Comparing 3D, 2.5D, and 2D Approaches to Brain Image Auto-Segmentation
Abstract
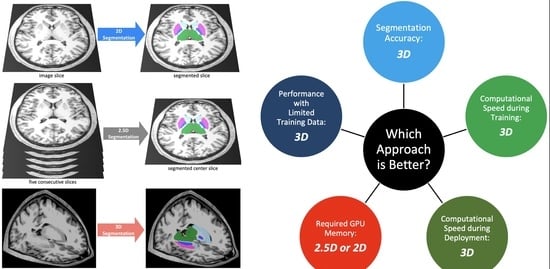
:1. Introduction
2. Methods
2.1. Dataset
2.2. Anatomic Segmentations
2.3. Image Pre-Processing
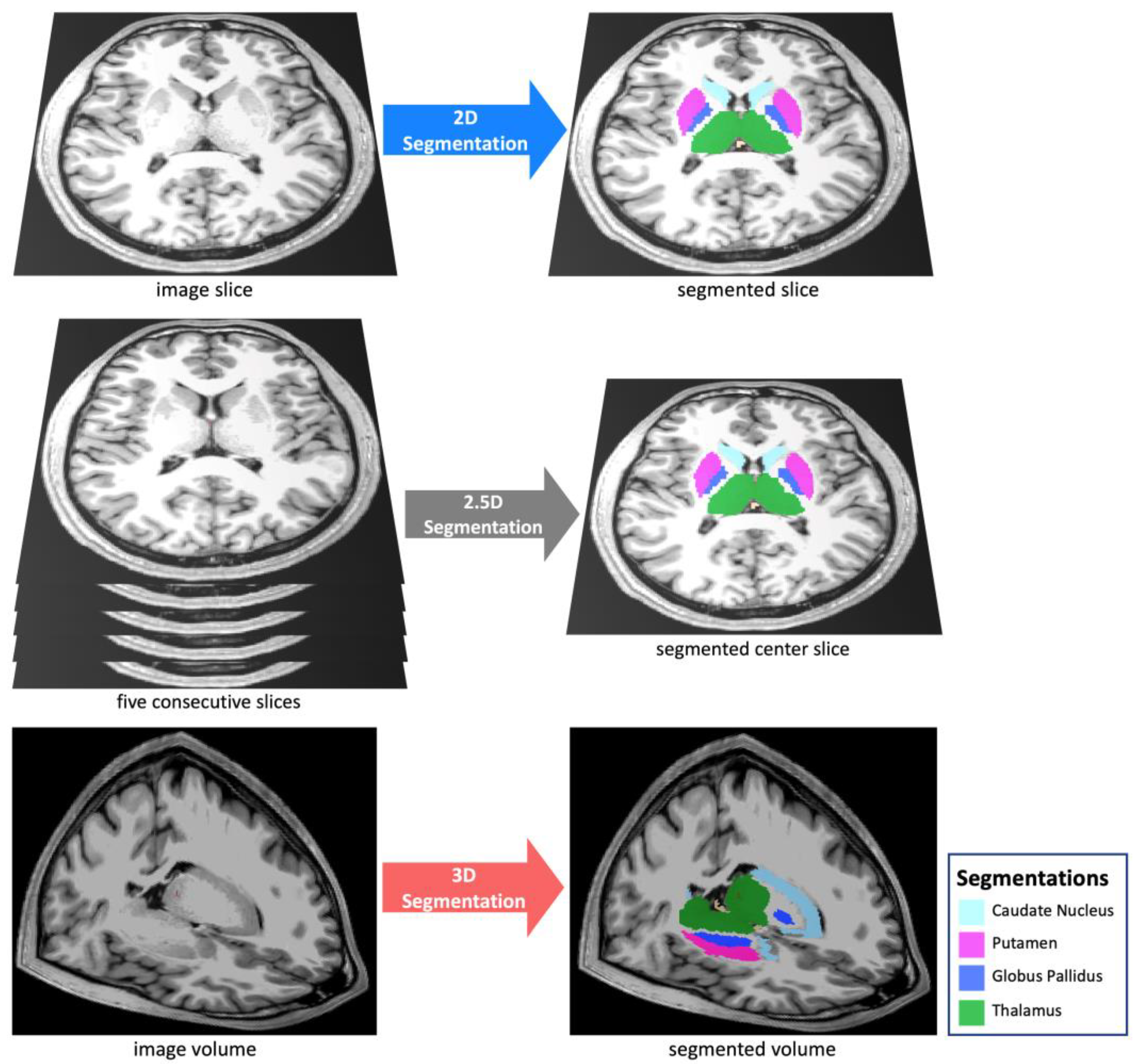
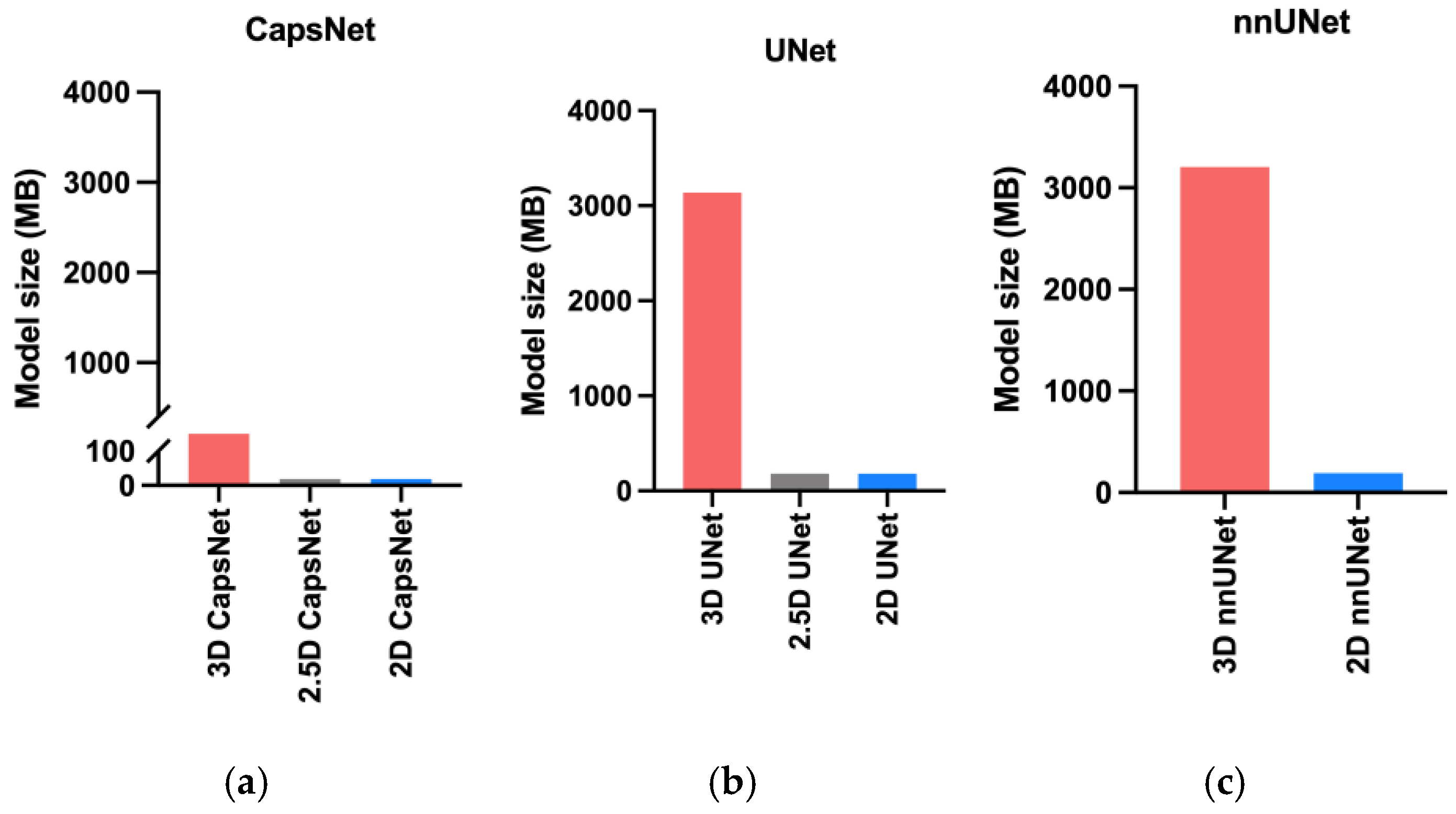
2.4. Auto-Segmentation Models
2.5. Training
2.6. Performance Metrics
2.7. Implementation
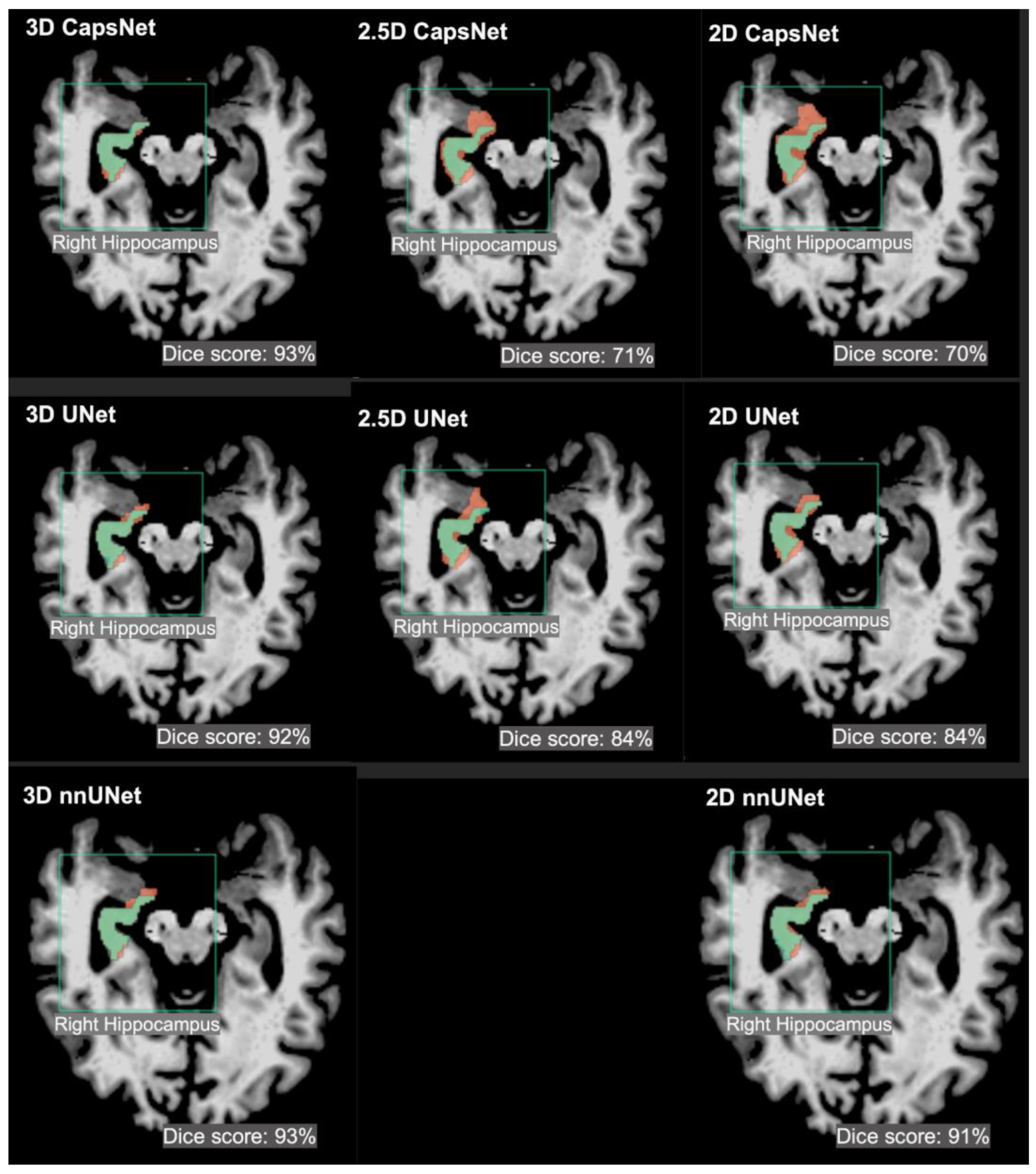
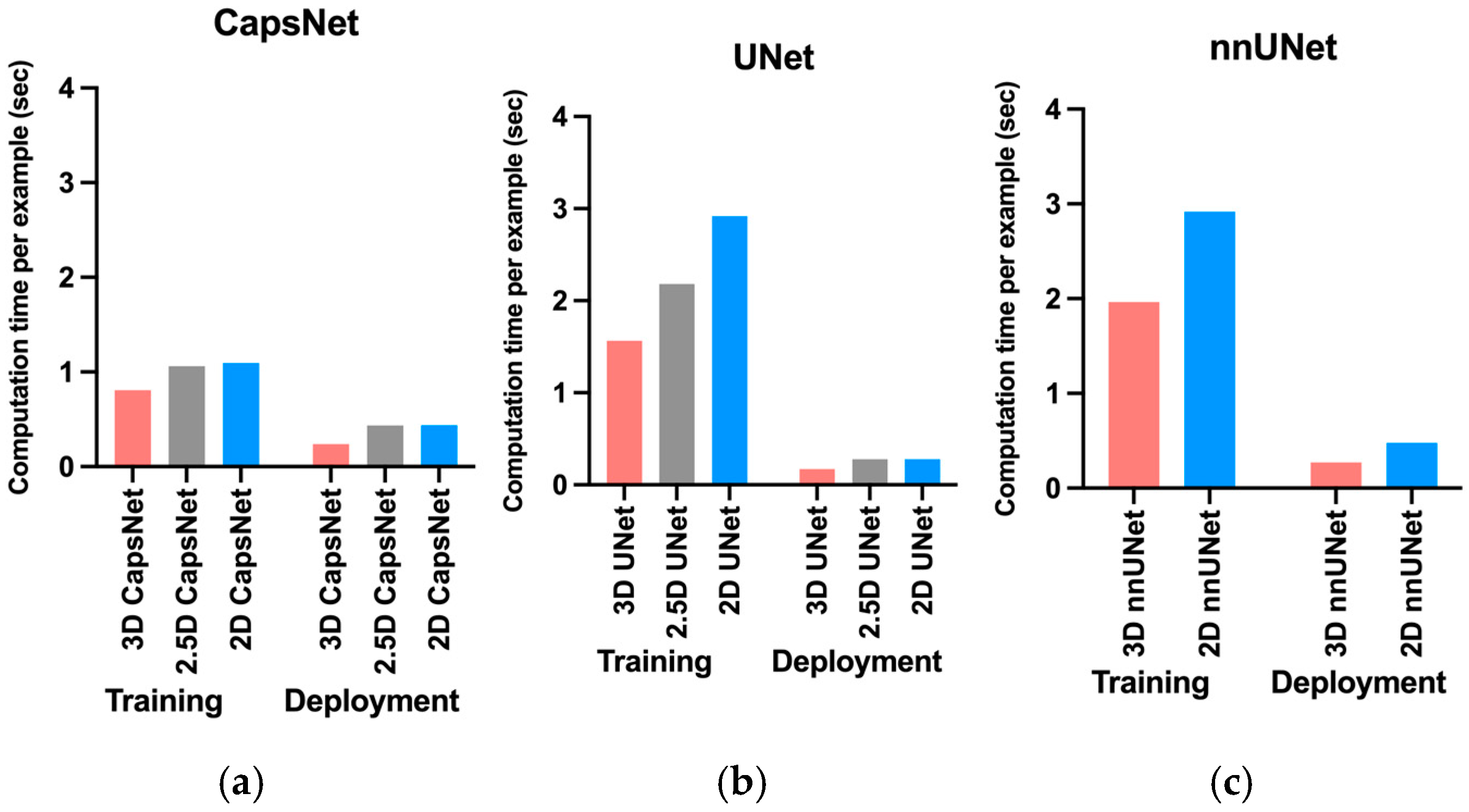
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2D segmentation | two-dimensional segmentation |
| 2.5D segmentation | enhanced two-dimensional segmentation |
| 3D segmentation | three-dimensional segmentation |
| ADNI | Alzheimer’s disease neuroimaging initiative |
| CapsNet | capsule network |
| CPU | central processing unit |
| CT | computed tomography |
| GB | giga-byte |
| GPU | graphics processing unit |
| MRI | magnetic resonance imaging |
References
- Feng, C.H.; Cornell, M.; Moore, K.L.; Karunamuni, R.; Seibert, T.M. Automated contouring and planning pipeline for hippocampal-avoidant whole-brain radiotherapy. Radiat. Oncol. 2020, 15, 251. [Google Scholar] [CrossRef] [PubMed]
- Dasenbrock, H.H.; See, A.P.; Smalley, R.J.; Bi, W.L.; Dolati, P.; Frerichs, K.U.; Golby, A.J.; Chiocca, E.A.; Aziz-Sultan, M.A. Frameless Stereotactic Navigation during Insular Glioma Resection using Fusion of Three-Dimensional Rotational Angiography and Magnetic Resonance Imaging. World Neurosurg. 