1. Introduction
An electrocardiogram (ECG) is a recording of the electrical response of a heart caused by its movement; thus, an ECG can be used to determine the activity of the heart [
1]. It is generally used to identify a patient’s health condition by classifying the status of each stage of cardiac activity through changes in the ECG signals’ form or value; hence, these signals can be the most intuitive data in determining heart disease. The task of analyzing an ECG and classifying a patient’s abnormal symptoms based on the ECG characteristics is very important in determining the health of the patient’s heart. A typical disease studied using ECG is arrhythmia, which refers to an irregular pattern or phenomenon occurring in an ECG. It is classified according to the type of occurrence: atrial fibrillation (AFIB), atrial flutter (AFL), and sinus bradycardia (SBR), which are rhythmic conditions that have been identified as major threats to a heart’s health [
2,
3].
Experts visually check the waveforms of ECG signals and classify them based on their experience and background knowledge of cardiac rhythms [
4]. However, due to the many patterns of heart activity and various environmental factors, diagnosis based on experience and background knowledge can lead to misjudgments, which hinders the timely application of an appropriate treatment method [
5]. Indeed, the number of patients suffering from AFIB has been increasing, and about 15% of deaths caused by heart disease worldwide are caused by such misjudgments [
6,
7,
8]. Therefore, if ECG signals are analyzed well, and cardiac rhythms are identified more accurately, an accurate diagnosis is likely possible.
In the past, ECG signals were analyzed by methods based on experts’ knowledge and finding known abnormal patterns. For example, ECG features (e.g., time elapsing between two consecutive R waves in an ECG, called the R–R interval) were calculated and judged by these methods. Meanwhile, a machine learning technique has been used to classify heart-related abnormal symptoms through learning based on collected data, which were accumulated gradually from patients [
9,
10]. However, previous studies have focused mainly on finding locations that correspond to R peaks in ECG signal patterns and classifying the types of beats found [
11,
12,
13].
Recently, methods using deep learning have been used in various fields and problems, and such methods have been used in analyzing ECG signals (e.g., classifying AFIB and a rhythm) [
14,
15,
16,
17,
18,
19,
20,
21,
22]. Nevertheless, a problem in analyzing an ECG is the difference in the number of data samples. Depending on the frequency of occurrence of rhythms, there could be few and many samples for rhythms with low and frequent occurrence, respectively. For this reason, an imbalance among rhythms exists in ECG databases such as the Massachusetts Institute Of Technology-Beth Israel Hospital (MIT-BIH) arrhythmia database, which is widely used and public [
23]. This imbalance is a hindrance when learning and distinguishing various ECG rhythms, and it causes difficulties during the classification of correct ECG rhythms. For these reasons, research on how to classify multiple rhythms is insufficient. However, as mentioned above, the limitations are the difficulty of classifying rhythms using relatively few samples from the measured data and properly distinguishing similar patterns or characteristics between rhythms [
24,
25,
26,
27].
In this paper, to compensate for these limitations, we maximized the information present in an ECG. In analyzing rhythms, we assumed that the arrangement of beats in ECG signals is correlated with ECG rhythms. We then used the arrangement pattern of ECG beats to differentiate the various rhythms. In addition, we developed a method that considers not only these patterns but also features such as the R–R interval related to rhythm classification.
Through the proposed method, we tried to solve the difficulties caused by rhythms with similar characteristics and the existing limitations of imbalanced data. To achieve these goals, two datasets were constructed to classify various rhythm labels by the proposed method using MIT-BIH arrhythmia data. Finally, we determined the performance of the proposed model. Our main contributions can be summarized as follows.
We consider ECG rhythm as an arrangement of beats and classify various ECG rhythms by utilizing the arrangement of this pattern of beats. For this purpose, a series of ECG beat segments divided by a specific criterion are obtained from each ECG rhythm and are employed to train a beat classification model. The prediction score vector for each beat segment in the classification model is then used to generate an arrangement pattern of beats for each ECG rhythm. In doing so, some changes in ECG rhythms of the same or different types can be reflected as much as possible through the prediction score vector of the beat classification model.
The arrangement pattern of beats for a given ECG signal is converted into a beat score map (BSM) image, of which a continuous wavelet transform (CWT) is then fed to a deep convolutional neural network (CNN) for rhythm classification. Unlike existing methods that mostly focus on the overall characteristics of various ECG rhythms in the time or frequency domain, we subdivide the rhythms into a series of beat segments and characterize each beat segment by the prediction score vector of the beat classification model. The prediction score vectors for a series of beat segments are aligned along with time interval padding, leading to the production of the BSM image.
The proposed method is effective in classifying various types of ECG rhythms with data imbalance problems. Particularly, certain types of ECG rhythms with few samples can be distinguished well from other types with many samples. In addition, our method can well distinguish different ECG rhythms of similar characteristics, such as between AFIB and AFL, which have been known to be difficult to classify in previous studies.
The remainder of this paper is organized as follows:
Section 2 presents existing recent research related to ECG rhythms.
Section 3 describes the overall details of the proposed method such as BSM image generation and structure of network. Experiments and results are shown in
Section 4 and
Section 5, respectively. Conclusions are provided with some discussion in
Section 6.
2. Related Works
Hand-crafted features based on background knowledge have been extracted and used to analyze ECG signals. Recently, feature extraction, which is not done by humans, was conducted for the desired purpose of using deep learning [
22,
28]. Through these studies, hand-crafted features based on existing background knowledge were obtained by using machine learning [
29,
30,
31]. In deep learning, the trend is toward automatically obtaining the most suitable features by methods such as CNN [
32,
33,
34], long-short term network (LSTM) [
11,
35,
36], and CNN autoencoder [
37]. In a recent study [
38], matrix images using the correlation between multi-channel EEG signals were created and analyzed using a deep CNN. As such, methods of imaging various physiological signals and analyzing them through CNN have been proposed.
Research has been undertaken on classifying ECG beats included in an ECG signal. In previous studies, QRS complexes that are features created based on knowledge of repetitive morphological patterns of ECG signals have been used [
39]. However, rather than quantifying and analyzing the repeated patterns in a beat classification, studies have suggested the learning of the ECG signal’s one-dimensional (1-D) data itself to suit beat classification [
40,
41,
42]. Alternatively, the ECG signal is converted to an image through transformation for use as an input of a 2D deep learning method [
3,
43]. Notably, the method of converting and utilizing ECG using CWT to compensate for possible limitations in an existing Fourier transform is performing well [
44,
45].
