Tuning Fatty Acid Profile and Yield in Pichia pastoris
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Media, and Cultivation Conditions
2.2. Strains and Synthetic Genes
2.3. Strain Design and Genetic Manipulations
2.4. Bioreactor Cultivations
2.5. Fed-Batch Feeding
2.6. Fatty Acid Methyl Ester (FAME) Analysis—Sample Preparation
2.7. GC Analysis
2.8. Glucose and Ethanol Concentration Analysis
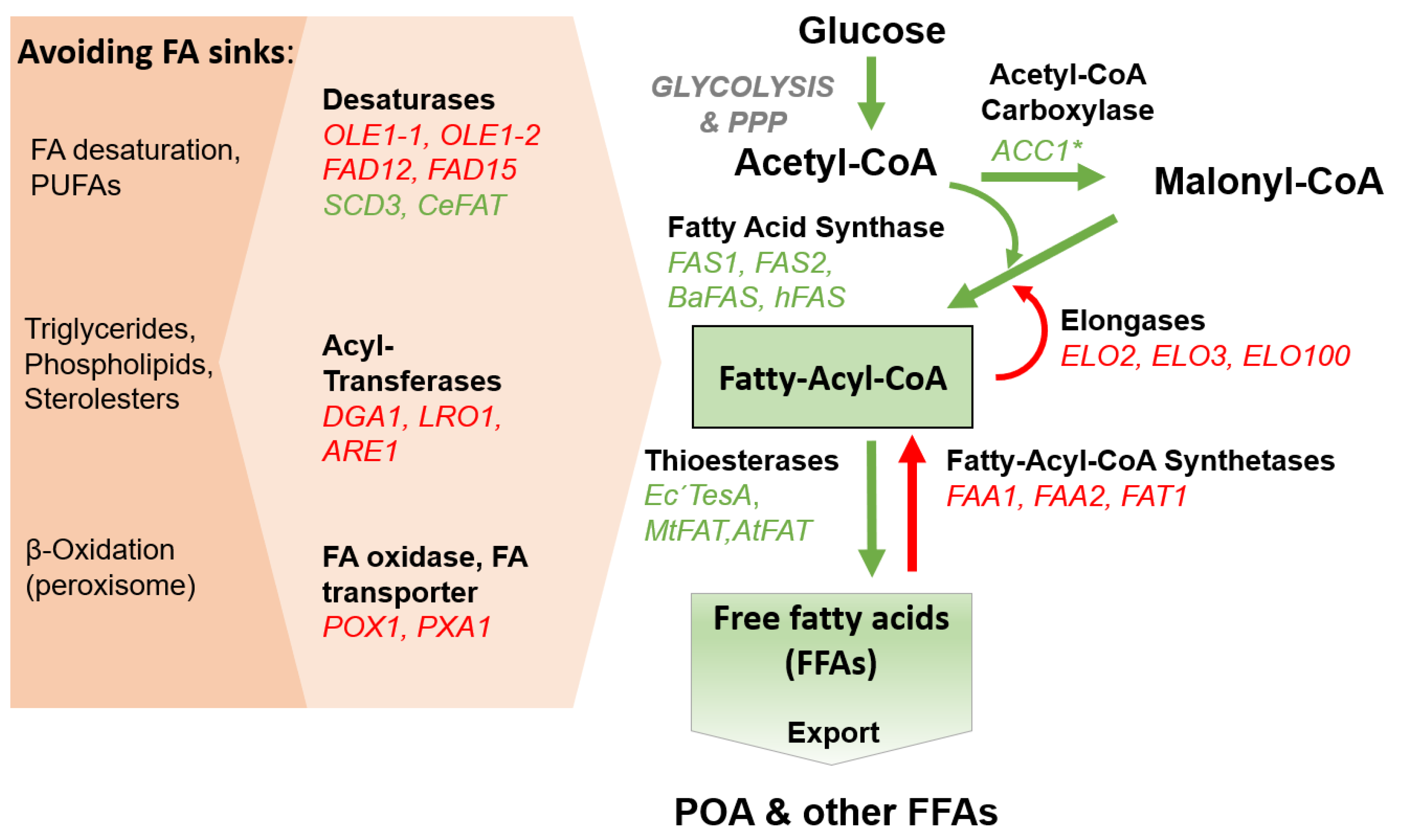
3. Results and Discussion
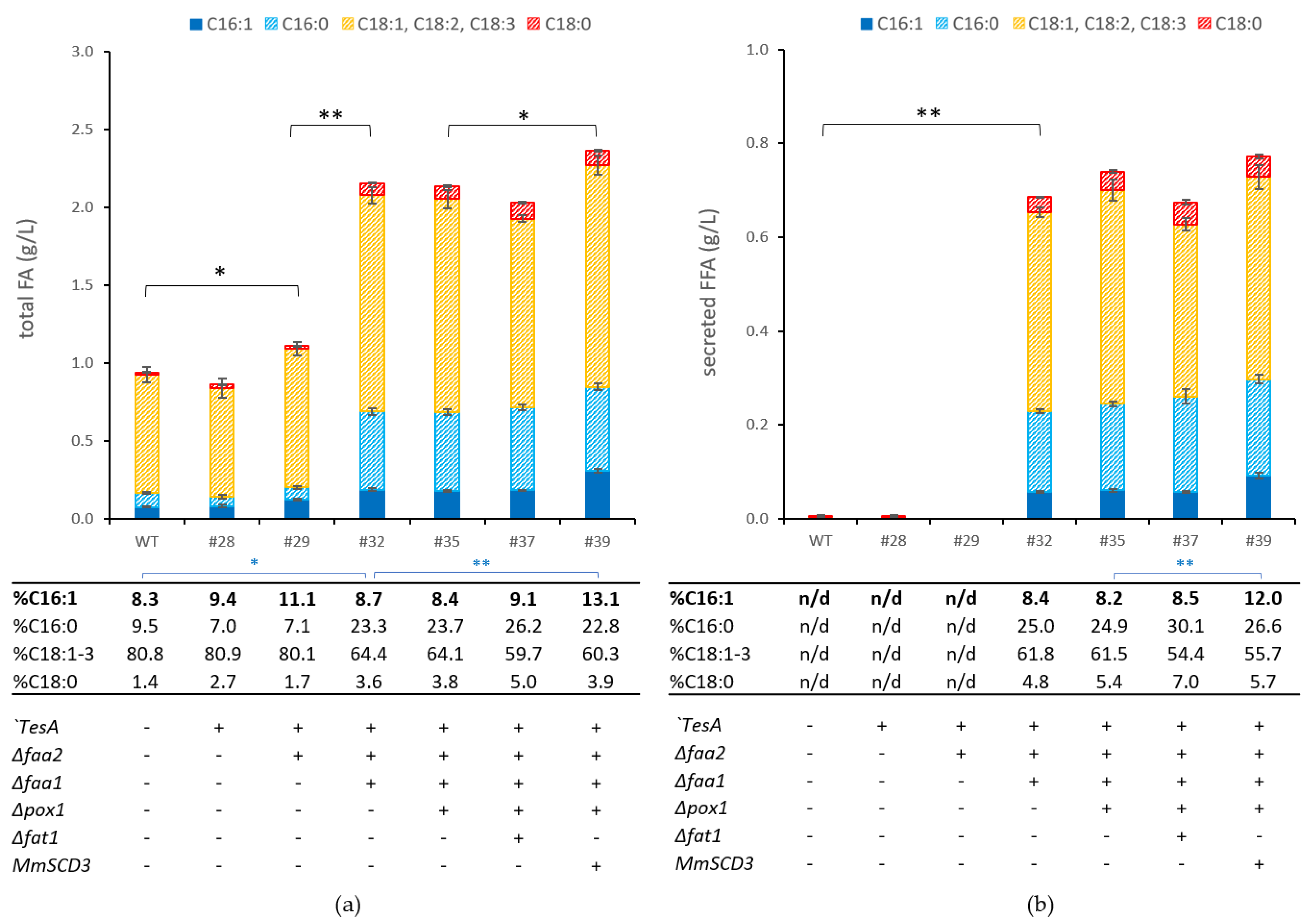
