Jointly Learning Non-Cartesian k-Space Trajectories and Reconstruction Networks for 2D and 3D MR Imaging through Projection
Abstract
1. Introduction
2. Materials and Methods
2.1. Data and Preprocessing
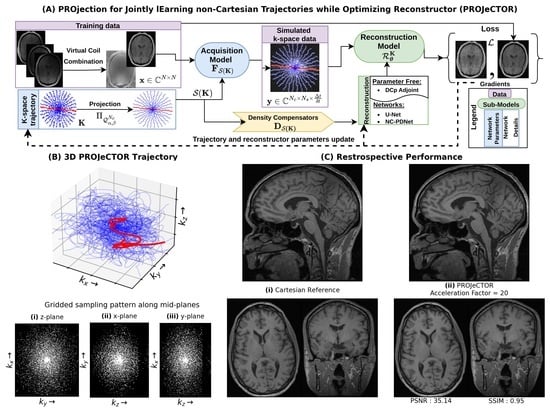
2.2. K-Space Trajectory (K)
2.3. Acquisition Model ()
2.4. Reconstruction Model: Deep Neural Network ()
2.5. Loss, Gradients and Optimizer
2.6. Multi-Resolution
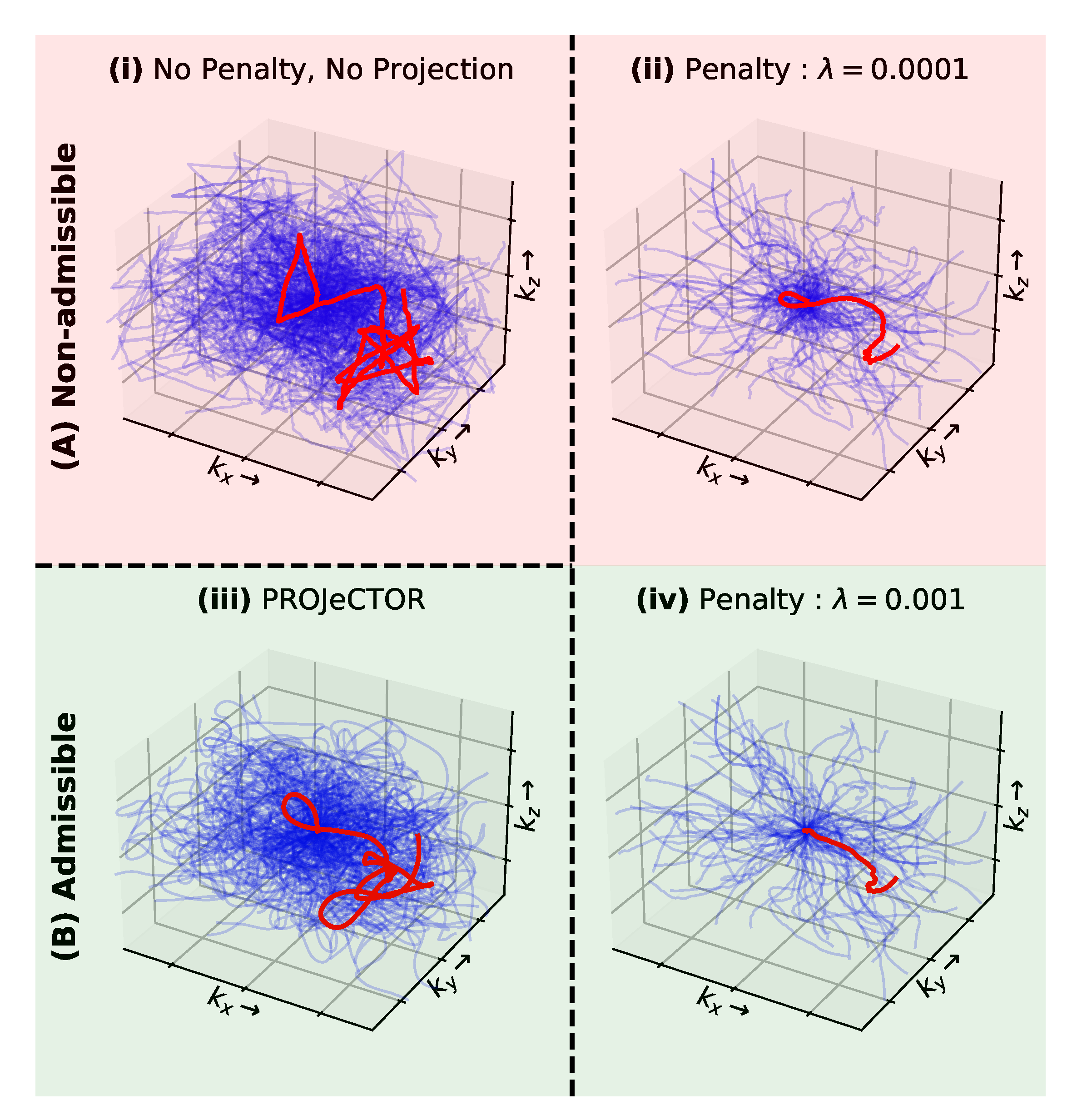
2.7. Constraints: Projection vs. Penalty
- Need for hyper-parameter tuning: Under the penalty based formulation, the hyper-parameters need to be tuned, which requires additional computation. Note that while we can view Equation (8) as an augmented Lagrangian form for the constrained optimization problem Equation (6), the corresponding Karush-Kuhn-Tucker (KKT) conditions are computationally complex to be solved. Further, as we do not satisfy the Slater’s conditions, as the reconstruction loss is non-convex, the solutions of the KKT conditions are not guaranteed to be global minima.
