar] [CrossRef]
- Salman, F.M.; Abu-Naser, S.S.; Alajrami, E.; Abu-Nasser, B.S.; Alashqar, M.A.M. COVID-19 detection using artificial intelligence. Int. J. Acad. Eng. Res. 2020, 4, 18–25. [Google Scholar]
- Kaissis, G.; Ziller, A.; Passerat-Palmbach, J.; Ryffel, T.; Usynin, D.; Trask, A.; Lima, I.; Mancuso, J.; Jungmann, F.; Steinborn, M.-M.; et al. End-to-end privacy preserving deep learning on multi-institutional medical imaging. Nat. Mach. Intell. 2021, 3, 473–484. [Google Scholar] [CrossRef]
- Flores, M.; Dayan, I.; Roth, H.; Zhong, A.; Harouni, A.; Gentili, A.; Abidin, A.; Liu, A.; Costa, A.; Wood, B.; et al. Federated Learning used for predicting outcomes in SARS-COV-2 patients. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Dou, Q.; So, T.Y.; Jiang, M.; Liu, Q.; Vardhanabhuti, V.; Kaissis, G.; Li, Z.; Si, W.; Lee, H.H.C.; Yu, K.; et al. Federated deep learning for detecting COVID-19 lung abnormalities in CT: A privacy-preserving multinational validation study. NPJ Digit. Med. 2021, 4, 60. [Google Scholar] [CrossRef]
- Yang, D.; Xu, Z.; Li, W.; Myronenko, A.; Roth, H.R.; Harmon, S.; Xu, S.; Turkbey, B.; Turkbey, E.; Wang, X.; et al. Federated semi-supervised learning for COVID region segmentation in chest CT using multi-national data from China, Italy, Japan. Med. Image Anal. 2021, 70, 101992. [Google Scholar] [CrossRef]
- Kumar, R.; Khan, A.A.; Kumar, J.; Zakria, A.; Golilarz, N.A.; Zhang, S.; Ting, Y.; Zheng, C.; Wang, W. Blockchain-federated-learning and deep learning models for COVID-19 detection using ct imaging. IEEE Sens. J. 2021, 21, 16301–16314. [Google Scholar] [CrossRef]
- Podder, P.; Das, S.R.; Mondal, M.R.H.; Bharati, S.; Maliha, A.; Hasan, J.; Piltan, F. LDDNet: A Deep Learning Framework for the Diagnosis of Infectious Lung Diseases. Sensors 2023, 23, 480. [Google Scholar] [CrossRef]
- Attallah, O. RADIC: A tool for diagnosing COVID-19 from chest CT and X-ray scans using deep learning and quad-radiomics. Chemom. Intell. Lab. Syst. 2023, 233, 104750. [Google Scholar] [CrossRef]
- El-Bana, S.; Al-Kabbany, A.; Sharkas, M. A multi-task pipeline with specialized streams for classification and segmentation of infection manifestations in COVID-19 scans. PeerJ Comput. Sci. 2020, 6, e303. [Google Scholar] [CrossRef]
- Shah, V.; Keniya, R.; Shridharani, A.; Punjabi, M.; Shah, J.; Mehendale, N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg. Radiol. 2021, 28, 497–505. [Google Scholar] [CrossRef]
- Attallah, O.; Samir, A. A wavelet-based deep learning pipeline for efficient COVID-19 diagnosis via CT slices. Appl. Soft Comput. 2022, 128, 109401. [Google Scholar] [CrossRef]
- Attallah, O. A computer-aided diagnostic framework for coronavirus diagnosis using texture-based radiomics images. Digit. Health 2022, 8, 20552076221092543. [Google Scholar] [CrossRef]
- Zhao, W.; Jiang, W.; Qiu, X. Deep learning for COVID-19 detection based on CT images. Sci. Rep. 2021, 11, 14353. [Google Scholar] [CrossRef]
- Shankar, K.; Perumal, E. A novel hand-crafted with deep learning features based fusion model for COVID-19 diagnosis and classification using chest X-ray images. Complex Intell. Syst. 2021, 7, 1277–1293. [Google Scholar] [CrossRef]
- Jingxin, L.; Mengchao, Z.; Yuchen, L.; Jinglei, C.; Yutong, Z.; Zhong, Z.; Lihui, Z. COVID-19 lesion detection and segmentation–A deep learning method. Methods 2022, 202, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.C.; Ding, M.; Pathirana, P.N.; Seneviratne, A.; Zomaya, A.Y. Federated learning for COVID-19 detection with generative adversarial networks in edge cloud computing. IEEE Internet Things J. 2021, 9, 10257–10271. [Google Scholar] [CrossRef]
