Myocardial Segmentation of Tagged Magnetic Resonance Images with Transfer Learning Using Generative Cine-To-Tagged Dataset Transformation
Abstract
1. Introduction
2. Materials and Methods
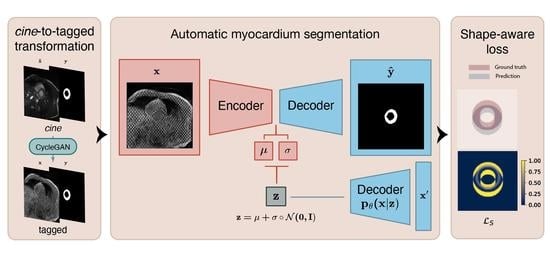
2.1. Unpaired and Unlabeled Cine to Tagged Image Transformation
2.1.1. Public Annotated Cine Datasets
2.1.2. Physics-Driven Cine to Tagged Data Transformation
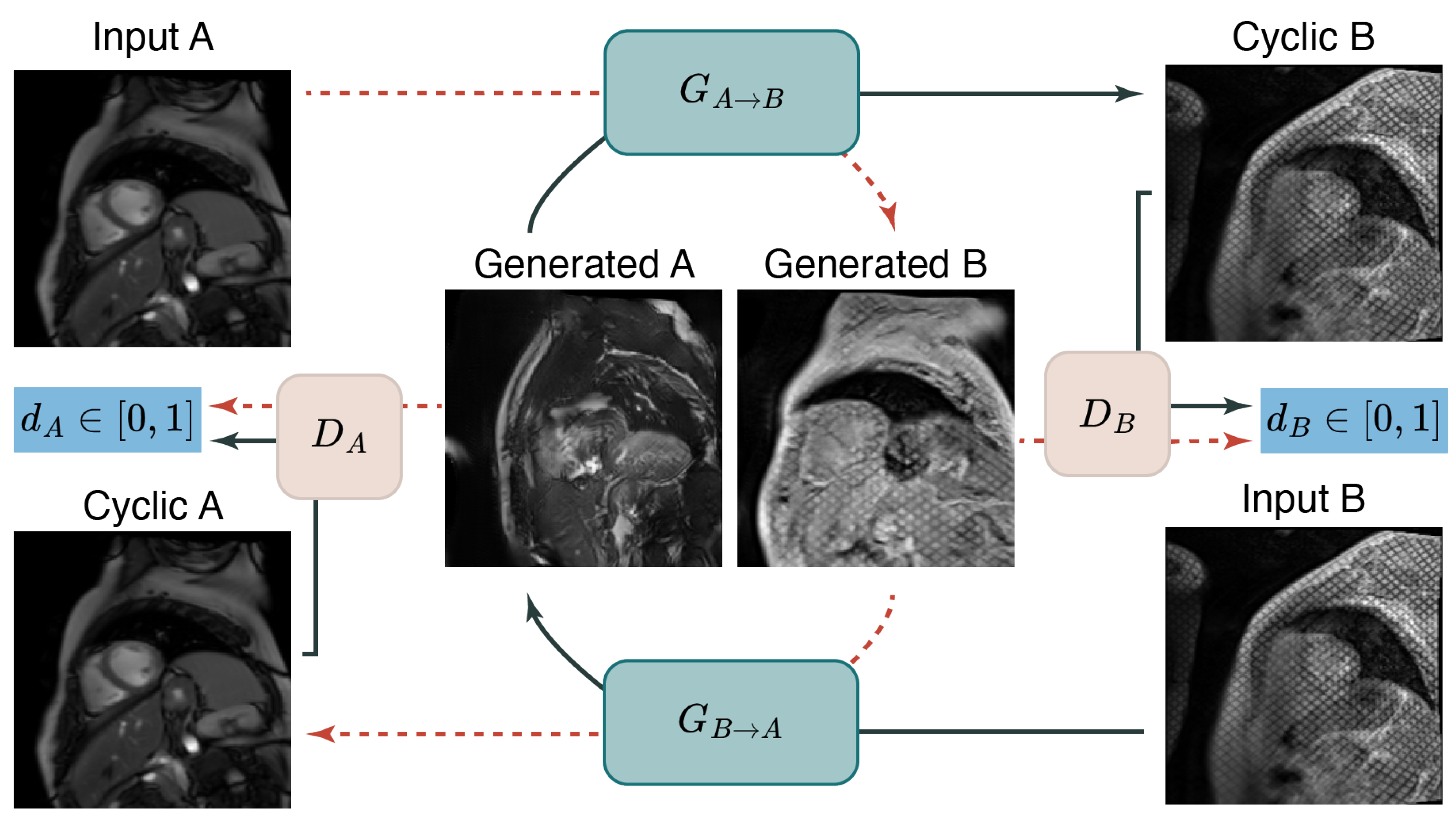
2.1.3. Deep Learning Based Cine to Tagged Data Transformation