- Influence of gradients and convergence: With the addition of penalty terms , the gradient updates involve added gradients from these penalties , which influence the overall trajectory development, and hence the final optimized k-space trajectories. Gradient updates with these additional gradient terms can no longer guarantee optimal image reconstruction by minimizing the reconstruction loss .
- Guarantee of admissibility: Finally, the optimization of the augmented Lagrangian form does not guarantee that the final optimized k-space trajectory K satisfies the constraints Equation (1).
2.8. Practical Implementations
3. Results
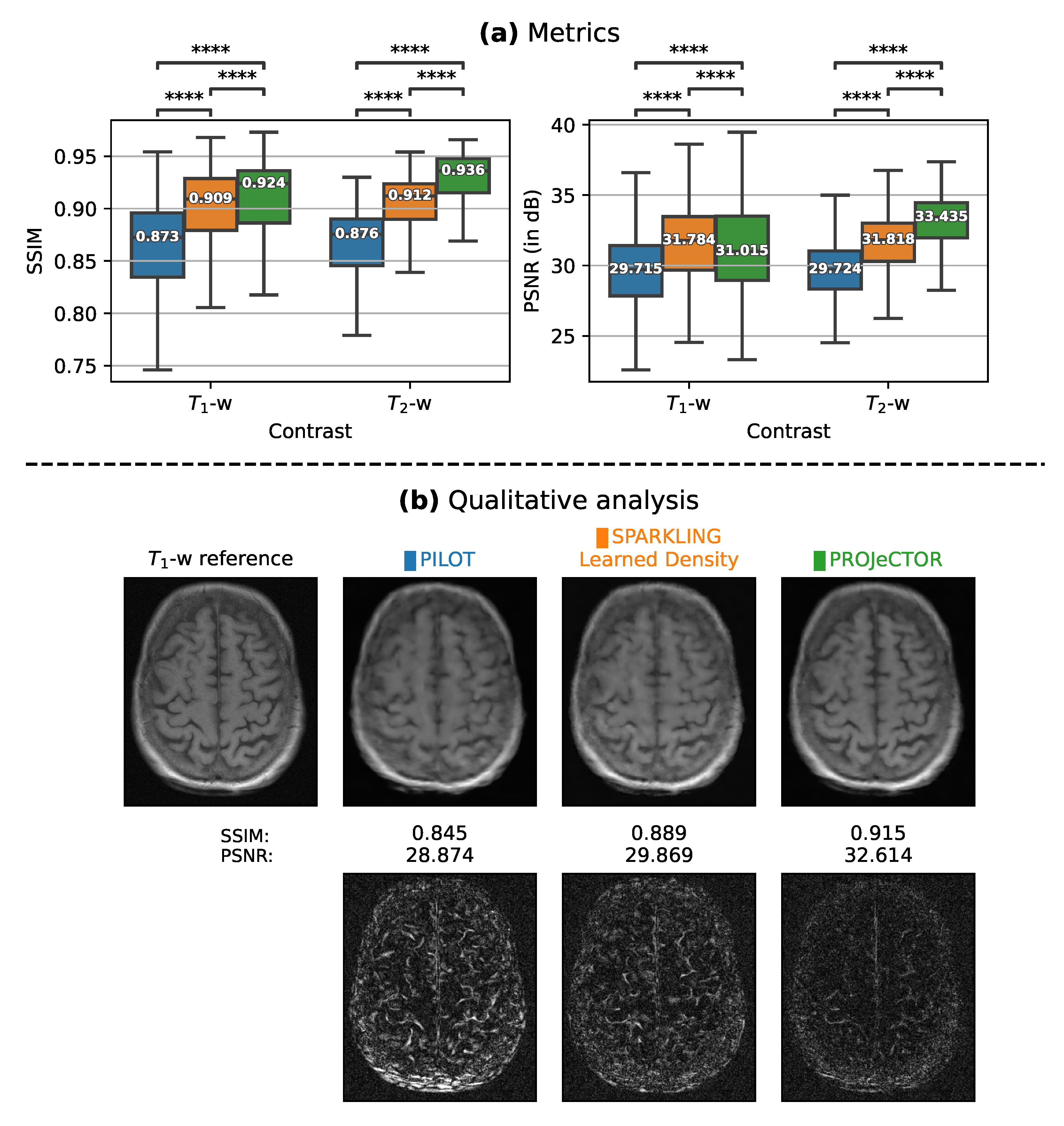
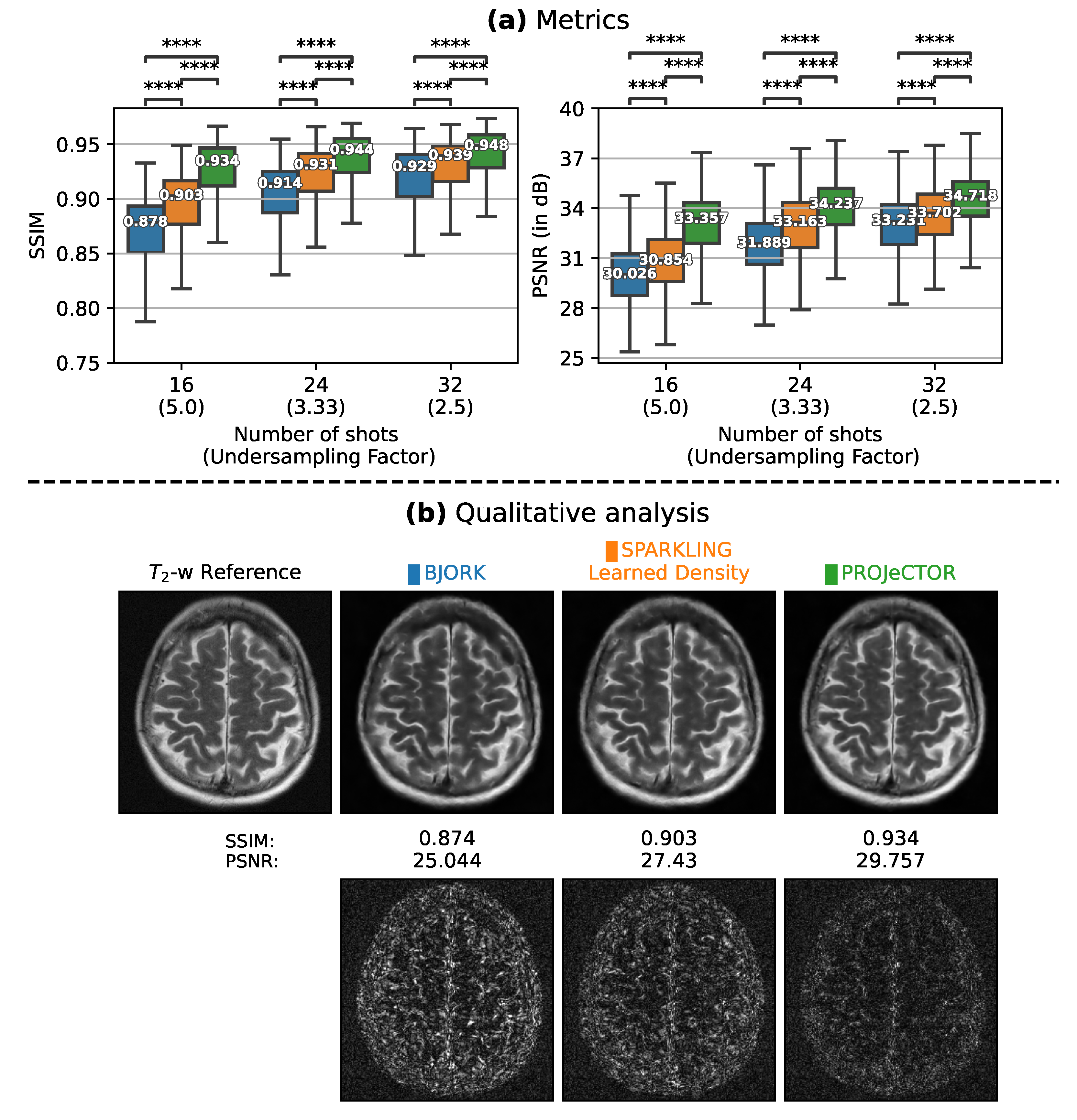
3.1. Comparison with State-of-the-Art Methods in 2D
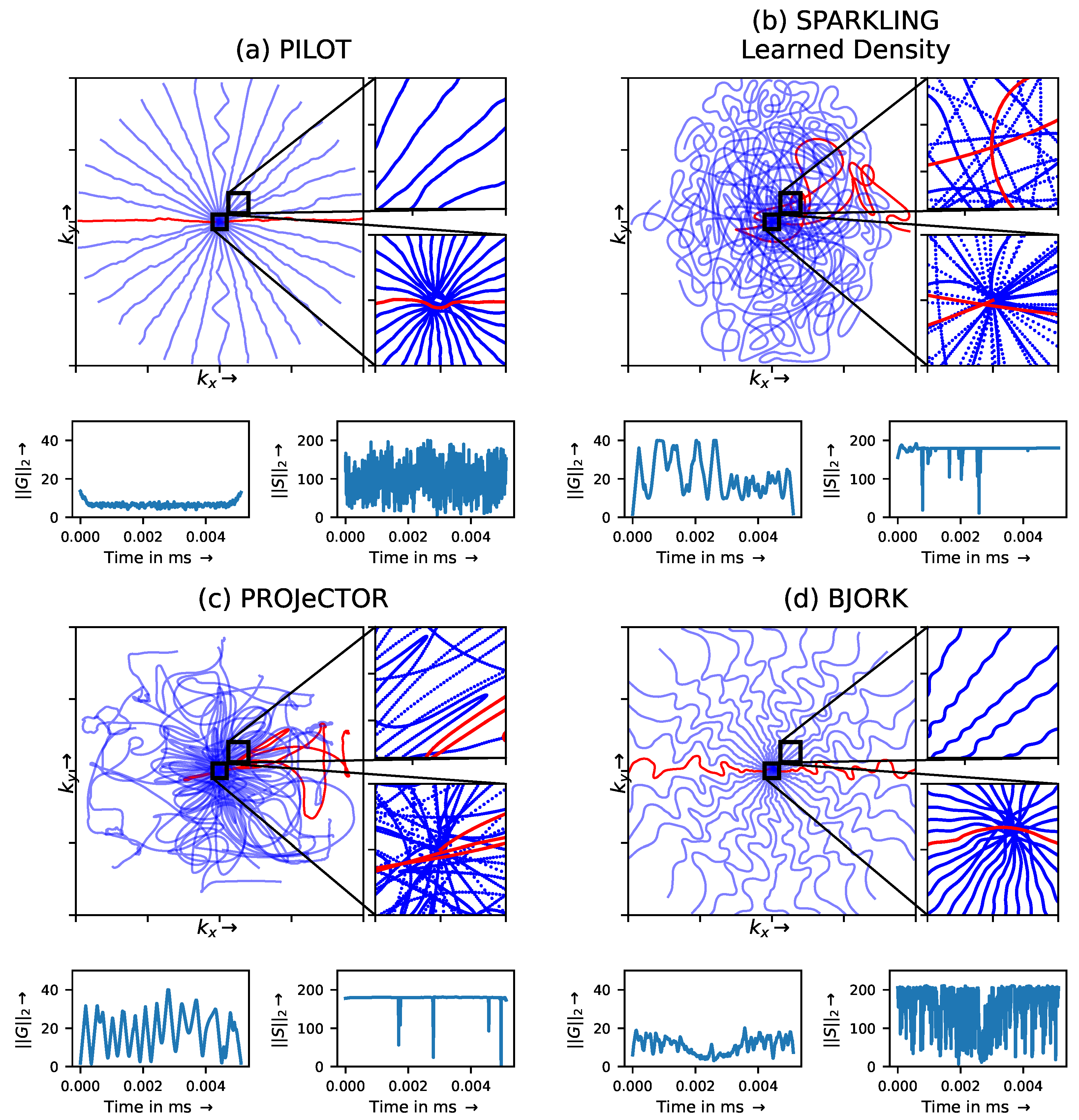
3.1.1. Trajectory Analysis
3.1.2. Retrospective Study
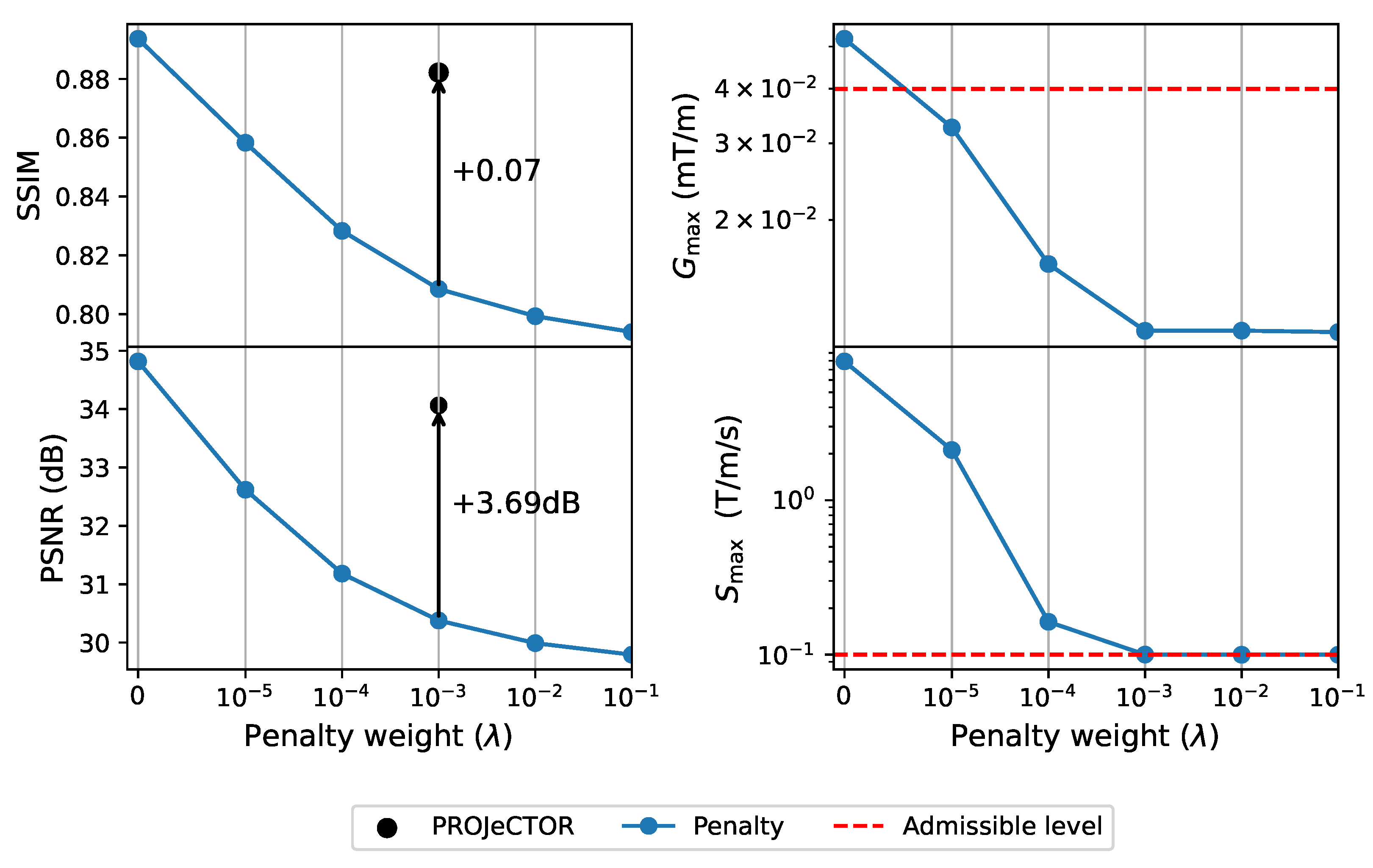
3.2. Hardware Constraints: Penalty vs. Projection
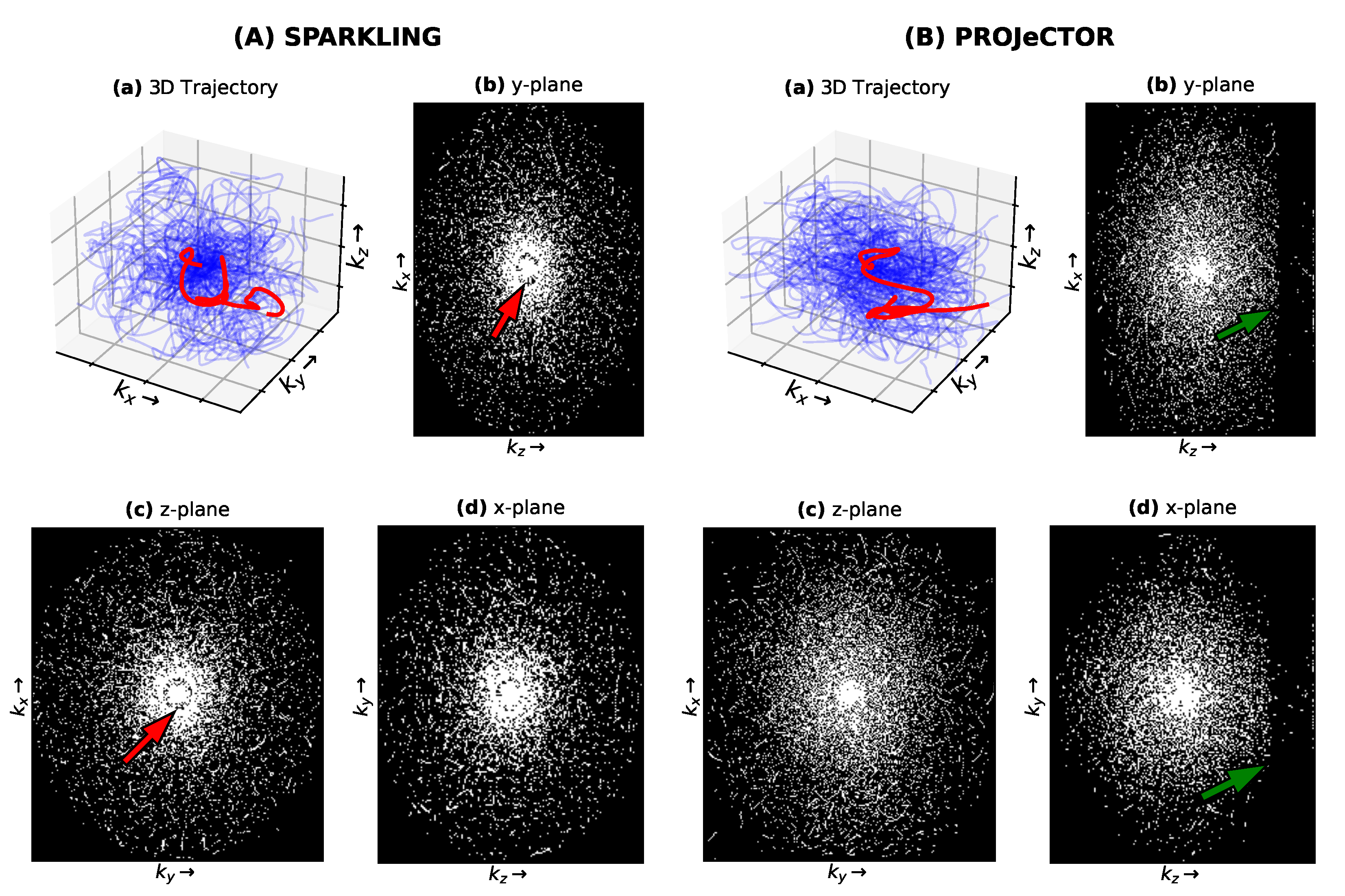
3.3. Comparison with SPARKLING in 3D
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADC | analog-to-digital converter |
| AF | Acceleration Factor |
| BJORK | B-spline parameterized Joint Optimization of Reconstruction and K-space |
| trajectories | |
| CS | Compressed Sensing |
| DCp | Density Compensators |
| KKT | Karush–Kuhn–Tucker |
| MRI | Magnetic Resonance Imaging |
| NUFFT | Non-Uniform Fast Fourier Transform |
| PILOT | Physics-informed learned optimal trajectories |
| PROJeCTOR | PROjection for Jointly lEarning non-Cartesian Trajectories while Optimizing |