2019, 126, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Dolati, P.; Gokoglu, A.; Eichberg, D.; Zamani, A.; Golby, A.; Al-Mefty, O. Multimodal navigated skull base tumor resection using image-based vascular and cranial nerve segmentation: A prospective pilot study. Surg. Neurol. Int. 2015, 6, 172. [Google Scholar] [CrossRef] [PubMed]
- Clerx, L.; Gronenschild, H.B.M.; Echavarri, C.; Aalten, P.; Jacobs, H.I.L. Can FreeSurfer Compete with Manual Volumetric Measurements in Alzheimer’s Disease? Curr. Alzheimer Res. 2015, 12, 358–367. [Google Scholar] [CrossRef]
- Bousabarah, K.; Ruge, M.; Brand, J.-S.; Hoevels, M.; Rueß, D.; Borggrefe, J.; Hokamp, N.G.; Visser-Vandewalle, V.; Maintz, D.; Treuer, H.; et al. Deep convolutional neural networks for automated segmentation of brain metastases trained on clinical data. Radiat. Oncol. 2020, 15, 87. [Google Scholar] [CrossRef]
- Nimsky, C.; Ganslandt, O.; Cerny, S.; Hastreiter, P.; Greiner, G.; Fahlbusch, R. Quantification of, visualization of, and compensation for brain shift using intraoperative magnetic resonance imaging. Neurosurgery 2000, 47, 1070–1079. [Google Scholar] [CrossRef]
- Gerard, I.J.; Kersten-Oertel, M.; Petrecca, K.; Sirhan, D.; Hall, J.A.; Collins, D.L. Brain shift in neuronavigation of brain tumors: A review. Med. Image Anal. 2017, 35, 403–420. [Google Scholar] [CrossRef]
- Lorenzen, E.L.; Kallehauge, J.F.; Byskov, C.S.; Dahlrot, R.H.; Haslund, C.A.; Guldberg, T.L.; Lassen-Ramshad, Y.; Lukacova, S.; Muhic, A.; Nyström, P.W.; et al. A national study on the inter-observer variability in the delineation of organs at risk in the brain. Acta Oncol. 2021, 60, 1548–1554. [Google Scholar] [CrossRef]