In addition to ECG beat classification, studies have been conducted to identify the rhythm of ECG signals [
28]. Among these studies, the most active is in the field of single rhythm classification using AFIB. AFIB is a common ECG rhythm in many databases; thus, it is suitable for research. Methods for AFIB classification calculate features related to AFIB, and they analyze changes in the calculated features [
46,
47,
48]. Current methods have preferred using deep learning over traditional features. Recent studies have integrated the use of CNNs and other network structures, and they obtained good performance for classifying ECG rhythms [
49,
50,
51,
52,
53]. In addition, research is being conducted on real-life applications to reduce computational costs while maintaining the performance of deep CNNs. [
54]. However, they have not paid much attention to the various ECG rhythms existing in real life. As such, studies on single rhythm classification have shown good performance but failed to distinguish well between several rhythms.
Meanwhile, many studies have been conducted to classify several rhythms [
26,
27,
55,
56,
57]. However, they have shown severe degradation in classifying some rhythms with data imbalance problems. Moreover, they had some difficulties in differentiating between ECG rhythms of similar characteristics, such as between AFIB and AFL.
Finally, many researchers have studied the beat and rhythm of ECG signals using various features and network structures [
58,
59,
60]; however, we have not found research on the correlation between the listings of beats and rhythms. Thus, we intend to solve the problem of the existing threshold by utilizing the association between the listings of beats and rhythm in the proposed method.
3. Methods
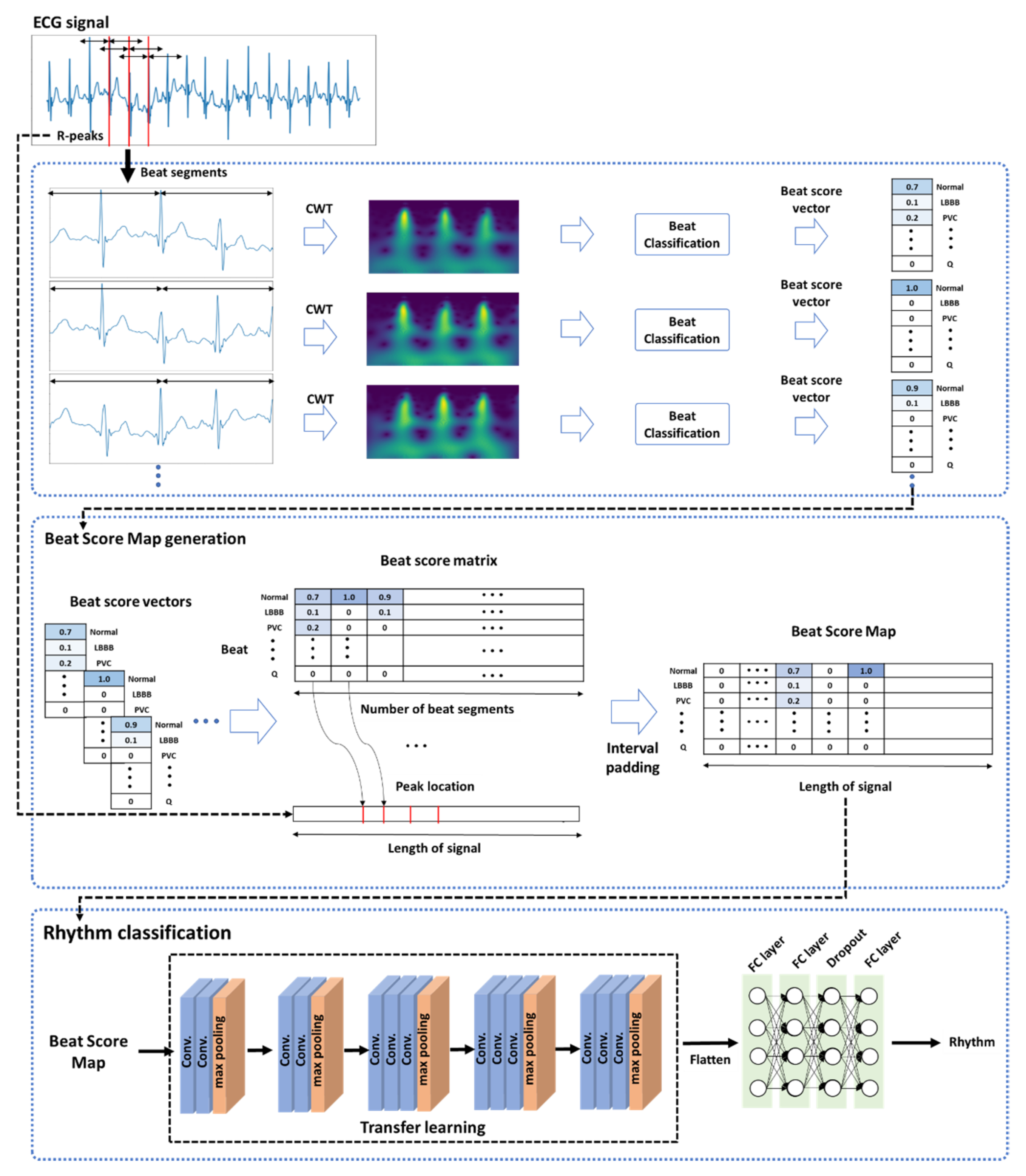
The proposed model for classifying ECG rhythms employs a 10-s ECG signal as an input. The proposed method is divided into three main parts: (1) beat unit analysis that trains a beat classification model using CWT and obtains a beat score vector from the trained model; (2) the creation of a BSM image by integrating the obtained beat score vector into a beat score matrix and applying time interval padding based on the R-peak position in the matrix; and (3) rhythm classification implemented by utilizing the information related to the existing feature and the correlation between the list of beats in an ECG signal and ECG rhythms.
A general overview of the proposed model is provided in
Figure 1. In the first part, a given ECG signal of an input is created with beat segments based on R peak detection, and the ECG signal of the created segments is then converted into an image by the CWT method. The converted image is applied to classify the beats contained in a segment. As a result, we construct a beat score vector by taking the predicted score values rather than the label values for each beat class to preserve information about differences in ECG signals. In the second part, the beat score vectors created from each segment are merged with interval padding to form a beat score matrix, where the interval padding is applied to consider the R–R interval in the created matrix. For this purpose, the location of the R peaks used to create the beat segments is utilized. Thus, beat score map (BSM) images conveying both listing patterns of beats and R–R intervals are created by this part of the method. In the third part, a two-dimensional (2-D) convolutional neural network (CNN) structure model is trained using the created BSM images by transfer learning, and the ECG rhythms are classified.