- Aswathy, A.L.; Vinod, C.S.S. Cascaded 3D UNet architecture for segmenting the COVID-19 infection from lung CT volume. Sci. Rep. 2022, 12, 3090. [Google Scholar]
- Singh, P.; Kaur, R. Implementation of the QoS framework using fog computing to predict COVID-19 disease at early stage. World J. Eng. 2021, 19, 80–89. [Google Scholar] [CrossRef]
- Kochgaven, C.; Mishra, P.; Shitole, S. Detecting Presence of COVID-19 with ResNet-18 using PyTorch. In Proceedings of the 2021 International Conference on Communication information and Computing Technology (ICCICT), Mumbai, India, 25–27 June 2021; pp. 1–6. [Google Scholar]
- Chen, X.; Shao, Y.; Xue, Z.; Yu, Z. Multi-Modal COVID-19 Discovery With Collaborative Federated Learning. In Proceedings of the 2021 IEEE 7th International Conference on Cloud Computing and Intelligent Systems (CCIS), Xi’an, China, 7–8 November 2021; pp. 52–56. [Google Scholar]
- Le Dinh, T.; Lee, S.-H.; Kwon, S.-G.; Kwon, K.-R. COVID-19 Chest X-ray Classification and Severity Assessment Using Convolutional and Transformer Neural Networks. Appl. Sci. 2022, 12, 4861. [Google Scholar] [CrossRef]
- Yang, X.; He, X.; Zhao, J.; Zhang, Y.; Zhang, S.; Xie, P. COVID-CT-dataset: A CT scan dataset about COVID-19. arXiv 2020, arXiv:2003.13865. [Google Scholar]
- Rahimzadeh, M.; Attar, A.; Sakhaei, S.M. A fully automated deep learning-based network for detecting COVID-19 from a new and large lung ct scan dataset. Biomed. Signal Process. Control 2021, 68, 102588. [Google Scholar] [CrossRef]
- Qorib, M.; Oladunni, T.; Denis, M.; Ososanya, E.; Cotae, P. COVID-19 vaccine hesitancy: Text mining, sentiment analysis and machine learning on COVID-19 vaccination twitter dataset. Expert Syst. Appl. 2023, 212, 118715. [Google Scholar] [CrossRef]
- Angelov, P.; Soares, E. Towards explainable deep neural networks (xDNN). Neural Netw. 2020, 130, 185–194. [Google Scholar] [CrossRef]
- Morozov, S.P.; Andreychenko, A.E.; Pavlov, N.A.; Vladzymyrskyy, A.V.; Ledikhova, N.V.; Gombolevskiy, V.A.; Blokhin, I.A.; Gelezhe, P.B.; Gonchar, A.V.; Chernina, V.Y. Mosmeddata: Chest ct scans with COVID-19 related findings dataset. arXiv 2020, arXiv:2005.06465. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, L.; Jiang, S.; Zou, J.; Fu, H.; Yang, S.; Wang, Y. Capsule networks showed excellent performance in the classification of hERG blockers/nonblockers. Front. Pharmacol. 2020, 10, 1631. [Google Scholar] [CrossRef]
- Malik, H.; Farooq, M.S.; Khelifi, A.; Abid, A.; Qureshi, J.N.; Hussain, M. A comparison of transfer learning performance versus health experts in disease diagnosis from medical imaging. IEEE Access 2020, 8, 139367–139386. [Google Scholar] [CrossRef]
- Malik, H.; Anees, T.; Din, M.Z. BDCNet: Multi-classification convolutional neural network model for classification of COVID-19, pneumonia, and lung cancer from chest radiographs. Multimed. Syst. 2022, 28, 815–829. [Google Scholar] [CrossRef]
- Naeem, A.; Anees, T.; Fiza, M.; Naqvi, R.A.; Lee, S.-W. SCDNet: A Deep Learning-Based Framework for the Multiclassification of Skin Cancer Using Dermoscopy Images. Sensors 2022, 22, 5652. [Google Scholar] [CrossRef]
- Saeed, H.; Malik, H.; Bashir, U.; Ahmad, A.; Riaz, S.; Ilyas, M.; Bukhari, W.A.; Khan, M.I.A. Blockchain technology in healthcare: A systematic review. PLoS ONE 2022, 17, e0266462. [Google Scholar] [CrossRef]
- Malik, H.; Bashir, U.; Ahmad, A. Multi-classification neural network model for detection of abnormal heartbeat audio signals. Biomed. Eng. Adv. 2022, 4, 100048. [Google Scholar] [CrossRef]