- Duong, M.; Rudie, J.; Wang, J.; Xie, L.; Mohan, S.; Gee, J.; Rauschecker, A. Convolutional Neural Network for Automated FLAIR Lesion Segmentation on Clinical Brain MR Imaging. Am. J. Neuroradiol. 2019, 40, 1282–1290. [Google Scholar] [CrossRef]
- Zettler, N.; Mastmeyer, A. Comparison of 2D vs. 3D U-Net Organ Segmentation in abdominal 3D CT images. arXiv 2021, arXiv:2107.04062. [Google Scholar]
- Ou, Y.; Yuan, Y.; Huang, X.; Wong, K.; Volpi, J.; Wang, J.Z.; Wong, S.T.C. LambdaUNet: 2.5D Stroke Lesion Segmentation of Diffusion-weighted MR Images. arXiv 2021, arXiv:arXiv:2104.13917. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Douglas, L.; Drukker, K.; Hu, Q.; Fuhrman, J.; Sheth, D.; Giger, M.L. Comparison of 2D and 3D U-Net breast lesion segmentations on DCE-MRI. In Medical Imaging 2021: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2021; Volume 11597, pp. 81–87. [Google Scholar]
- Kern, D.; Klauck, U.; Ropinski, T.; Mastmeyer, A. 2D vs. 3D U-Net abdominal organ segmentation in CT data using organ bounds. In Medical Imaging 2021: Imaging Informatics for Healthcare, Research, and Applications; SPIE: Bellingham, WA, USA, 2021; Volume 11601, pp. 192–200. [Google Scholar]
- Kulkarni, A.; Carrion-Martinez, I.; Dhindsa, K.; Alaref, A.A.; Rozenberg, R.; van der Pol, C.B. Pancreas adenocarcinoma CT texture analysis: Comparison of 3D and 2D tumor segmentation techniques. Abdom. Imaging 2020, 46, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Crawford, K.L.; Neu, S.C.; Toga, A.W. The Image and Data Archive at the Laboratory of Neuro Imaging. Neuroimage 2016, 124, 1080–1083. [Google Scholar] [CrossRef] [PubMed]