| Reconstructor | |
| PSNR | Peak Signal-to-Noise Ratio |
| SNR | Signal-to-Noise Ratio |
| SPARKLING | Spreading Projection Algorithm for Rapid K-space sampLING |
| SSIM | Structural Similarity Index |
| TE | Echo Time |
| TSD | Target Sampling Density |
| UF | Undersampling Factor |
| VDS | Variable Density Sampling |
References
- Lustig, M.; Donoho, D.; Pauly, J.M. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. 2007, 58, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Puy, G.; Vandergheynst, P.; Wiaux, Y. On variable density compressive sampling. IEEE Signal Process. Lett. 2011, 18, 595–598. [Google Scholar] [CrossRef]
- Chauffert, N.; Ciuciu, P.; Weiss, P. Variable density compressed sensing in MRI. Theoretical vs heuristic sampling strategies. In Proceedings of the 2013 IEEE 10th International Symposium on Biomedical Imaging, San Francisco, CA, USA, 7–11 April 2013; pp. 298–301. [Google Scholar]
- Chauffert, N.; Ciuciu, P.; Kahn, J.; Weiss, P. Variable density sampling with continuous trajectories. Application to MRI. SIAM J. Imag. Sci. 2014, 7, 1962–1992. [Google Scholar] [CrossRef]
- Adcock, B.; Hansen, A.C.; Poon, C.; Roman, B. Breaking the coherence barrier: Asymptotic incoherence and asymptotic sparsity in compressed sensing. arXiv 2013, arXiv:1302.0561. [Google Scholar]
- Boyer, C.; Bigot, J.; Weiss, P. Compressed sensing with structured sparsity and structured acquisition. Appl. Comput. Harmon. Anal. 2019, 46, 312–350. [Google Scholar] [CrossRef]