3.1. Preprocessing
We carry out the removal of baseline drift and high-frequency noise in a signal by discrete wavelet transform. For removing baseline drift, the decomposition scale was set to 9 in Daubichies-4 (db4). For high-frequency noise filtering, the decomposition scale was set to 6 in the same db-4, and the frequencies ranging from 50 Hz to 100 kHz were filtered. In rhythm classification, each window was slid every 1 s, and the rhythm label corresponding to a sequence in each 10-s unit was recorded.
3.2. BSM Image Generation
To compensate for known limitations, BSM image generation is designed to utilize beat prediction scores obtained in the beat classification process and information from the R–R interval through interval padding.
3.2.1. Beat Classification
Beat classification was implemented based on the structure of a recently studied method that creates a spectrogram image using ECG signal CWT [
44]. To create images by CWT, a 2.4-s chunk for each R-peak position was created, and each chunk consisted of ECG signals from 1.2 s before to 1.2 s after an R peak. Using the created images as input, the model was trained via images created for the classification of the given beat.
3.2.2. Interval Padding and Resizing
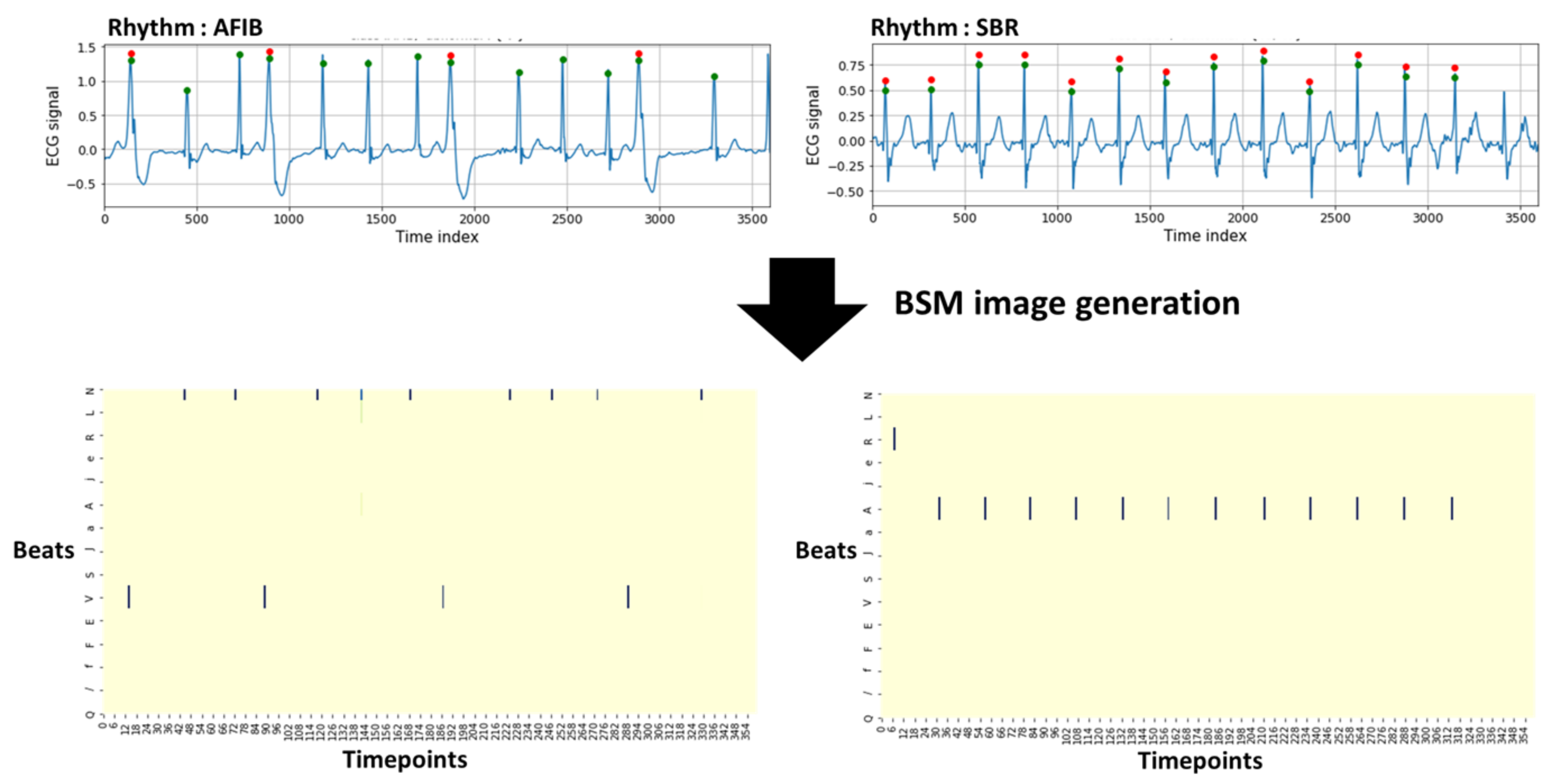
A beat score vector for each beat was obtained using the prediction score value from the learned beat classification model. All vectors included in a 10-s ECG segment were merged to form a beat score matrix. If the vectors contained in an ECG signal were combined simply to form a matrix, the association between the beats and rhythms was considered; however, information on the R–R interval associated with the rhythm classification was lost. Thus, a BSM image was created by applying interval padding in which the beat score vector was entered at the location where the R peak exists, while the rest were filled with zero. Information such as the location of the overall R peaks within a unit ECG signal and the R–R interval obtained therefrom were also included in the BSM image.
BSM images were produced initially in a 3600 × 15 structure because the data used in the experiment had a sampling rate of 360 Hz, resulting in 3600 time points in 10 s. The size was reduced by one-tenth and adjusted to 360. In a value corresponding to 15, the number of labels for all beats in the provided data is given. The created matrices resized to 360 × 150 are suitable for CNN learning. The structure of a created 360 × 15 matrix is shown in
Figure 2. The x- and y-axis represent the 360 time points and each beat label, respectively. The figure confirms that the beat interval, beat type, and prediction score value for each beat are configured differently for each rhythm class.