- Weiner, M.; Petersen, R.; Aisen, P. Alzheimer’s Disease Neuroimaging Initiative 2014. Available online: https://clinicaltrials.gov/ct2/show/NCT00106899 (accessed on 21 March 2022).
- Ochs, A.L.; Ross, D.E.; Zannoni, M.D.; Abildskov, T.J.; Bigler, E.D.; Alzheimer’s Disease Neuroimaging Initiative. Comparison of Automated Brain Volume Measures obtained with NeuroQuant® and FreeSurfer. J. Neuroimaging 2015, 25, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Fischl, B. FreeSurfer. NeuroImage 2012, 62, 774–781. [Google Scholar] [CrossRef]
- Fischl, B.; Salat, D.H.; Busa, E.; Albert, M.; Dieterich, M.; Haselgrove, C.; van der Kouwe, A.; Killiany, R.; Kennedy, D.; Klaveness, S.; et al. Whole Brain Segmentation: Automated Labeling of Neuroanatomical Structures in the Human Brain. Neuron 2002, 33, 341–355. [Google Scholar] [CrossRef]
- Ganzetti, M.; Wenderoth, N.; Mantini, D. Quantitative Evaluation of Intensity Inhomogeneity Correction Methods for Structural MR Brain Images. Neuroinformatics 2015, 14, 5–21. [Google Scholar] [CrossRef]
- Somasundaram, K.; Kalaiselvi, T. Automatic brain extraction methods for T1 magnetic resonance images using region labeling and morphological operations. Comput. Biol. Med. 2011, 41, 716–725. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Avesta, A.; Hui, Y.; Aboian, M.; Duncan, J.; Krumholz, H.M.; Aneja, S. 3D Capsule Networks for Brain MRI Segmentation. medRxiv 2021. [Google Scholar] [CrossRef]