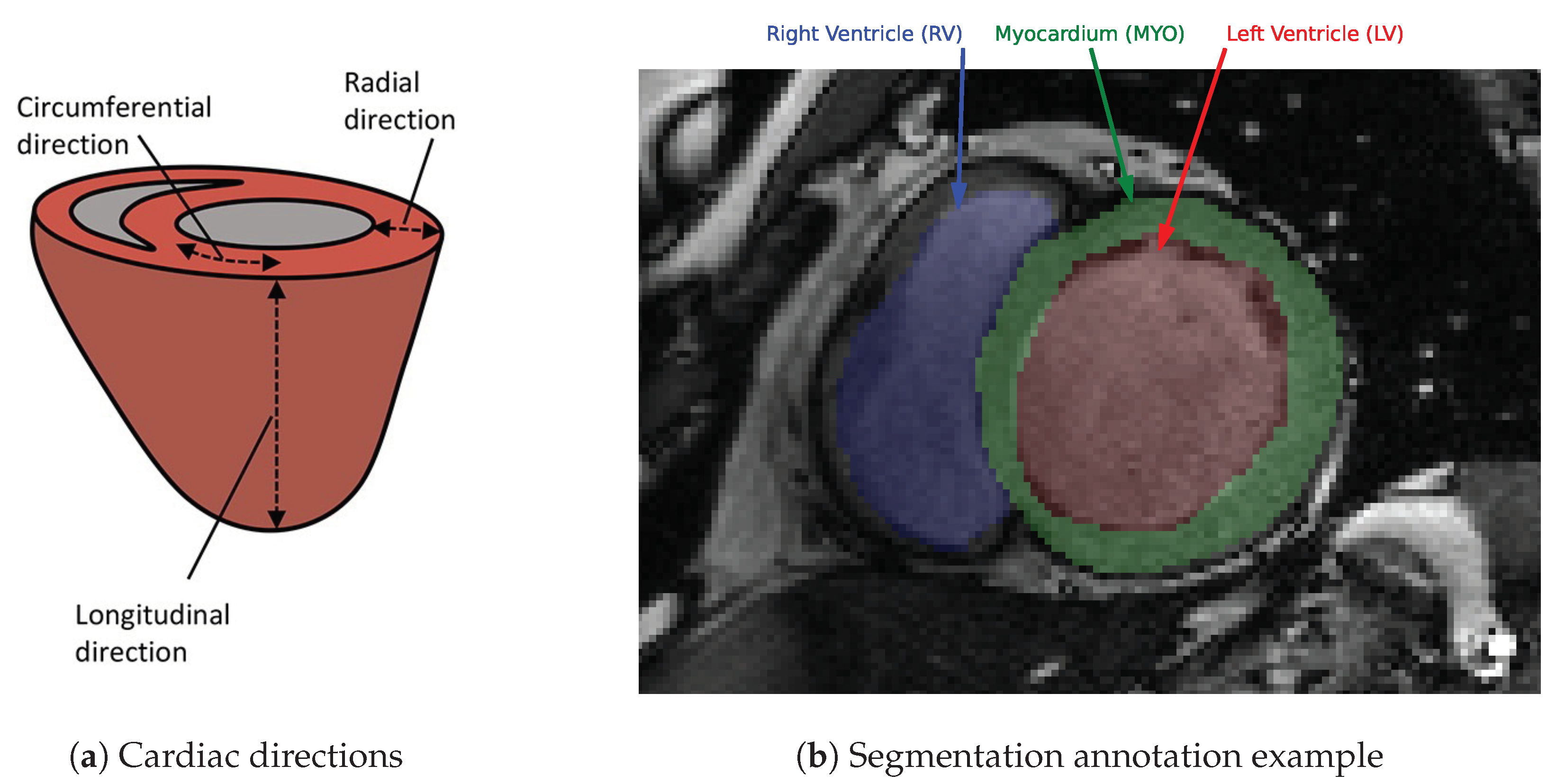
2.2. Segmentation of the Myocardium in Tagged CMR Images
2.2.1. Dataset
2.2.2. Training Strategy
- Train from scratch.
- Pretrain with public datasets of cine images.
- Pretrain with public datasets of cine images, transformed into a simulated tagged domain with the physics-driven method.
- Pretrain with public datasets of cine images, transformed into a simulated tagged domain with the .