3.3. BSM Image Classification
Based on the created BSM image, rhythm classification was implemented by a 2D CNN. For image classification by transfer learning, the commonly used 2D CNN structure of VGG16 [
61] was utilized. Therefore, the pre-trained weight was the initial value, and the weight was learned newly with a given image. The network consisted of five convolution blocks and three layers that were connected fully in a large structure.
4. Experiments
The proposed method used labels for the rhythm of ECG records obtained from the MIT-BIH arrhythmia database and the locations of R peaks. We constructed two ECG rhythm datasets using the given rhythm labels, and the performance of the method was evaluated for each dataset.
4.1. Dataset
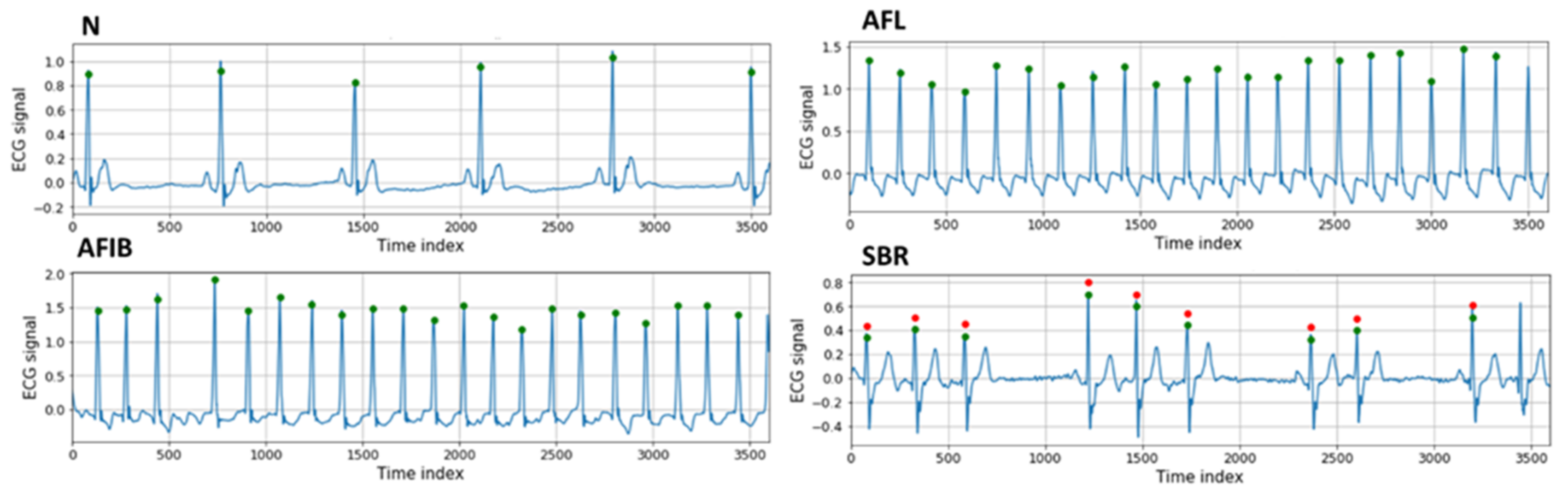
In our experiment, we used the MIT-BIH arrhythmia database containing 48 half-hour records with various labels. The database was measured in MLII and V5 at a sampling rate of 360 Hz from two leads. The experiment was conducted using only the MLII among the two leads. Annotations created by two or more experts are provided for each record, and they provide the locations of beats based on the R peak. The labels of the beats in 16 categories are also provided, which also provide the labels for ECG rhythms within each record. Sections corresponding to each rhythm in the database were defined as the label of the rhythm. The beat label ‘?’ was excluded because it was not found; thus, only 15 beat labels were used in the experiment. The rhythm labels were the following: normal sinus rhythm (N), AFIB, AFL, SBR, supraventricular tachyarrhythmia (SVTA), ventricular bigeminy (B), ventricular trigeminy (T), and paced rhythms (P). Representative 10-s ECG segments for four rhythm labels, N, AFL, AFIB, and SBR, are presented in
Figure 3. The green and red circles represent the R-peak locations and beats (not normal beats), respectively.
We configured two ECG rhythm sets for various ECG rhythms. First, we classified five rhythm classes: N, AFIB, SVTA, B, and T, which aimed to evaluate the performance of the model for rhythms with relatively few samples. Then, we evaluated AFIB and AFL, which are difficult to classify owing to their similar ECG rhythms. The second dataset consisted of six ECG rhythm classes: N, AFIB, B, P, AFL, and SBR. The number of samples per rhythm in each experimental dataset is summarized in
Table 1 and
Table 2.
Frequently occurring rhythms contain many data samples, while relatively rare T and SVTA contain fewer data samples. These result in data imbalance; thus, when configuring the data of a mini-batch in the learning process, different sampling weights were designated for each rhythm. Each mini-batch was obtained by weighted random sampling, and the value of the batch size was 16.
4.2. Hyperparameters and Settings
CNN weights were learned by transfer learning; hence, after a pre-trained initialized weight was called, the learning was carried out using an Adam optimizer. To search for the optimal values for hyperparameters, such as the learning rate and the number of epochs, a grid search was performed on the validation dataset. The learning rate was chosen as the best value in the range of 0.1 to 0.0001, and the number of epochs was chosen as the best value in the range of 10 to 50. As a result, the optimal performance on the validation dataset was at a learning rate of 0.001 and an epoch of 30. We also set the batch size to 4, which is the largest value that can be selected in the experimental environment settings. Five-fold cross-validation was conducted on all experiments.
5. Results
N and AFIB are studied often; thus, they were included. Two ECG rhythm datasets were created, and an experiment was conducted on these datasets. The first dataset consisted of rhythms with fewer samples than N and AFIB. Through this, we evaluated the ability of the proposed algorithm to distinguish N and AFIB simultaneously and differentiate rhythm classes with relatively few samples. The second rhythm dataset, which was created to assess the ability of the proposed algorithm to distinguish between AFIB and AFL, is discussed as a limitation of existing studies. For this goal, AFIB, AFL, N, and three additional ECG rhythms were included. We analyzed the ability of the proposed model for this rhythm dataset to classify AFIB and AFL.