- Yin, X.-X.; Sun, L.; Fu, Y.; Lu, R.; Zhang, Y. U-Net-Based Medical Image Segmentation. J. Healthc. Eng. 2022, 2022, 4189781. [Google Scholar] [CrossRef] [PubMed]
- Rudie, J.D.; Weiss, D.A.; Colby, J.B.; Rauschecker, A.M.; Laguna, B.; Braunstein, S.; Sugrue, L.P.; Hess, C.P.; Villanueva-Meyer, J.E. Three-dimensional U-Net Convolutional Neural Network for Detection and Segmentation of Intracranial Metastases. Radiol. Artif. Intell. 2021, 3, e200204. [Google Scholar] [CrossRef] [PubMed]
- LaLonde, R.; Xu, Z.; Irmakci, I.; Jain, S.; Bagci, U. Capsules for biomedical image segmentation. Med. Image Anal. 2020, 68, 101889. [Google Scholar] [CrossRef] [PubMed]
- Rauschecker, A.M.; Gleason, T.J.; Nedelec, P.; Duong, M.T.; Weiss, D.A.; Calabrese, E.; Colby, J.B.; Sugrue, L.P.; Rudie, J.D.; Hess, C.P. Interinstitutional Portability of a Deep Learning Brain MRI Lesion Segmentation Algorithm. Radiol. Artif. Intell. 2022, 4, e200152. [Google Scholar] [CrossRef]
- Rudie, J.D.; Weiss, D.A.; Saluja, R.; Rauschecker, A.M.; Wang, J.; Sugrue, L.; Bakas, S.; Colby, J.B. Multi-Disease Segmentation of Gliomas and White Matter Hyperintensities in the BraTS Data Using a 3D Convolutional Neural Network. Front. Comput. Neurosci. 2019, 13, 84. [Google Scholar] [CrossRef]
- Weiss, D.A.; Saluja, R.; Xie, L.; Gee, J.C.; Sugrue, L.P.; Pradhan, A.; Bryan, R.N.; Rauschecker, A.M.; Rudie, J.D. Automated multiclass tissue segmentation of clinical brain MRIs with lesions. NeuroImage Clin. 2021, 31, 102769. [Google Scholar] [CrossRef]