- Ahn, C.B.; Kim, J.H.; Cho, Z.H. High-speed spiral-scan echo planar NMR imaging-I. IEEE Trans. Med. Imag. 1986, 5, 2–7. [Google Scholar] [CrossRef]
- Meyer, C.H.; Hu, B.S.; Nishimura, D.G.; Macovski, A. Fast spiral coronary artery imaging. Magn. Reson. Med. 1992, 28, 202–213. [Google Scholar] [CrossRef]
- Jackson, J.I.; Nishimura, D.G.; Macovski, A. Twisting radial lines with application to robust magnetic resonance imaging of irregular flow. Magn. Reson. Med. 1992, 25, 128–139. [Google Scholar] [CrossRef]
- Noll, D.C. Multishot rosette trajectories for spectrally selective MR imaging. IEEE Trans. Med. Imag. 1997, 16, 372–377. [Google Scholar] [CrossRef]
- Law, C.S.; Glover, G.H. Interleaved spiral-in/out with application to functional MRI (fMRI). Magn. Reson. Med. 2009, 62, 829–834. [Google Scholar] [CrossRef]
- Lustig, M.; Lee, J.; Donoho, D.; Pauly, J. Faster imaging with randomly perturbed, under-sampled spirals and ℓ1 reconstruction. In Proceedings of the 13th ISMRM, Miami Beach, FL, USA, 7–13 May 2005; p. 685. [Google Scholar]
- Lazarus, C.; Weiss, P.; Chauffert, N.; Mauconduit, F.; Gueddari, L.E.; Destrieux, C.; Zemmoura, I.; Vignaud, A.; Ciuciu, P. SPARKLING: Variable-density k-space filling curves for accelerated T2*-weighted MRI. Magn. Reson. Med. 2019, 81, 3643–3661. [Google Scholar] [CrossRef]
- Lazarus, C.; Weiss, P.; Gueddari, L.E.; Mauconduit, F.; Massire, A.; Ripart, M.; Vignaud, A.; Ciuciu, P. 3D variable-density SPARKLING trajectories for high-resolution T2*-weighted magnetic resonance imaging. NMR Biomed. 2020, 33, e4349. [Google Scholar] [CrossRef]