- Naeem, A.; Anees, T.; Naqvi, R.A.; Loh, W.-K. A Comprehensive Analysis of Recent Deep and Federated-Learning-Based Methodologies for Brain Tumor Diagnosis. J. Pers. Med. 2022, 12, 275. [Google Scholar] [CrossRef]
- Komal, A.; Malik, H. Transfer Learning Method with Deep Residual Network for COVID-19 Diagnosis Using Chest Radiographs Images. In Proceedings of the International Conference on Information Technology and Applications; Springer: Singapore, 2022; pp. 145–159. [Google Scholar]
- Malik, H.; Anees, T.; Din, M.; Naeem, A. CDC_Net: Multi-classification convolutional neural network model for detection of COVID-19, pneumothorax, pneumonia, lung Cancer, and tuberculosis using chest X-rays. Multimed. Tools Appl. 2022, 1–26. [Google Scholar] [CrossRef]
- Liang, N.-Y.; Huang, G.-B.; Saratchandran, P.; Sundararajan, N. A fast and accurate online sequential learning algorithm for feedforward networks. IEEE Trans. Neural Netw. 2006, 17, 1411–1423. [Google Scholar] [CrossRef]
- Wang, B.; Huang, S.; Qiu, J.; Liu, Y.; Wang, G. Parallel online sequential extreme learning machine based on MapReduce. Neurocomputing 2015, 149, 224–232. [Google Scholar] [CrossRef]
- Mascarenhas, S.; Agarwal, M. A comparison between VGG16, VGG19 and ResNet50 architecture frameworks for Image Classification. In Proceedings of the 2021 International Conference on Disruptive Technologies for Multi-Disciplinary Research and Applications (CENTCON), Bengaluru, India, 19–21 November 2021; Volume 1, pp. 96–99. [Google Scholar]
- Sitaula, C.; Hossain, M.B. Attention-based VGG-16 model for COVID-19 chest X-ray image classification. Appl. Intell. 2021, 51, 2850–2863. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-H.; Zhang, Y.-D. DenseNet-201-based deep neural network with composite learning factor and precomputation for multiple sclerosis classification. ACM Trans. Multimed. Comput. Commun. Appl. 2020, 16, 1–19. [Google Scholar] [CrossRef]
- Deng, J. A large-scale hierarchical image database. In Proceedings of the IEEE Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009. [Google Scholar]
- Targ, S.; Almeida, D.; Lyman, K. Resnet in resnet: Generalizing residual architectures. arXiv 2016, arXiv:1603.08029. [Google Scholar]
- Maymounkov, P.; Mazieres, D. Kademlia: A peer-to-peer information system based on the xor metric. In International Workshop on Peer-to-Peer Systems; Springer: Berlin/Heidelberg, Germany, 2002; pp. 53–65. [Google Scholar]
- Cintrón-Arias, A.; Banks, H.T.; Capaldi, A.; Lloyd, A.L. A sensitivity matrix based methodology for inverse problem formulation. J. Inverse Ill-Posed Probl. 2009, 17, 545–564. [Google Scholar] [CrossRef]
- Lund, O.; Nielsen, M.; Kesmir, C.; Pedersen, A.G.; Justesen, S.; Lundegaard, C.; Worning, P.; Sylvester-Hvid, C.; Lamberth, K. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics 2004, 55, 797–810. [Google Scholar] [CrossRef]
- Pang, J.; Huang, Y.; Xie, Z.; Han, Q.; Cai, Z. Realizing the heterogeneity: A self-organized federated learning framework for IoT. IEEE Internet Things J. 2020, 8, 3088–3098. [Google Scholar] [CrossRef]
- Kuzdeuov, A.; Baimukashev, D.; Karabay, A.; Ibragimov, B.; Mirzakhmetov, A.; Nurpeiissov, M.; Lewis, M.; Varol, H.A. A network-based stochastic epidemic simulator: Controlling COVID-19 with region-specific policies. IEEE J. Biomed. Health Inform. 2020, 24, 2743–2754. [Google Scholar] [CrossRef]
- Wang, S.; Kang, B.; Ma, J.; Zeng, X.; Xiao, M.; Guo, J.; Cai, M.; Yang, J.; Li, Y.; Meng, X.; et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). Eur. Radiol. 2021, 31, 6096–6104. [Google Scholar] [CrossRef]