- Yaqub, M.; Feng, J.; Zia, M.; Arshid, K.; Jia, K.; Rehman, Z.; Mehmood, A. State-of-the-Art CNN Optimizer for Brain Tumor Segmentation in Magnetic Resonance Images. Brain Sci. 2020, 10, 427. [Google Scholar] [CrossRef]
- Sun, Y.C.; Hsieh, A.T.; Fang, S.T.; Wu, H.M.; Kao, L.W.; Chung, W.Y.; Chen, H.-H.; Liou, K.-D.; Lin, Y.-S.; Guo, W.-Y.; et al. Can 3D artificial intelligence models outshine 2D ones in the detection of intracranial metastatic tumors on magnetic resonance images? J. Chin. Med. Assoc. JCMA 2021, 84, 956–962. [Google Scholar] [CrossRef]
- Nemoto, T.; Futakami, N.; Yagi, M.; Kumabe, A.; Takeda, A.; Kunieda, E.; Shigematsu, N. Efficacy evaluation of 2D, 3D U-Net semantic segmentation and atlas-based segmentation of normal lungs excluding the trachea and main bronchi. J. Radiat Res. 2020, 61, 257–264. [Google Scholar] [CrossRef]
- Tran, M.; Vo-Ho, V.-K.; Le, N.T.H. 3DConvCaps: 3DUnet with Convolutional Capsule Encoder for Medical Image Segmentation. arXiv 2022, arXiv:arXiv:2205.09299. [Google Scholar] [CrossRef]
- Tran, M.; Ly, L.; Hua, B.-S.; Le, N. SS-3DCapsNet: Self-supervised 3D Capsule Networks for Medical Segmentation on Less La-beled Data. arXiv 2022, arXiv:arXiv:2201.05905. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Nguyen, T.; Hua, B.-S.; Le, N. 3D UCaps: 3D Capsule Unet for Volumetric Image Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; Springer International Publishing: Cham, Switzerland, 2021; pp. 548–558. [Google Scholar]
- Bonheur, S.; Štern, D.; Payer, C.; Pienn, M.; Olschewski, H.; Urschler, M. Matwo-CapsNet: A Multi-label Semantic Segmentation Capsules Network. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; Volume 11768, pp. 664–672. [Google Scholar]