2.2.3. Network Architectures
2.2.4. Train-Time Data Augmentation
2.2.5. Optimization
2.2.6. Evaluation Metrics
3. Results
3.1. Unpaired and Unlabeled Cine to Tagged Image Transformation
3.2. Segmentation of the Myocardium in Tagged CMR Images
3.2.1. Model Architecture
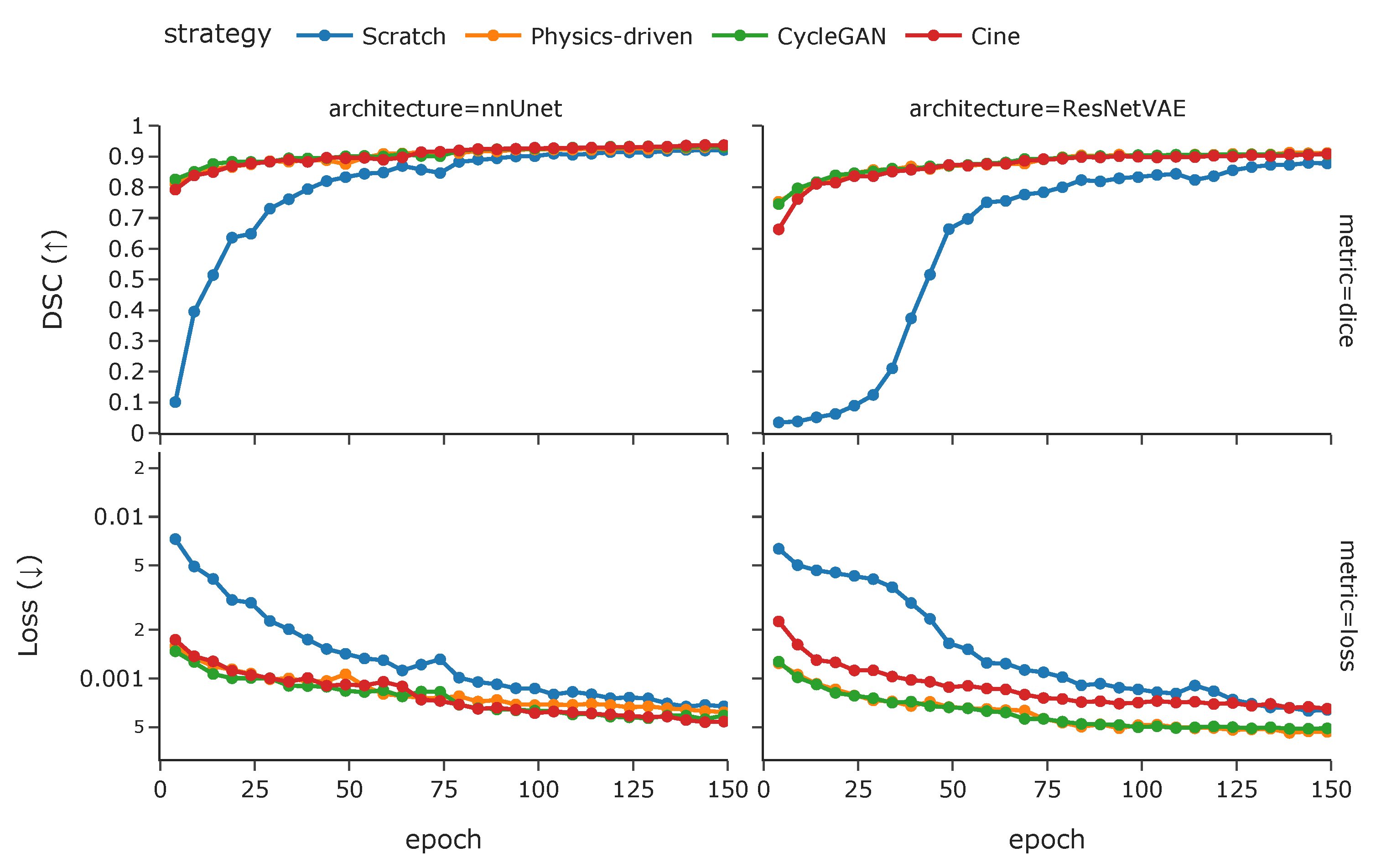
3.2.2. Training Strategies
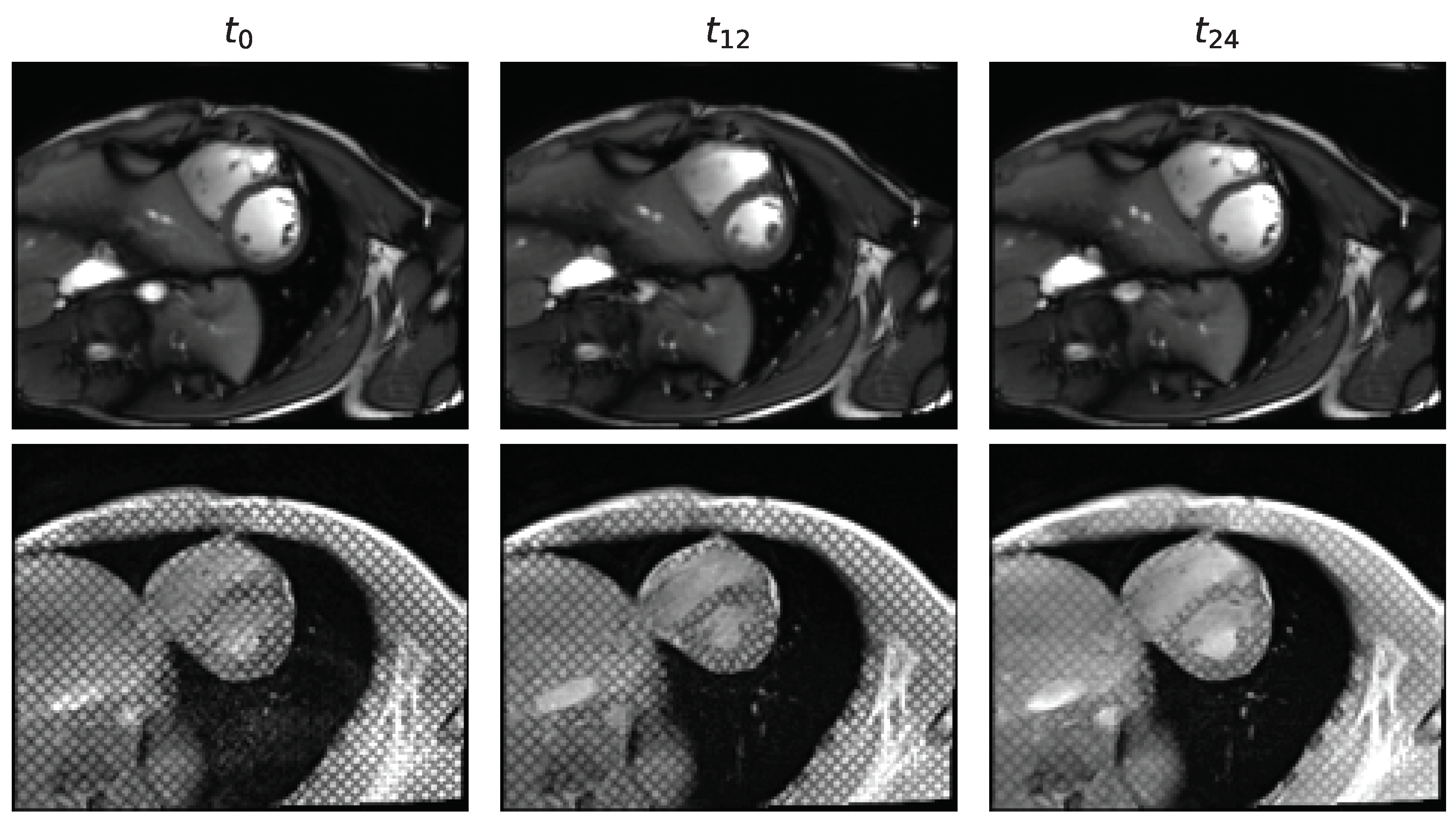
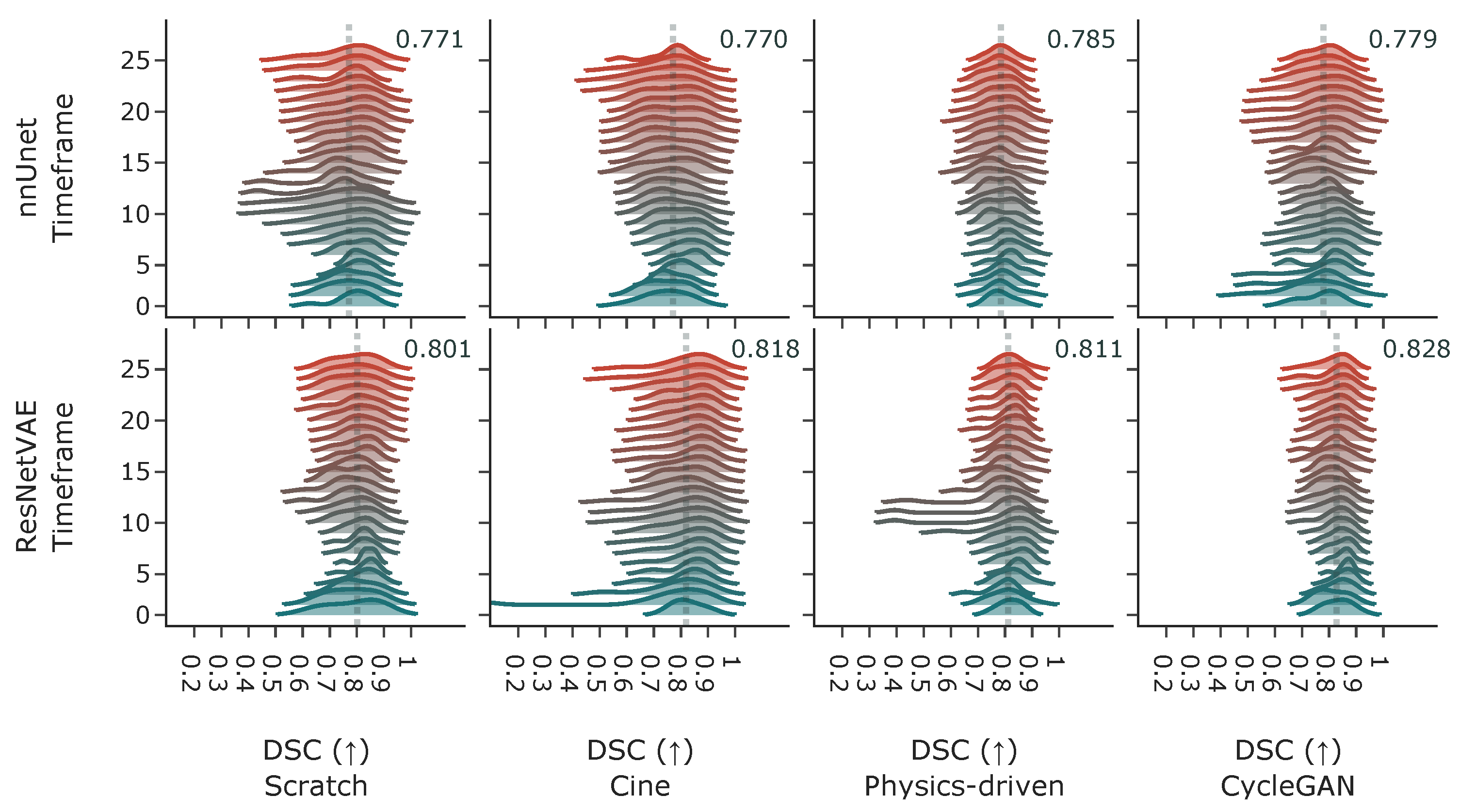
3.2.3. Acquisition Time-Frame
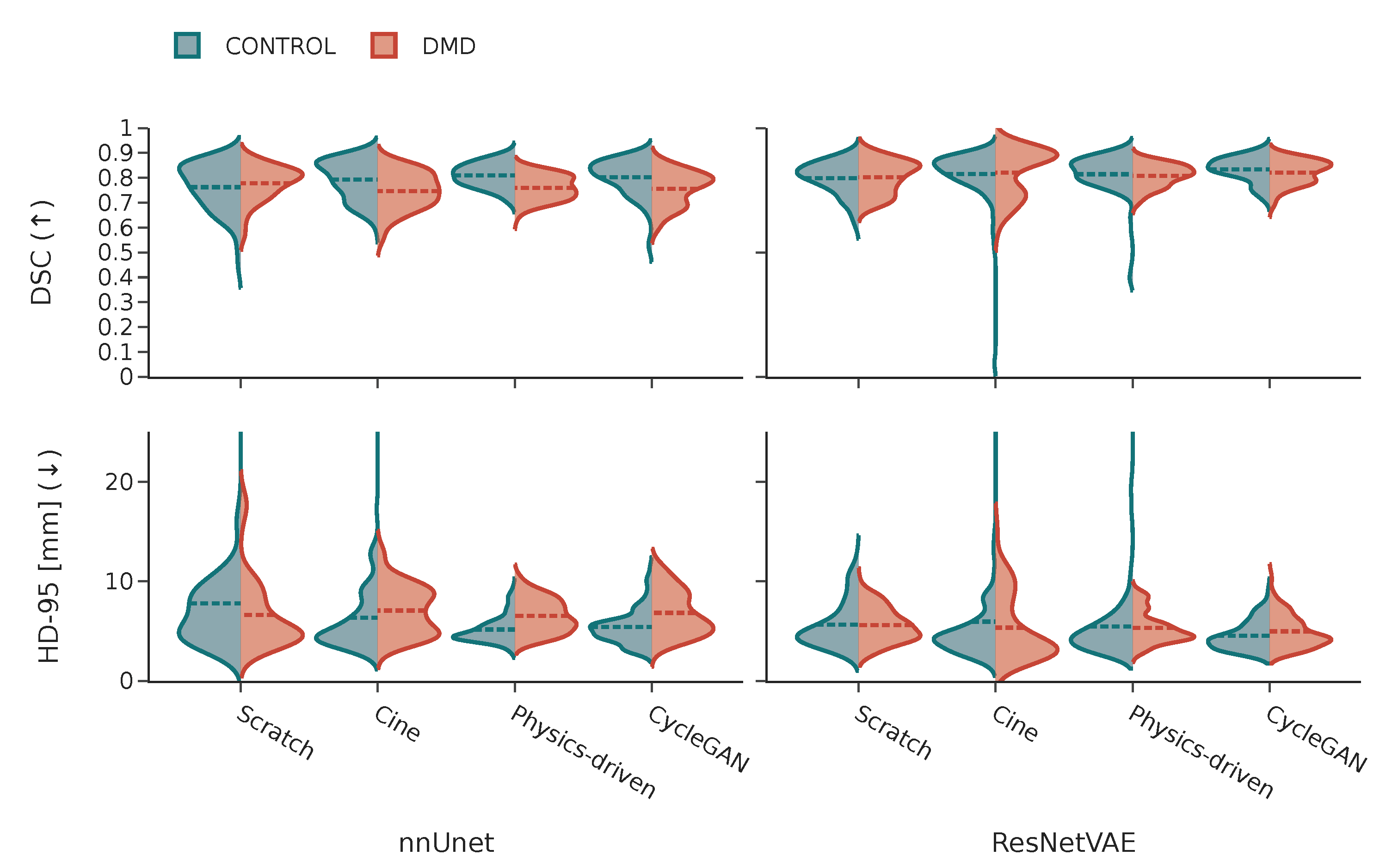
3.2.4. Pathology-Wise Performance
3.2.5. Shape-Aware Loss
4. Discussion
4.1. Analysis of the Cine to Tagged Image Transformation Models
4.2. Analysis of the DL-Based Segmentation Network
4.3. Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amzulescu, M.S.; De Craene, M.; Langet, H.; Pasquet, A.; Vancraeynest, D.; Pouleur, A.C.; Vanoverschelde, J.L.; Gerber, B.L. Myocardial strain imaging: Review of general principles, validation, and sources of discrepancies. Eur. Heart J.