3.1. Establishing Free Fatty Acid Secretion in P. pastoris
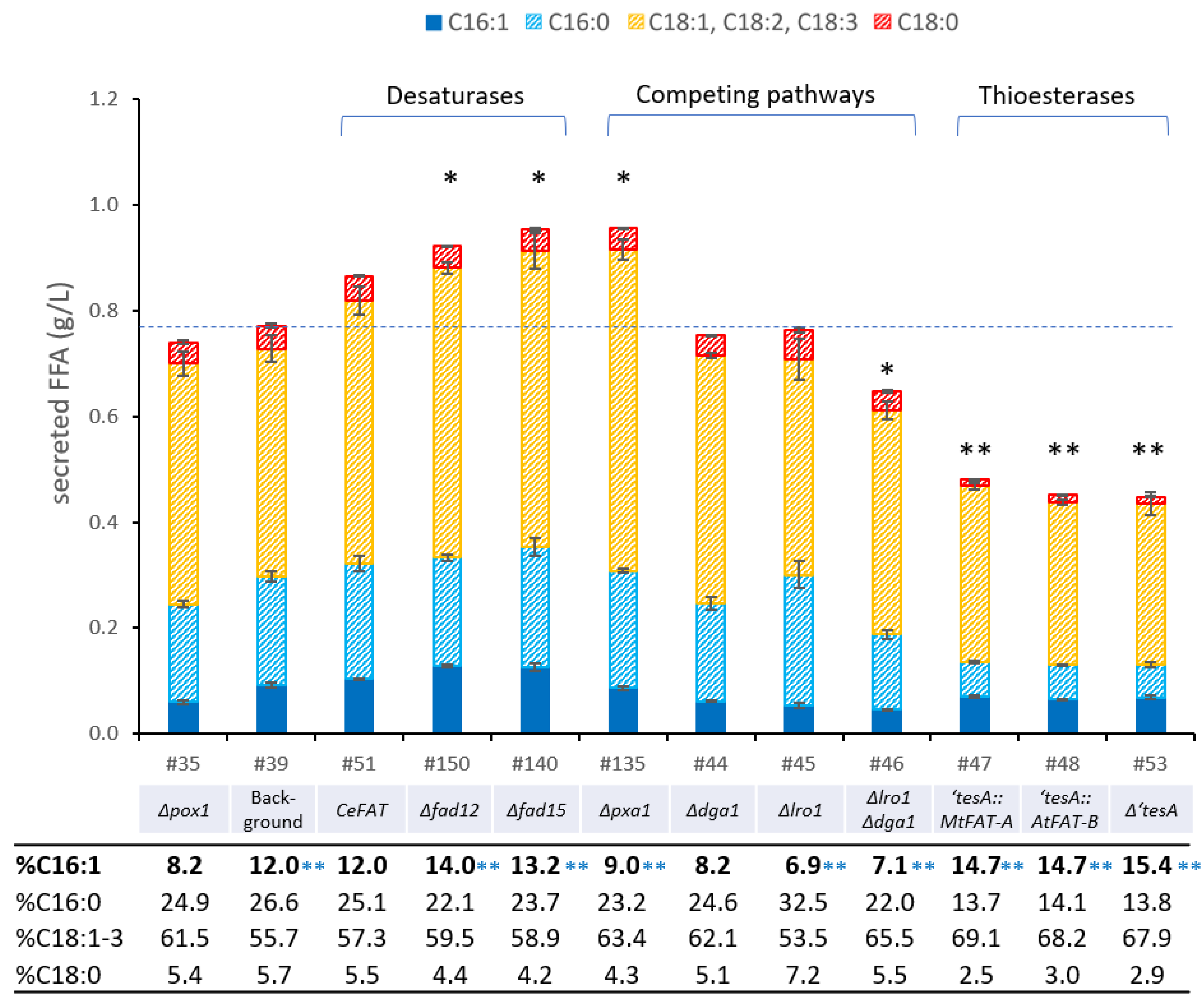
3.2. Engineering the Fatty Acid Desaturase System in P. pastoris
3.3. Analysis of Growth Conditions and Nutrient Manipulation
3.4. Down-Regulation of Competing Pathways