- Dong, J.; Liu, C.; Yang, C.; Lin, N.; Cao, Y. Robust Segmentation of the Left Ventricle from Cardiac MRI via Capsule Neural Network. In Proceedings of the 2nd International Symposium on Image Computing and Digital Medicine, ISICDM 2018, New York, NY, USA, 23 October 2019; pp. 88–91. [Google Scholar]
- Angermann, C.; Haltmeier, M. Random 2.5D U-net for Fully 3D Segmentation. In Machine Learning and Medical Engineering for Cardiovascular Health and Intravascular Imaging and Computer Assisted Stenting; Springer: Cham, Switzerland, 2019; Volume 11794, pp. 158–166. [Google Scholar]
- Li, J.; Liao, G.; Sun, W.; Sun, J.; Sheng, T.; Zhu, K.; von Deneen, K.M.; Zhang, Y. A 2.5D semantic segmentation of the pancreas using attention guided dual context embedded U-Net. Neurocomputing 2022, 480, 14–26. [Google Scholar] [CrossRef]

| Data Partitions | Number of MRIs | Number of Patients | Age (Mean ± SD) | Gender † | Diagnosis †† |
|---|---|---|---|---|---|
| Training set | 3199 | 841 | 76 ± 7 | 42% F, 58% M | 29% CN, 54% MCI, 17% AD |
| Validation set | 117 | 30 | 75 ± 6 | 30% F, 70% M | 21% CN, 59% MCI, 20% AD |
| Test set | 114 | 30 | 77 ± 7 | 33% F, 67% M | 27% CN, 47% MCI, 26% AD |
| CapsNet | |||
|---|---|---|---|
| Brain Structure | 3D Dice (95% CI) | 2.5D Dice (95% CI) | 2D Dice (95% CI) |
| 3rd ventricle | 95% (94 to 96) | 90% (89 to 91) | 90% (88 to 92) |
| Thalamus | 94% (93 to 95) | 76% (74 to 78) | 75% (72 to 78) |
| Hippocampus | 92% (91 to 93) | 73% (71 to 75) | 71% (68 to 74) |