-Cardiovasc. Imaging 2019, 20, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, E.S.H. Myocardial tagging by cardiovascular magnetic resonance: Evolution of techniques–pulse sequences, analysis algorithms, and applications. J. Cardiovasc. Magn. Reson. 2011, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Jeung, M.Y.; Germain, P.; Croisille, P.; Ghannudi, S.E.; Roy, C.; Gangi, A. Myocardial Tagging with MR Imaging: Overview of Normal and Pathologic Findings. Radiographics 2012, 32, 1381–1398. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Sands, G.B.; LeGrice, I.J.; Young, A.A.; Ennis, D.B. Myocardial mesostructure and mesofunction. Am. J. Physiol.-Heart Circ. Physiol. 2022, 323, H257–H275. [Google Scholar] [CrossRef] [PubMed]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Gonzalez Ballester, M.A.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Loecher, M.; Hannum, A.J.; Perotti, L.E.; Ennis, D.B. Arbitrary Point Tracking with Machine Learning to Measure Cardiac Strains in Tagged MRI. In International Conference on Functional Imaging and Modeling of the Heart; Ennis, D.B., Perotti, L.E., Wang, V.Y., Eds.; Springer: Cham, Switzerland, 2021; pp. 213–222. [Google Scholar] [CrossRef]
- Ferdian, E.; Suinesiaputra, A.; Fung, K.; Aung, N.; Lukaschuk, E.; Barutcu, A.; Maclean, E.; Paiva, J.; Piechnik, S.K.; Neubauer, S.; et al. Fully Automated Myocardial Strain Estimation from Cardiovascular MRI–tagged Images Using a Deep Learning Framework in the UK Biobank. Radiol. Cardiothorac. Imaging 2020, 2, e190032. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Myronenko, A. 3D MRI bRain Tumor Segmentation Using Autoencoder Regularization. Available online: http://xxx.lanl.gov/abs/1810.11654 (accessed on 15 March 2012).