- Chaithya, G.R.; Weiss, P.; Daval-Frérot, G.; Massire, A.; Vignaud, A.; Ciuciu, P. Optimizing full 3D SPARKLING trajectories for high-resolution T2*-weighted Magnetic Resonance imaging. IEEE Trans. Med. Imaging, 2021; under review. [Google Scholar]
- Chaithya, G.R.; Ramzi, Z.; Ciuciu, P. Learning the sampling density in 2D SPARKLING MRI acquisition for optimized image reconstruction. In Proceedings of the 2021 29th European Signal Processing Conference (EUSIPCO), Dublin, Ireland, 23–27 August 2021. [Google Scholar]
- Bahadir, C.D.; Wang, A.Q.; Dalca, A.V.; Sabuncu, M.R. Deep-learning-based optimization of the under-sampling pattern in MRI. IEEE Trans. Comput. Imaging 2020, 6, 1139–1152. [Google Scholar] [CrossRef]
- Weiss, T.; Senouf, O.; Vedula, S.; Michailovich, O.; Zibulevsky, M.; Bronstein, A. PILOT: Physics-Informed Learned Optimized Trajectories for Accelerated MRI. arXiv 2019, arXiv:1909.05773. [Google Scholar] [CrossRef]
- Wang, G.; Luo, T.; Nielsen, J.F.; Noll, D.C.; Fessler, J.A. B-spline parameterized joint optimization of reconstruction and k-space trajectories (BJORK) for accelerated 2d MRI. arXiv 2021, arXiv:2101.11369. [Google Scholar] [CrossRef]
- Vedula, S.; Senouf, O.; Bronstein, A. 3D FLAT: Feasible Learned Acquisition Trajectories for Accelerated MRI. In Proceedings of the MLMIR WS at MICCAI, Lima, Peru, 4–8 October 2020; p. 3. [Google Scholar]