- Jin, C.; Chen, W.; Cao, Y.; Xu, Z.; Tan, Z.; Zhang, X.; Deng, L.; Zheng, C.; Zhou, J.; Shi, H.; et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat. Commun. 2020, 11, 5088. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, X.; Ma, C.; Du, P.; Li, X.; Lv, S.; Yu, L.; Chen, Y.; Su, J.; Lang, G. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering 2020, 6, 1122–1129. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, S.; Li, L.; Zhang, X.; Zhang, X.; Huang, Z.; Chen, J.; Zhao, H.; Jie, Y.; Wang, R. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 2775–2780. [Google Scholar] [CrossRef]
- Wang, B.; Jin, S.; Yan, Q.; Xu, H.; Luo, C.; Wei, L.; Zhao, W.; Hou, X.; Ma, W.; Xu, Z.; et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system. Appl. Soft Comput. 2021, 98, 106897. [Google Scholar] [CrossRef]
- Durga, R.; Poovammal, E. FLED-Block: Federated Learning Ensembled Deep Learning Blockchain Model for COVID-19 Prediction. Front. Public Health 2022, 10, 892499. [Google Scholar] [CrossRef]
- Zheng, C.; Deng, X.; Fu, Q.; Zhou, Q.; Feng, J.; Ma, H.; Liu, W.; Wang, X. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv 2020. [Google Scholar] [CrossRef]
- Shi, F.; Xia, L.; Shan, F.; Song, B.; Wu, D.; Wei, Y.; Yuan, H.; Jiang, H.; He, Y.; Gao, Y.; et al. Large-scale screening to distinguish between COVID-19 and community-acquired pneumonia using infection size-aware classification. Phys. Med. Biol. 2021, 66, 065031. [Google Scholar] [CrossRef]
- Chen, J.; Wu, L.; Zhang, J.; Zhang, L.; Gong, D.; Zhao, Y.; Chen, Q.; Huang, S.; Yang, M.; Yang, X.; et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020, 10, 19196. [Google Scholar] [CrossRef]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology 2020, 200905. [Google Scholar]
- Naeem, A.; Anees, T.; Ahmed, K.T.; Naqvi, R.A.; Ahmad, S.; Whangbo, T. Deep learned vectors’ formation using auto-correlation, scaling, and derivations with CNN for complex and huge image retrieval. Complex Intell. Syst. 2022, 1–23. [Google Scholar] [CrossRef]



| Ref | Models | Objective | Image Type | FL | Outcomes | |
|---|---|---|---|---|---|---|
| CXR | CT | |||||
| [37] | EMR CXR AI | To identify the COVID-19-infected patients. | ✓ | × | ✓ | AUC = 0.92 |
| [38] | FL + CNN | To detect lung abnormalities that occur due to COVID-19. | × | ✓ | ✓ | Specificity = 95.27% |
| [39] | FL + Semi-supervised Learning | To segment the COVID-19-infected regions of the lungs. | × | ✓ | ✓ | Accuracy = 0.71 |
| [40] | Capsule Network | To find the COVID-19 infection in the lungs. | × | ✓ | ✓ | Precision = 0.83 |
| [50] | FED-GAN | To identify the COVID-19-infected individuals. | × | ✓ | ✓ | Precision = 96.6% |
| [51] | 3D UNET | To segment the infected areas of the lungs. | × | ✓ | ✓ | Accuracy = 98.07% |
| [52] | RF | To predict COVID-19 virus activity | × | ✓ | ✓ | Accuracy = 81.80% |
| [53] | ResNet-18 | To find evidence of COVID-19’s existence in the lungs. | ✓ | ✓ | ✓ | Accuracy = 97.78% |
| [54] | AlexNet | To identify COVID-19 from pneumonia. | × | ✓ | ✓ | Accuracy = 96.29% |
| [55] | Multiple CNN models | To forecast the COVID-19 illness | × | ✓ | × | Accuracy = 94.00% |
| Datasets | No. of Patients | COVID-19 CT Scans | Normal CT Scans | Image Format | Type of CT Scans | Total Number of CT Scans |
|---|---|---|---|---|---|---|
| Dataset 1 [56] | 1000 | 35,635 | 9367 | DICOM | 2D | 45,002 |
| Dataset 2 [57] | 89 | 28,395 | 5611 | PNG | 3D | 34,006 |