- Campello, V.M.; Gkontra, P.; Izquierdo, C.; Martín-Isla, C.; Sojoudi, A.; Full, P.M.; Maier-Hein, K.; Zhang, Y.; He, Z.; Ma, J.; et al. Multi-Centre, Multi-Vendor and Multi-Disease Cardiac Segmentation: The M&Ms Challenge. IEEE Trans. Med. Imaging 2021, 40, 3543–3554. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. Available online: http://xxx.lanl.gov/abs/1703.10593 (accessed on 15 March 2012).
- Zhu, J.Y.; Krähenbühl, P.; Shechtman, E.; Efros, A.A. Generative Visual Manipulation on the Natural Image Manifold. arXiv 2018, arXiv:1609.03552. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv 2016, arXiv:1511.06434. [Google Scholar]
- Wang, S.; Chen, Z.; Yu, W.; Lei, B. Brain Stroke Lesion Segmentation Using Consistent Perception Generative Adversarial Network. arXiv 2020, arXiv:2008.13109. [Google Scholar] [CrossRef]
- Radau, P.; Lu, Y.; Connelly, K.; Paul, G.; Dick, A.J.; Wright, G.A. Evaluation Framework for Algorithms Segmenting Short Axis Cardiac MRI. MIDAS J.-Card. MR Left Ventricle Segmentation Chall. 2009, 49, 4. [Google Scholar] [CrossRef]
- Loecher, M.; Perotti, L.E.; Ennis, D.B. Using synthetic data generation to train a cardiac motion tag tracking neural network. Med. Image Anal. 2021, 74, 102223. [Google Scholar] [CrossRef] [PubMed]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics. JMLR Workshop and Conference Proceedings, Sardinia, Italy, 13–15 May 2021; pp. 249–256, ISSN 1938-7228. [Google Scholar]
- Consortium, T.M. Project MONAI. 2020. Available online: https://zenodo.org/record/4323059# (accessed on 4 March 2022).
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Maier-Hein, L.; Reinke, A.; Christodoulou, E.; Glocker, B.; Godau, P.; Isensee, F.; Kleesiek, J.; Kozubek, M.; Reyes, M.; Riegler, M.A.; et al. Metrics reloaded: Pitfalls and recommendations for image analysis validation. arXiv 2022, arXiv:2206.01653. [Google Scholar]