- Zbontar, J.; Knoll, F.; Sriram, A.; Murrell, T.; Huang, Z.; Muckley, M.J.; Defazio, A.; Stern, R.; Johnson, P.; Bruno, M.; et al. fastMRI: An open dataset and benchmarks for accelerated MRI. arXiv 2018, arXiv:1811.08839. [Google Scholar]
- Guanhua, W.; Douglas, C.N.; Fessler, J.A. Efficient NUFFT Backpropagation for Stochastic Sampling Optimization in MRI. In Proceedings of the 29th ISMRM Society, Online, 15–20 May 2021. Number 0913. [Google Scholar]
- Ramzi, Z.; Chaithya, G.R.; Starck, J.L.; Ciuciu, P. NC-PDNet: A Density-Compensated Unrolled Network for 2D and 3D non-Cartesian MRI Reconstruction. IEEE TMI 2022, 41, 1625–1638. [Google Scholar] [CrossRef]
- Chauffert, N.; Weiss, P.; Kahn, J.; Ciuciu, P. A projection algorithm for gradient waveforms design in MRI. IEEE Trans. Med. Imag. 2016, 35, 2026–2039. [Google Scholar] [CrossRef]
- de Gournay, F.; Kahn, J.; Lebrat, L.; Weiss, P. Optimal Transport Approximation of 2-Dimensional Measures. SIAM J. Imag. Sci. 2019, 12, 762–787. [Google Scholar]
- Parker, D.L.; Payne, A.; Todd, N.; Hadley, J.R. Phase reconstruction from multiple coil data using a virtual reference coil. Magn. Reson. Med. 2014, 72, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.; Lucena, O.; Garrafa, J.; Gobbi, D.; Saluzzi, M.; Appenzeller, S.; Rittner, L.; Frayne, R.; Lotufo, R. An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. NeuroImage 2018, 170, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Fessler, J.; Sutton, B. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Trans. Signal Process. 2003, 51, 560–574. [Google Scholar] [CrossRef]