| Dataset 3 [58] | NA | 1252 | 1230 | PNG | 2D | 2482 |
| Dataset 4 [59] | 1110 | 125 | 254 | 3D CT scans | 3D | 379 |
| Dataset 5 [60] | 216 | 349 | 463 | PNG | 2D | 812 |
| Steps | Process | CapsNet | Traditional ANN |
|---|---|---|---|
| 1 | Output | Vector (Vj) | Scaler (Sj) |
| 2 | Affine Transformation | NA | |
| 3 | Weighted Sum | ||
| 4 | Activation Function |
| Models | Nodes | Pre-Trained | ACU | PRE | REC | SPF | F1-Score |
|---|---|---|---|---|---|---|---|
| VGG-16 | MLP | ImageNet | 91.75% | 91.42% | 91.47% | 91.29% | 91.22% |
| VGG-19 | MLP | ImageNet | 92.21% | 92.09% | 92.12% | 92.08% | 92.14% |
| ResNet-101 | MLP | ImageNet | 94.81% | 94.21% | 94.32% | 94.27% | 94.22% |
| DenseNet-169 | MLP | ImageNet | 94.99% | 95.01% | 94.96% | 94.97% | 95.00% |
| DenseNet-201 | MLP | ImageNet | 95.45% | 95.54% | 95.47% | 95.43% | 95.48% |
| Proposed Models | Capsule Network and IELMs | - | 98.99% | 98.96% | 98.97% | 98.95% | 98.96% |
| Models | ACU | PRE | REC | SPF | F1-Score | DSC |
|---|---|---|---|---|---|---|
| UNET | 84.15% | 84.42% | 84.99% | 84.75% | 84.22% | 84.19% |
| UNET++ | 86.10% | 86.99% | 86.09% | 86.99% | 86.02% | 86.01% |
| Proposed Model | 95.81% | 95.49% | 95.32% | 95.57% | 95.51% | 95.50% |
| Ref | Models | Disease Classification | Accuracy (%) | Data Sharing | BCT Privacy Protection |
|---|---|---|---|---|---|
| [82] | 3D-ResNet + Attention | COVID-19, Pneumonia, and Normal | 93.30 | No | No |
| [83] | CNN | COVID-19 and Normal | 82.90 | No | No |
| [40] | Capsule Network | COVID-19 and Normal | 98.0 | Yes | Yes |
| [84] | 2D-CNN | COVID-19, Pneumonia, and Normal | 94.41 | No | No |
| [85] | 2D-CNN | COVID-19, Influenza, and Normal | 86.70 | No | No |
| [86] | ResNet-50 | COVID-19, Pneumonia, and Normal | 86.00 | No | No |
| [87] | UNET++ | COVID-19, Pneumonia, and Normal | 97.40 | No | No |
| [88] | Deep learning | COVID-19 | 98.20 | Yes | Yes |
| [89] | UNET and CNN | COVID-19, Pneumonia, and Normal | 90.70 | No | No |
| [90] | RF | COVID-19, Pneumonia (bacterial and viral), and Normal | 87.90 | No | No |
| [91] | UNET++ and CNN | COVID-19, Pneumonia, and Normal | 95.20 | No | No |
| [92] | ResNet-50 | COVID-19, Pneumonia, and Normal | 90.00 | No | No |
| Ours | CapsNet with IELMs | COVID-19 and Normal | 98.99 | Yes | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malik, H.; Anees, T.; Naeem, A.; Naqvi, R.A.; Loh, W.-K. Blockchain-Federated and Deep-Learning-Based Ensembling of Capsule Network with Incremental Extreme Learning Machines for Classification of COVID-19 Using CT Scans. Bioengineering 2023, 10, 203. https://doi.org/10.3390/bioengineering10020203
Malik H, Anees T, Naeem A, Naqvi RA, Loh W-K. Blockchain-Federated and Deep-Learning-Based Ensembling of Capsule Network with Incremental Extreme Learning Machines for Classification of COVID-19 Using CT Scans. Bioengineering. 2023; 10(2):203. https://doi.org/10.3390/bioengineering10020203
Chicago/Turabian StyleMalik, Hassaan, Tayyaba Anees, Ahmad Naeem, Rizwan Ali Naqvi, and Woong-Kee Loh. 2023. "Blockchain-Federated and Deep-Learning-Based Ensembling of Capsule Network with Incremental Extreme Learning Machines for Classification of COVID-19 Using CT Scans" Bioengineering 10, no. 2: 203. https://doi.org/10.3390/bioengineering10020203
APA StyleMalik, H., Anees, T., Naeem, A., Naqvi, R. A., & Loh, W.-K. (2023). Blockchain-Federated and Deep-Learning-Based Ensembling of Capsule Network with Incremental Extreme Learning Machines for Classification of COVID-19 Using CT Scans. Bioengineering, 10(2), 203. https://doi.org/10.3390/bioengineering10020203