- Kong, F.; Shadden, S.C. A Generalizable Deep-Learning Approach for Cardiac Magnetic Resonance Image Segmentation Using Image Augmentation and Attention U-Net. In Statistical Atlases and Computational Models of the Heart. M&Ms and EMIDEC Challenges; Series Title: Lecture Notes in Computer Science; Puyol Anton, E., Pop, M., Sermesant, M., Campello, V., Lalande, A., Lekadir, K., Suinesiaputra, A., Camara, O., Young, A., Eds.; Springer International Publishing: Cham, Switzerland, 2021; Volume 12592, pp. 287–296. [Google Scholar] [CrossRef]


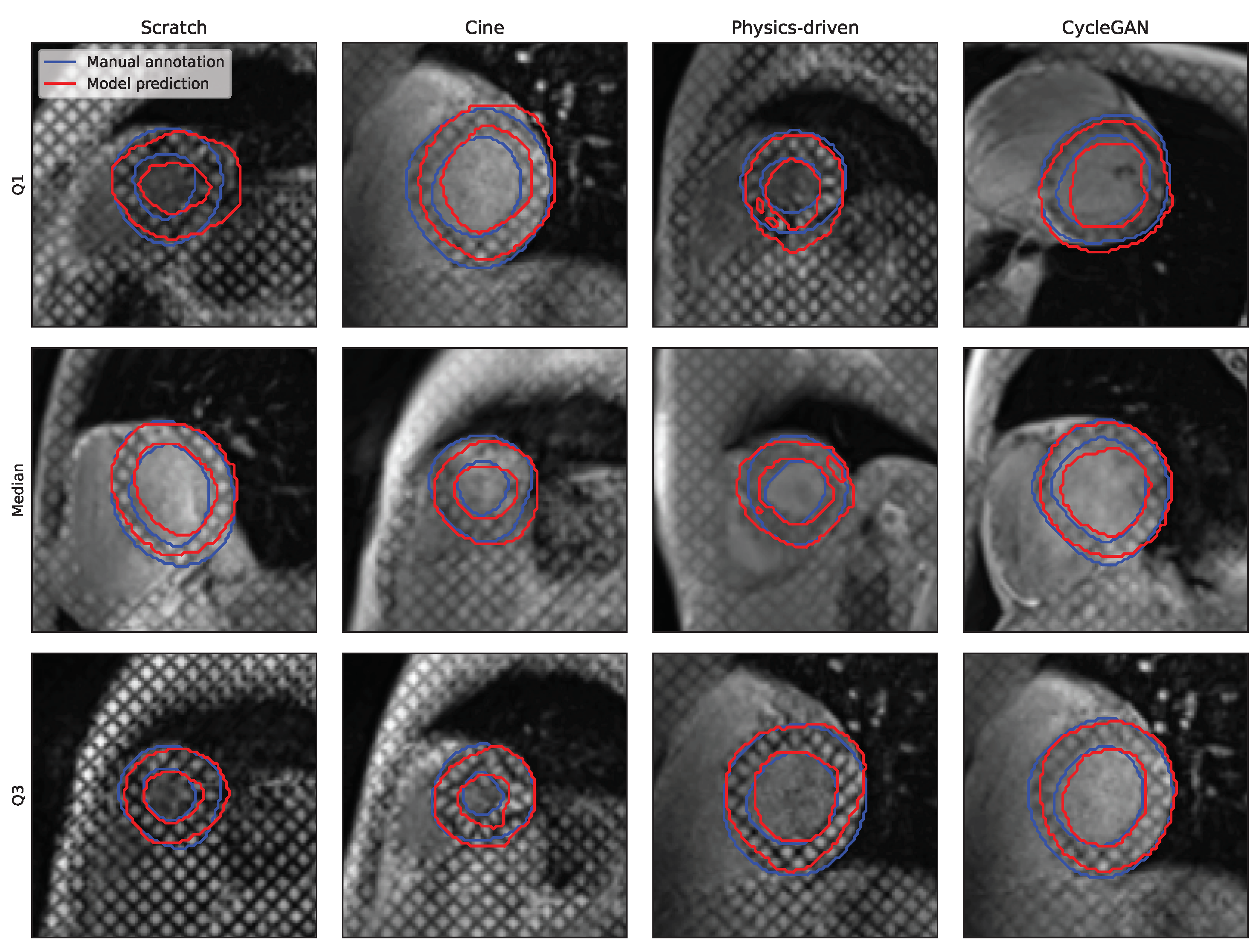
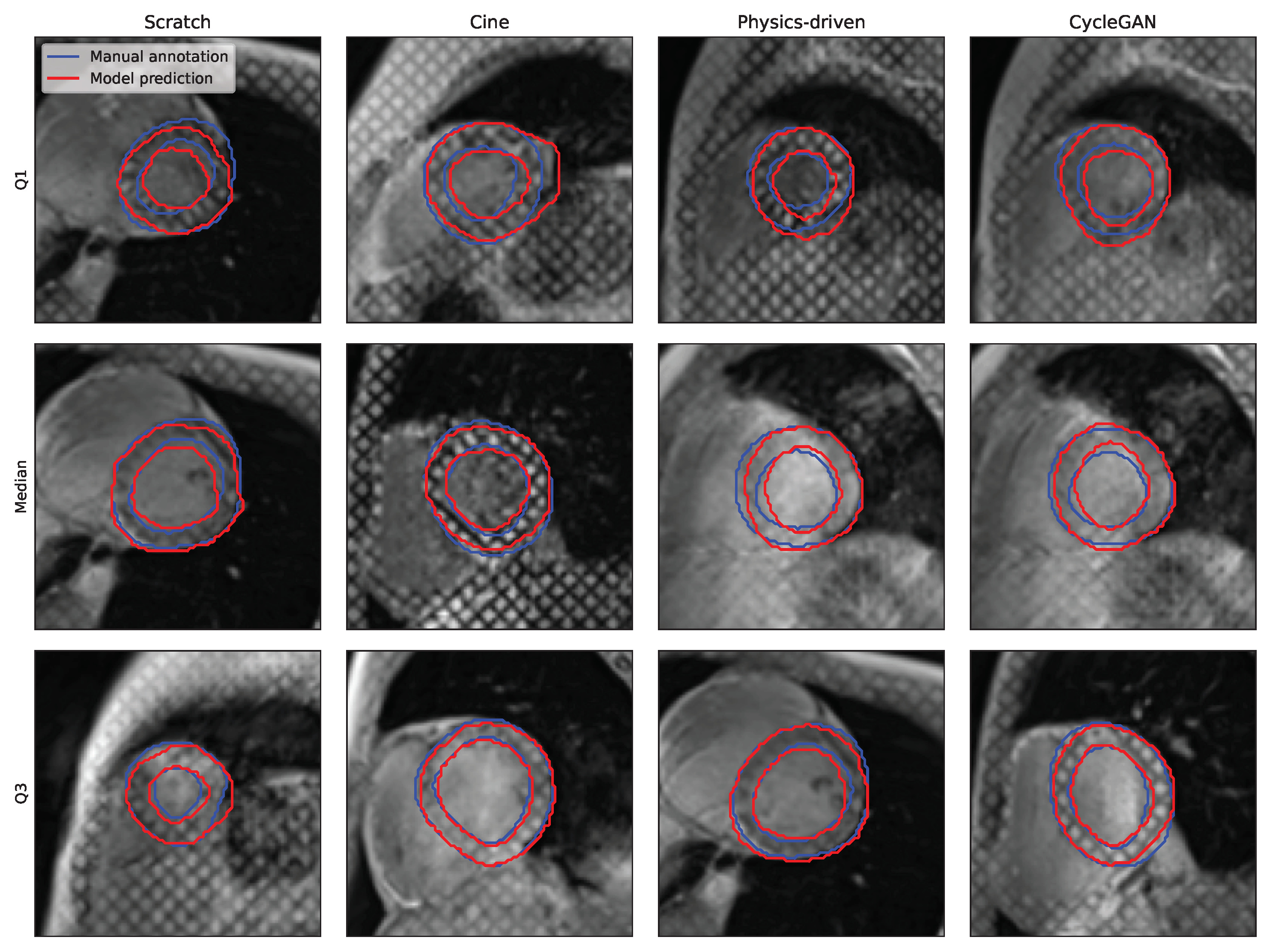

| Name | Date | Subjects | Pathologies | Centres | Vendors | Images | Labels |
|---|---|---|---|---|---|---|---|
| ACDC [5] | 2017 | 150 | 4 | 1 | 1 | 1828 | LV, RV, MYO |
| M&Ms [10] | 2020 | 375 | 9 | 6 | 4 | 2468 | LV, RV, MYO |
| SCD [15] | 2009 | 15 | 3 | 1 | 1 | 282 | LV, MYO |
| Architecture | No. Parameters | Training Time | Inference Time |
|---|---|---|---|
| ResNetVAE | 3555891 | 64.3 s/epoch | 112 ms ± 28.7 ms |
| nnUnet | 34164258 | 163.2 s/epoch | 661 ms ± 274 ms |
| DSC (↑) | HD-95 [mm] (↓) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Split | Train | Test | Train | Test | |||||
| Model | Strategy | Avg. | Std. | Avg. | Std. | Avg. | Std. | Avg. | Std. |
| nnUnet | Scratch | 0.894 | 0.021 | 0.771 | 0.084 | 2.519 | 0.554 | 7.219 | 5.075 |
| Cine | 0.895 | 0.020 | 0.770 | 0.078 | 2.325 | 0.545 | 6.700 | 3.435 | |
| Physics-driven | 0.895 | 0.020 | 0.785 | 0.054 | 3.115 | 0.871 | 5.846 | 1.625 | |
| CycleGAN | 0.894 | 0.020 | 0.779 | 0.074 | 2.385 | 0.569 | 6.117 | 2.087 | |
| ResNetVAE | Scratch | 0.872 | 0.027 | 0.801 | 0.065 | 3.301 | 0.746 | 5.616 | 1.989 |
| Cine | 0.820 | 0.100 | 0.818 | 0.096 | 6.621 | 8.741 | 5.647 | 5.549 | |
| Physics-driven | 0.895 | 0.023 | 0.811 | 0.068 | 2.517 | 0.564 | 5.407 | 3.350 | |
| CycleGAN | 0.898 | 0.022 | 0.828 | 0.049 | 2.481 | 0.572 | 4.745 | 1.537 | |