- Pipe, J.G.; Menon, P. Sampling density compensation in MRI: Rationale and an iterative numerical solution. Magn. Reson. Med. 1999, 41, 179–186. [Google Scholar] [CrossRef]
- Pezzotti, N.; Yousefi, S.; Elmahdy, M.S.; Van Gemert, J.H.F.; Schuelke, C.; Doneva, M.; Nielsen, T.; Kastryulin, S.; Lelieveldt, B.P.; Van Osch, M.J.; et al. An Adaptive Intelligence Algorithm for Undersampled Knee MRI Reconstruction. IEEE Access 2020, 8, 204825–204838. [Google Scholar] [CrossRef]
- Wang, Z.; Simoncelli, E.; Bovik, A. Multiscale structural similarity for image quality assessment. In Proceedings of the Thrity-Seventh Asilomar Conference on Signals, Systems Computers, Pacific Grove, CA, USA, 9–12 November 2003; Volume 2, pp. 1398–1402. [Google Scholar] [CrossRef]
- de Gournay, F.; Gossard, A.; Weiss, P. Spurious minimizers in non uniform Fourier sampling optimization. arXiv 2022, arXiv:2207.10323. [Google Scholar]
- Wang, G.; Nielsen, J.F.; Fessler, J.A.; Noll, D.C. Stochastic Optimization of 3D Non-Cartesian Sampling Trajectory (SNOPY). arXiv 2022, arXiv:2209.11030. [Google Scholar] [CrossRef]
- Montalt Tordera, J. TensorFlow NUFFT. Available online: https://doi.org/10.5281/zenodo.7377974 (accessed on 18 December 2022).
- Shih, Y.H.; Wright, G.; Andén, J.; Blaschke, J.; Barnett, A.H. cuFINUFFT: A load-balanced GPU library for general-purpose nonuniform FFTs. In Proceedings of the 2021 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), Portland, OR, USA, 17–21 June 2021. [Google Scholar] [CrossRef]