The performance of the proposed algorithm was analyzed with respect to its accuracy (Acc), precision (Pre, known as PPV), sensitivity (Sen, known as recall), specificity (Spec), and F1 score.
5.1. Experiment to Classify ECG Rhythm with Few Samples
First, an experiment was conducted on the first rhythm dataset, and the five rhythm classes used were N, AFIB, SVTA, B, and T. The number of data samples for each rhythm is shown in
Table 1. The results of the experiment are summarized in
Table 3. The overall Acc was 99.08%, and the proposed method is suitable for rhythm classes with a relatively small number of data samples (e.g., B, T, and SVTA). Notably, the model was able to distinguish SVTA with the smallest number of samples with 100% accuracy.
A confusion matrix verifying the classification performance for each class is presented in
Table 4. According to the matrix, T is often misclassified as N because the definition of T is the ECG signal that generates a prematurity ventricular contraction after two normal beats in this class. Thus, a significant number of normal beats are included in the sequence, which makes it difficult for the model to differentiate between T and N.
5.2. Experiment for ECG Rhythms That Are Difficult to Distinguishable
The following is an experiment evaluating the performance of the proposed method using the second rhythm dataset. A total of 6 six rhythms, N, AFIB, P, SBR, B, and AFL, were tested. The number of samples for each rhythm is summarized in
Table 2.
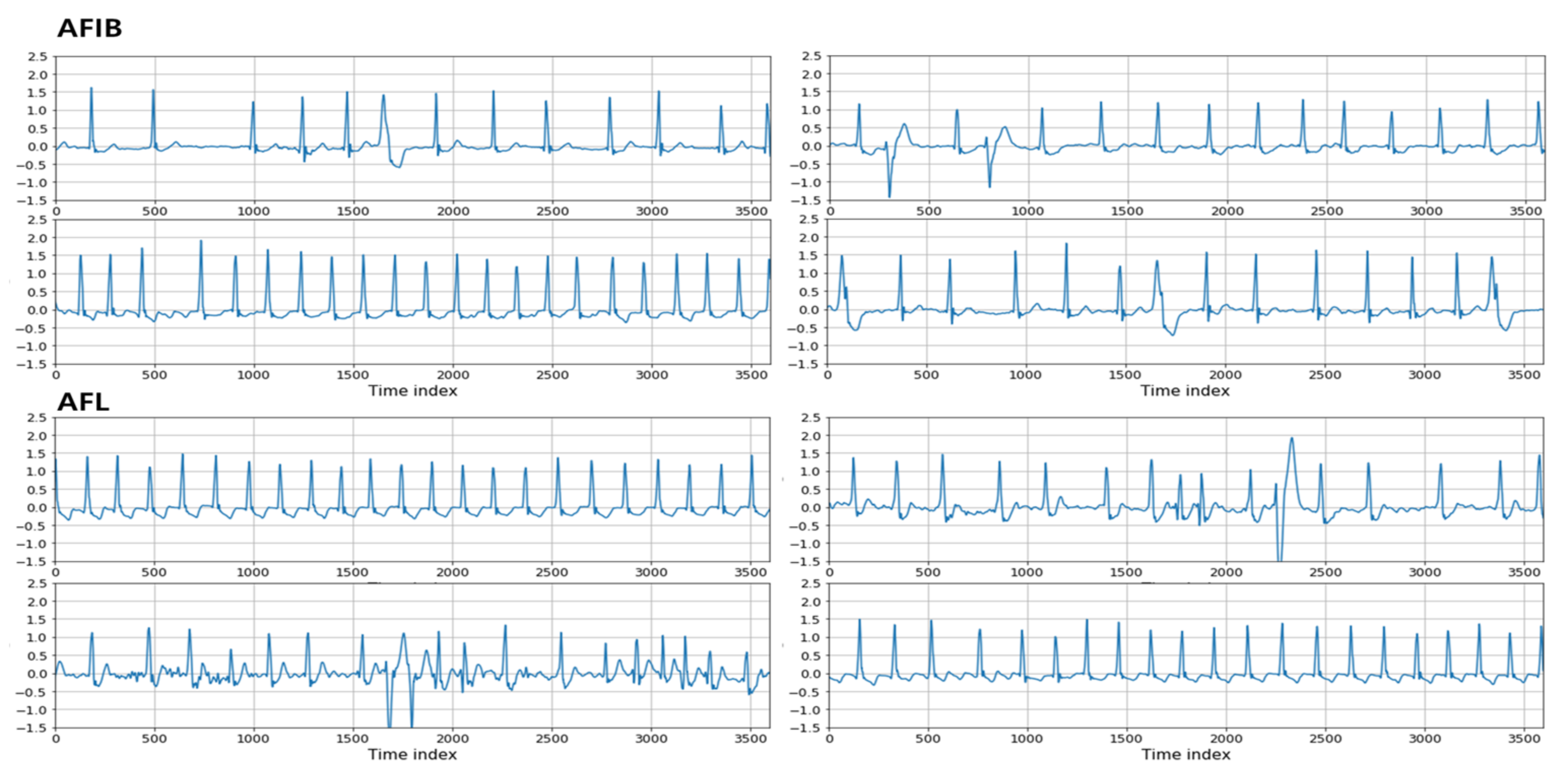
The purposes of this experiment include evaluating the ability of the proposed method to classify different ECG rhythms, including AFIB and AFL, with similar characteristics and determining whether the proposed method can be supplemented well. First, the appearance of the ECG waveforms of AFIB and AFL was confirmed in
Figure 4. After comparing AFIB and AFL, we found that some parts are different, while many parts are similar. Thus, we tried to distinguish AFIB and AFL, which are hard to separate, by distinguishing various ECG rhythms with the proposed algorithm.
The results of this experiment are summarized in
Table 5, which demonstrates that the proposed method was able to classify all the considered rhythms. The overall accuracy was 99.24%, while the F-1 score was ~99%, except for the AFL. Based on the confusion matrix shown in
Table 6, we found that the proposed algorithm was successful in classifying AFIB and AFL. Therefore, all rhythms except AFL are well classified. In the case of AFL, ~9% of AFL rhythms were misclassified as AFIB. Nevertheless, the proposed method shows significant performance for AFIB and AFL.