| DSC (↑) | HD-95 [mm] (↓) | Rank | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Split | Train | Test | Train | Test | |||||
| Avg. | Std. | Avg. | Std. | Avg. | Std. | Avg. | Std. | ||
| 0.000 | 0.880 | 0.025 | 0.768 | 0.094 | 3.082 | 0.668 | 7.701 | 8.967 | 7 |
| 0.001 | 0.815 | 0.025 | 0.771 | 0.045 | 3.375 | 0.771 | 5.358 | 5.946 | 2 |
| 0.005 | 0.886 | 0.023 | 0.770 | 0.102 | 2.976 | 0.658 | 7.628 | 9.414 | 4 |
| 0.050 | 0.872 | 0.027 | 0.801 | 0.065 | 3.301 | 0.746 | 5.616 | 1.989 | 1 |
| 0.100 | 0.865 | 0.026 | 0.798 | 0.083 | 3.547 | 0.849 | 6.423 | 5.314 | 3 |
| 0.500 | 0.789 | 0.023 | 0.736 | 0.050 | 3.496 | 0.958 | 6.449 | 10.825 | 6 |
| 1.000 | 0.585 | 0.035 | 0.556 | 0.041 | 4.838 | 3.672 | 5.715 | 1.975 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhaene, A.P.; Loecher, M.; Wilson, A.J.; Ennis, D.B. Myocardial Segmentation of Tagged Magnetic Resonance Images with Transfer Learning Using Generative Cine-To-Tagged Dataset Transformation. Bioengineering 2023, 10, 166. https://doi.org/10.3390/bioengineering10020166
Dhaene AP, Loecher M, Wilson AJ, Ennis DB. Myocardial Segmentation of Tagged Magnetic Resonance Images with Transfer Learning Using Generative Cine-To-Tagged Dataset Transformation. Bioengineering. 2023; 10(2):166. https://doi.org/10.3390/bioengineering10020166
Chicago/Turabian StyleDhaene, Arnaud P., Michael Loecher, Alexander J. Wilson, and Daniel B. Ennis. 2023. "Myocardial Segmentation of Tagged Magnetic Resonance Images with Transfer Learning Using Generative Cine-To-Tagged Dataset Transformation" Bioengineering 10, no. 2: 166. https://doi.org/10.3390/bioengineering10020166
APA StyleDhaene, A. P., Loecher, M., Wilson, A. J., & Ennis, D. B. (2023). Myocardial Segmentation of Tagged Magnetic Resonance Images with Transfer Learning Using Generative Cine-To-Tagged Dataset Transformation. Bioengineering, 10(2), 166. https://doi.org/10.3390/bioengineering10020166