- Muckley, M.J.; Riemenschneider, B.; Radmanesh, A.; Kim, S.; Jeong, G.; Ko, J.; Jun, Y.; Shin, H.; Hwang, D.; Mostapha, M.; et al. Results of the 2020 fastMRI Challenge for Machine Learning MR Image Reconstruction. IEEE TMI 2021, 40, 2306–2317. [Google Scholar] [CrossRef]
- Daval-Frérot, G.; Massire, A.; Mailhe, B.; Nadar, M.; Vignaud, A.; Ciuciu, P. Iterative static field map estimation for off-resonance correction in non-Cartesian susceptibility weighted imaging. Magn. Reson. Med. 2022, 88, 1592–1607. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radhakrishna, C.G.; Ciuciu, P. Jointly Learning Non-Cartesian k-Space Trajectories and Reconstruction Networks for 2D and 3D MR Imaging through Projection. Bioengineering 2023, 10, 158. https://doi.org/10.3390/bioengineering10020158
Radhakrishna CG, Ciuciu P. Jointly Learning Non-Cartesian k-Space Trajectories and Reconstruction Networks for 2D and 3D MR Imaging through Projection. Bioengineering. 2023; 10(2):158. https://doi.org/10.3390/bioengineering10020158
Chicago/Turabian StyleRadhakrishna, Chaithya Giliyar, and Philippe Ciuciu. 2023. "Jointly Learning Non-Cartesian k-Space Trajectories and Reconstruction Networks for 2D and 3D MR Imaging through Projection" Bioengineering 10, no. 2: 158. https://doi.org/10.3390/bioengineering10020158
APA StyleRadhakrishna, C. G., & Ciuciu, P. (2023). Jointly Learning Non-Cartesian k-Space Trajectories and Reconstruction Networks for 2D and 3D MR Imaging through Projection. Bioengineering, 10(2), 158. https://doi.org/10.3390/bioengineering10020158