5.3. Comparison with a Recent Study
To compare the performance of the proposed method with existing methods, we refer to a recent paper that studied the same data and rhythms. The previous paper, for comparison with the proposed method in the first rhythm dataset, implemented a rhythm classification that combines beat unit and spectrogram unit features of an ECG signal [
27]. A comparison of the proposed and previous models relative to small ECG rhythm samples is shown in
Table 7. The F1-score was used to evaluate their performance. The proposed method showed better overall performance, and the performance for SVTA rhythms has improved particularly by >20%. The poor performance of the previous method is due to overfitting in N and AFIB, whereas our method classified the rhythms without overfitting due to the presence of many samples.
To evaluate the performance of the second rhythm dataset, a previous paper proposed a method merging the 1D signal and R–R interval as inputs of CNN using an ECG signal in one dimension and network, classifying rhythms through this approach [
26]. A comparison of the performance obtained for the previous and proposed methods on each class using the F1-score is shown in
Table 8. The performance of the proposed method for classifying AFIB and AFL improved by about 37% compared with the previous study. Additionally, the performance of other rhythms is ameliorated by 2%–3%.
To investigate the effect of noise in ECG signals on the performance of the proposed method, we performed additional experiments. To this end, some randomly generated noise was added to ECG signals at two different signal-to-noise ratio (SNR) levels of 6 dB and −6 dB, respectively. These noise-added ECG signals were used to produce BSM images and train a beat classification model. The F1-score results of rhythm classification with noise ECG signals are shown in
Table 9. Some noises up to the SNR level of 6 dB do not appear to affect the rhythm classification performance of our proposed method. On the other hand, as the SNR level increases to −6 dB, the overall performance degrades significantly. The overall accuracy also dropped from 99.24% to 94.03% at an SNR level of −6 dB.
6. Conclusions and Discussion
We classified various ECG rhythms through the proposed algorithm. We solved the problems caused by differences in the number of data samples and distinguished between AFIB and AFL, which have similar characteristics. Our method converts ECG signals into a new type of image, which we refer to as a BSM image, and it classifies the created image through CNN. The BSM image was designed to consider previously used features, such as the R–R interval and the listing pattern of ECG beats. The proposed method can be used in classifying difficult rhythms with few samples. It can also be used to distinguish AFIB and AFL with similar characteristics. Based on this, it seems that the method can be helpful in distinguishing rhythms that are less frequent but dangerous and similar but different.
There are some limitations of this study that need to be explored in future work. For example, the locations of the R peaks obtained from the database were used to create the BSM image; hence, when using databases that do not have information about the locations of R peaks, peak detection is required separately. If there are many incorrect detection results, the BSM image will be affected; thus, the overall performance of the method may decrease.
Author Contributions
Conceptualization, J.L. and M.S.; methodology, J.L. and M.S.; software, J.L.; validation, J.L.; investigation, J.L. and M.S.; writing—original draft preparation, J.L. and M.S.; writing—review and editing, J.L. and M.S.; visualization, J.L.; supervision, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was mainly supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education. (2022R1I1A3054343), and this work was partly supported by an NRF grant funded by the Korean government (MSIT: Ministry of Science and ICT) (No. 2020R1F1A1069984).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Acknowledgments
We would like to thank Physionet, which provided the data used in the experiments. We would like to thank the anonymous reviewers and the journal editors for their time and constructive comments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Homaeinezhad, M.; Atyabi, S.; Tavakkoli, E.; Toosi, H.; Ghaffari, A.; Ebrahimpour, R. ECG arrhythmia recognition via a neuro-SVM–KNN hybrid classifier with virtual QRS image-based geometrical features. Expert Syst. Appl. 2012, 39, 2047–2058. [Google Scholar] [CrossRef]
- Brenyo, A.; Aktas, M.K. Review of Complementary and Alternative Medical Treatment of Arrhythmias. Am. J. Cardiol. 2014, 113, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Madan, P.; Singh, V.; Singh, D.P.; Diwakar, M.; Pant, B.; Kishor, A. A Hybrid Deep Learning Approach for ECG-Based Arrhythmia Classification. Bioengineering 2022, 9, 152. [Google Scholar] [CrossRef] [PubMed]
- Al-Khatib, S.M.; Arshad, A.; Balk, E.M.; Das, S.R.; Hsu, J.C.; Joglar, J.A.; Page, R.L. Risk stratification for arrhythmic events in patients with asymptomatic pre-excitation: A systematic review for the 2015 ACC/AHA/HRS guideline for the management of adult patients with supraventricular tachycardia: A report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Circulation 2016, 133, e575–e586. [Google Scholar]
- Wu, M.; Lu, Y.; Yang, W.; Wong, S.Y. A Study on Arrhythmia via ECG Signal Classification Using the Convolutional Neural Network. Front. Comput. Neurosci. 2021, 14, 564015. [Google Scholar] [CrossRef]
- Kirchhof, P.; Bax, J.; Blomstrom-Lundquist, C.; Calkins, H.; Camm, A.J.; Cappato, R.; Cosio, F.; Crijns, H.; Diener, H.-C.; Goette, A.; et al. Early and comprehensive management of atrial fibrillation: Proceedings from the 2nd AFNET/EHRA consensus conference on atrial fibrillation entitled ‘research perspectives in atrial fibrillation’. Europace 2009, 11, 860–885. [Google Scholar] [CrossRef]