| UNet | |||
| Brain Structure | 3D Dice (95% CI) | 2.5D Dice (95% CI) | 2D Dice (95% CI) |
| 3rd ventricle | 96% (95 to 97) | 92% (91 to 93) | 91% (89 to 91) |
| Thalamus | 95% (94 to 96) | 92% (91 to 93) | 90% (88 to 92) |
| Hippocampus | 93% (92 to 94) | 86% (84 to 88) | 88% (86 to 90) |
| nnUNet | nnUNet | nnUNet | nnUNet |
| Brain Structure | Brain Structure | Brain Structure | Brain Structure |
| 3rd ventricle | 3rd ventricle | 3rd ventricle | 3rd ventricle |
| Thalamus | Thalamus | Thalamus | Thalamus |
| Hippocampus | Hippocampus | Hippocampus | Hippocampus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avesta, A.; Hossain, S.; Lin, M.; Aboian, M.; Krumholz, H.M.; Aneja, S. Comparing 3D, 2.5D, and 2D Approaches to Brain Image Auto-Segmentation. Bioengineering 2023, 10, 181. https://doi.org/10.3390/bioengineering10020181
Avesta A, Hossain S, Lin M, Aboian M, Krumholz HM, Aneja S. Comparing 3D, 2.5D, and 2D Approaches to Brain Image Auto-Segmentation. Bioengineering. 2023; 10(2):181. https://doi.org/10.3390/bioengineering10020181
Chicago/Turabian StyleAvesta, Arman, Sajid Hossain, MingDe Lin, Mariam Aboian, Harlan M. Krumholz, and Sanjay Aneja. 2023. "Comparing 3D, 2.5D, and 2D Approaches to Brain Image Auto-Segmentation" Bioengineering 10, no. 2: 181. https://doi.org/10.3390/bioengineering10020181
APA StyleAvesta, A., Hossain, S., Lin, M., Aboian, M., Krumholz, H. M., & Aneja, S. (2023). Comparing 3D, 2.5D, and 2D Approaches to Brain Image Auto-Segmentation. Bioengineering, 10(2), 181. https://doi.org/10.3390/bioengineering10020181