- Mehra, R. Global public health problem of sudden cardiac death. J. Electrocardiol. 2007, 40, S118–S122. [Google Scholar] [CrossRef]
- Strodthoff, N.; Wagner, P.; Schaeffter, T.; Samek, W. Deep Learning for ECG Analysis: Benchmarks and Insights from PTB-XL. IEEE J. Biomed. Health Inform. 2020, 25, 1519–1528. [Google Scholar] [CrossRef]
- Polat, K.; Güneş, S. Detection of ECG Arrhythmia using a differential expert system approach based on principal component analysis and least square support vector machine. Appl. Math. Comput. 2007, 186, 898–906. [Google Scholar] [CrossRef]
- Melgani, F.; Bazi, Y. Classification of electrocardiogram signals with support vector machines and particle swarm optimization. IEEE Trans. Inf. Technol. Biomed. 2008, 12, 667–677. [Google Scholar] [CrossRef]
- Oh, S.L.; Ng, E.Y.; San Tan, R.; Acharya, U.R. Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput. Biol. Med. 2018, 102, 278–287. [Google Scholar] [CrossRef]
- Mathews, S.M.; Kambhamettu, C.; Barner, K.E. A novel application of deep learning for single-lead ECG classification. Comput. Biol. Med. 2018, 99, 53–62. [Google Scholar] [CrossRef]
- Li, D.; Zhang, J.; Zhang, Q.; Wei, X. Classification of ECG Signals Based on 1D Convolution Neural Network. In Proceedings of the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), Dalian, China, 12–15 October 2017; pp. 1–6. [Google Scholar]
- Plesinger, F.; Nejedly, P.; Viscor, I.; Halamek, J.; Jurak, P. Automatic Detection of Atrial Fibrillation and Other Arrhythmias in Holter ECG Recordings Using Rhythm Features and Neural Networks. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Rubin, J.; Parvaneh, S.; Rahman, A.; Conroy, B.; Babaeizadeh, S. Densely connected convolutional networks for detection of atrial fibrillation from short single-lead ECG recordings. J. Electrocardiol. 2018, 51, S18–S21. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, Y.; Deng, Y.; Zhang, X. ECG authentication system design incorporating a convolutional neural network and generalized S-Transformation. Comput. Biol. Med. 2018, 102, 168–179. [Google Scholar] [CrossRef]
- Kamaleswaran, R.; Mahajan, R.; Akbilgic, O. A robust deep convolutional neural network for the classification of abnormal cardiac rhythm using single lead electrocardiograms of variable length. Physiol. Meas. 2018, 39, 035006. [Google Scholar] [CrossRef]
- Nannavecchia, A.; Girardi, F.; Fina, P.R.; Scalera, M.; Dimauro, G. Personal heart health monitoring based on 1D convolutional neural network. J. Imaging 2021, 7, 26. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, H.; Lu, P.; Wang, Z. An effective LSTM recurrent network to detect arrhythmia on imbalanced ECG dataset. J. Healthc. Eng. 2019, 2019, 6320651. [Google Scholar] [CrossRef]
- Yildirim, O.; Talo, M.; Ciaccio, E.J.; San Tan, R.; Acharya, U.R. Accurate deep neural network model to detect cardiac arrhythmia on more than 10,000 individual subject ECG records. Comput. Methods Programs Biomed. 2020, 197, 105740. [Google Scholar] [CrossRef]
- Luo, X.; Yang, L.; Cai, H.; Tang, R.; Chen, Y.; Li, W. Multi-classification of arrhythmias using a HCRNet on imbalanced ECG datasets. Comput. Methods Programs Biomed. 2021, 208, 106258. [Google Scholar] [CrossRef]
- Darmawahyuni, A.; Nurmaini, S.; Rachmatullah, M.N.; Tutuko, B.; Sapitri, A.I.; Firdaus, F.; Fansyuri, A.; Predyansyah, A. Deep learning-based electrocardiogram rhythm and beat features for heart abnormality classification. PeerJ Comput. Sci. 2022, 8, e825. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, A.; Ganesan, M.; Lavanya, R. Arrhythmia Classification on ECG Using Deep Learning. In Proceedings of the 2019 5th International Conference on Advanced Computing & Communication Systems (ICACCS), Coimbatore, India, 15–16 March 2019; pp. 365–369. [Google Scholar]
- Yildirim, O.; Baloglu, U.B.; Tan, R.-S.; Ciaccio, E.J.; Acharya, U.R. A new approach for arrhythmia classification using deep coded features and LSTM networks. Comput. Methods Programs Biomed. 2019, 176, 121–133. [Google Scholar] [CrossRef] [PubMed]
- CChen, C.; Hua, Z.; Zhang, R.; Liu, G.; Wen, W. Automated arrhythmia classification based on a combination network of CNN and LSTM. Biomed. Signal Process. Control 2019, 57, 101819. [Google Scholar] [CrossRef]
- Pokaprakarn, T.; Kitzmiller, R.R.; Moorman, J.R.; Lake, D.E.; Krishnamurthy, A.K.; Kosorok, M.R. Sequence to Sequence ECG Cardiac Rhythm Classification Using Convolutional Recurrent Neural Networks. IEEE J. Biomed. Health Inform. 2021, 26, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, Z.; Loni, M.; Daneshtalab, M.; Gharehbaghi, A. A Review on Deep Learning Methods for ECG Arrhythmia Classification. Expert Syst. Appl. X 2020, 7, 100033. [Google Scholar] [CrossRef]
- Sufi, F.; Khalil, I. Diagnosis of cardiovascular abnormalities from compressed ECG: A data mining-based approach. IEEE Trans. Inf. Technol. Biomed. 2010, 15, 33–39. [Google Scholar] [CrossRef]
- Martis, R.J.; Acharya, U.R.; Prasad, H.; Chua, C.K.; Lim, C.M.; Suri, J.S. Application of higher order statistics for atrial arrhythmia classification. Biomed. Signal Process. Control 2013, 8, 888–900. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Adam, M.; Lih, O.S.; Hong, T.J.; Sudarshan, V.K.; Koh, J.E. Automated characterization of arrhythmias using nonlinear features from tachycardia ECG beats. In Proceedings of the 2016 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Budapest, Hungary, 9–12 October 2016; pp. 000533–000538. [Google Scholar]
- Al Rahhal, M.M.; Bazi, Y.; Al Zuair, M.; Othman, E.; BenJdira, B. Convolutional Neural Networks for Electrocardiogram Classification. J. Med. Biol. Eng. 2018, 38, 1014–1025. [Google Scholar] [CrossRef]
- Fan, X.; Yao, Q.; Cai, Y.; Miao, F.; Sun, F.; Li, Y. Multiscaled Fusion of Deep Convolutional Neural Networks for Screening Atrial Fibrillation From Single Lead Short ECG Recordings. IEEE J. Biomed. Health Inform. 2018, 22, 1744–1753. [Google Scholar] [CrossRef]
- Zhong, W.; Liao, L.; Guo, X.; Wang, G. A deep learning approach for fetal QRS complex detection. Physiol. Meas. 2018, 39, 045004. [Google Scholar] [CrossRef]
- Chu, J.; Wang, H.; Lu, W. A novel two-lead arrhythmia classification system based on CNN and LSTM. J. Mech. Med. Biol. 2019, 19, 1950004. [Google Scholar] [CrossRef]
- He, R.; Liu, Y.; Wang, K.; Zhao, N.; Yuan, Y.; Li, Q.; Zhang, H. Automatic Cardiac Arrhythmia Classification Using Combination of Deep Residual Network and Bidirectional LSTM. IEEE Access 2019, 7, 102119–102135. [Google Scholar] [CrossRef]
- Ochiai, K.; Takahashi, S.; Fukazawa, Y. Arrhythmia Detection from 2-Lead ECG Using Convolutional Denoising Autoencoders. In Proceedings of the KDD 2018 Deep Learning Day, London, UK, 20 August 2018; pp. 1–7. [Google Scholar]
- Hou, Y.; Jia, S.; Lun, X.; Hao, Z.; Shi, Y.; Li, Y.; Zeng, R.; Lv, J. GCNs-net: A graph convolutional neural network approach for decoding time-resolved eeg motor imagery signals. IEEE Trans. Neural Netw. Learn. Syst. 2022, 1–12. [Google Scholar] [CrossRef]
- Queiroz, J.A.; Azoubel, L.M.A.; Barros, A.K. Support system for classification of beat-to-beat arrhythmia based on variability and morphology of electrocardiogram. EURASIP J. Adv. Signal Process. 2019, 2019, 16. [Google Scholar] [CrossRef]
- Ihsanto, E.; Ramli, K.; Sudiana, D.; Gunawan, T.S. An efficient algorithm for cardiac arrhythmia classification using ensemble of depthwise separable convolutional neural networks. Appl. Sci. 2020, 10, 483. [Google Scholar] [CrossRef]
- Xia, Y.; Zhang, H.; Xu, L.; Gao, Z.; Zhang, H.; Liu, H.; Li, S. An automatic cardiac arrhythmia classification system with wearable electrocardiogram. IEEE Access 2018, 6, 16529–16538. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2015, 63, 664–675. [Google Scholar] [CrossRef]
- Li, W.; Li, J. Local Deep Field for Electrocardiogram Beat Classification. IEEE Sens. J. 2017, 18, 1656–1664. [Google Scholar] [CrossRef]
- Rashed-Al-Mahfuz, M.; Moni, M.A.; Lio’, P.; Islam, S.M.S.; Berkovsky, S.; Khushi, M.; Quinn, J.M. Deep convolutional neural networks based ECG beats classification to diagnose cardiovascular conditions. Biomed. Eng. Lett. 2021, 11, 147–162. [Google Scholar] [CrossRef]
- Wu, Z.; Feng, X.; Yang, C. A Deep Learning Method to Detect Atrial Fibrillation Based on Continuous Wavelet Transform. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 1908–1912. [Google Scholar]
- Gliner, V.; Yaniv, Y. Identification of Features for Machine Learning Analysis for Automatic Arrhythmogenic Event Classification. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Faust, O.; Shenfield, A.; Kareem, M.; San, T.R.; Fujita, H.; Acharya, U.R. Automated detection of atrial fibrillation using long short-term memory network with RR interval signals. Comput. Biol. Med. 2018, 102, 327–335. [Google Scholar] [CrossRef]
- Behar, J.A.; Rosenberg, A.; Yaniv, Y.; Oster, J. Rhythm and Quality Classification from Short ECGs Recorded Using a Mobile Device. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Zhai, X.; Tin, C. Automated ECG Classification Using Dual Heartbeat Coupling Based on Convolutional Neural Network. IEEE Access 2018, 6, 27465–27472. [Google Scholar] [CrossRef]
- Zihlmann, M.; Perekrestenko, D.; Tschannen, M. Convolutional Recurrent Neural Networks for Electrocardiogram Classification. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Zhao, Z.; Särkkä, S.; Rad, A.B. Kalman-based Spectro-Temporal ECG Analysis using Deep Convolutional Networks for Atrial Fibrillation Detection. J. Signal Process. Syst. 2020, 92, 621–636. [Google Scholar] [CrossRef]
- Hsieh, C.-H.; Li, Y.-S.; Hwang, B.-J.; Hsiao, C.-H. Detection of Atrial Fibrillation Using 1D Convolutional Neural Network. Sensors 2020, 20, 2136. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Shin, M. Learning Explainable Time-Morphology Patterns for Automatic Arrhythmia Classification from Short Single-Lead ECGs. Sensors 2021, 21, 4331. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Sun, Y.; Yan, H.; Wu, X. ECG signal classification with binarized convolutional neural network. Comput. Biol. Med. 2020, 121, 103800. [Google Scholar] [CrossRef]
- Sepahvand, M.; Abdali-Mohammadi, F. A novel method for reducing arrhythmia classification from 12-lead ECG signals to single-lead ECG with minimal loss of accuracy through teacher-student knowledge distillation. Inf. Sci. 2022, 593, 64–77. [Google Scholar] [CrossRef]
- Yao, Q.; Wang, R.; Fan, X.; Liu, J.; Li, Y. Multi-class arrhythmia detection from 12-lead varied-length ECG using attention-based time-incremental convolutional neural network. Inf. Fusion 2020, 53, 174–182. [Google Scholar] [CrossRef]
- Jin, Y.; Li, Z.; Liu, Y.; Liu, J.; Qin, C.; Zhao, L.; Liu, C. Multi-class 12-lead ECG automatic diagnosis based on a novel subdomain adaptive deep network. Sci. China Technol. Sci. 2022, 65, 2617–2630. [Google Scholar] [CrossRef]
- Asgharzadeh-Bonab, A.; Amirani, M.C.; Mehri, A. Spectral entropy and deep convolutional neural network for ECG beat classification. Biocybern. Biomed. Eng. 2020, 40, 691–700. [Google Scholar] [CrossRef]
- Murat, F.; Yildirim, O.; Talo, M.; Demir, Y.; Tan, R.-S.; Ciaccio, E.J.; Acharya, U.R. Exploring deep features and ECG attributes to detect cardiac rhythm classes. Knowl.-Based Syst. 2021, 232, 107473. [Google Scholar] [CrossRef]
- Panganiban, E.B.; Paglinawan, A.C.; Chung, W.Y.; Paa, G.L.S. ECG diagnostic support system (EDSS): A deep learning neural network based classification system for detecting ECG abnormal rhythms from a low-powered wearable biosensors. Sens. Bio-Sens. Res. 2021, 31, 100398. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).