3.5. Expression of Heterologous Thioesterases
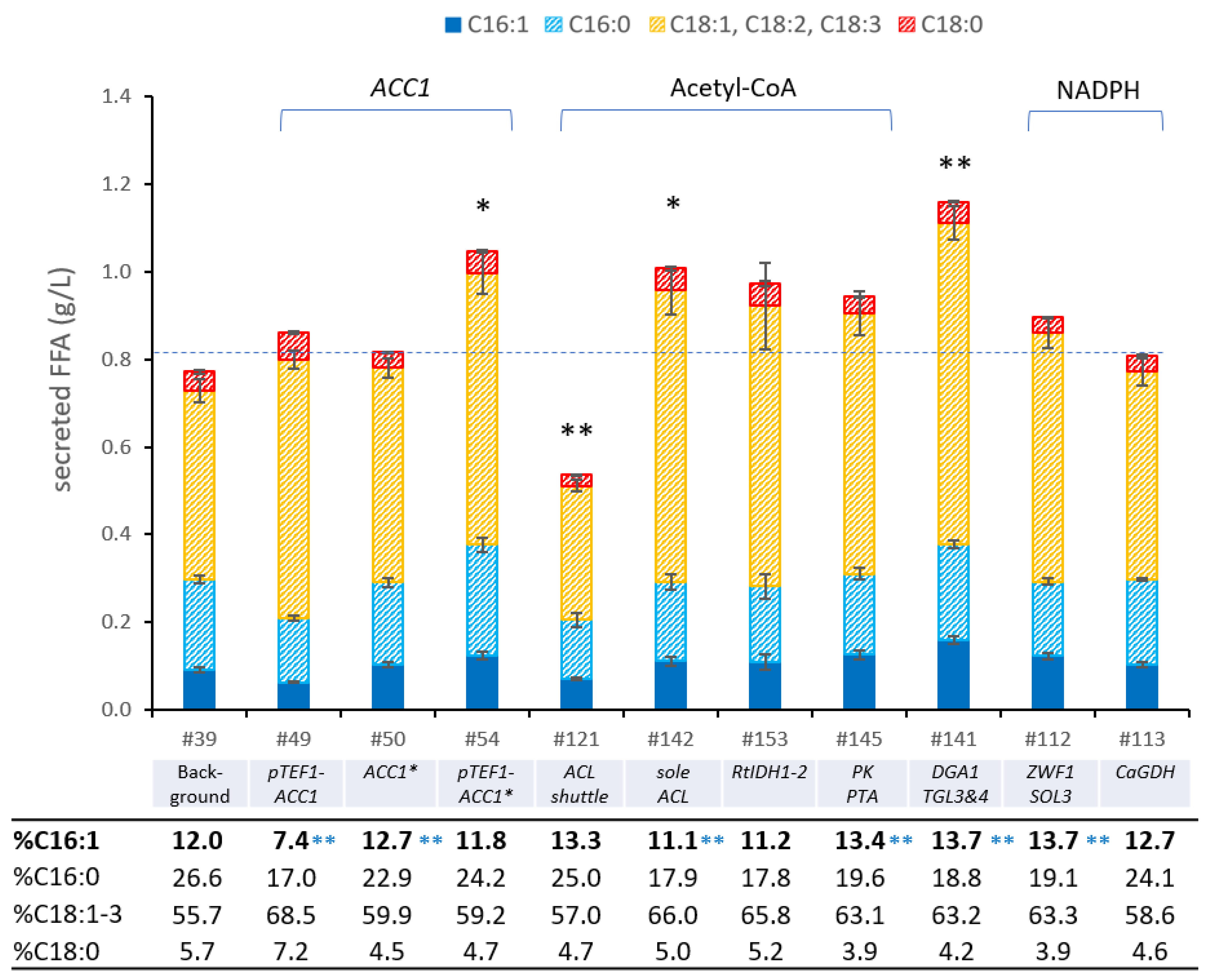
3.6. Enhancing the Availablity of Fatty Acid Precursor Molecules
3.7. Expression of Heterologous Fatty Acid Synthases
3.8. Engineering the Endogenous Fatty Acid Elongase System
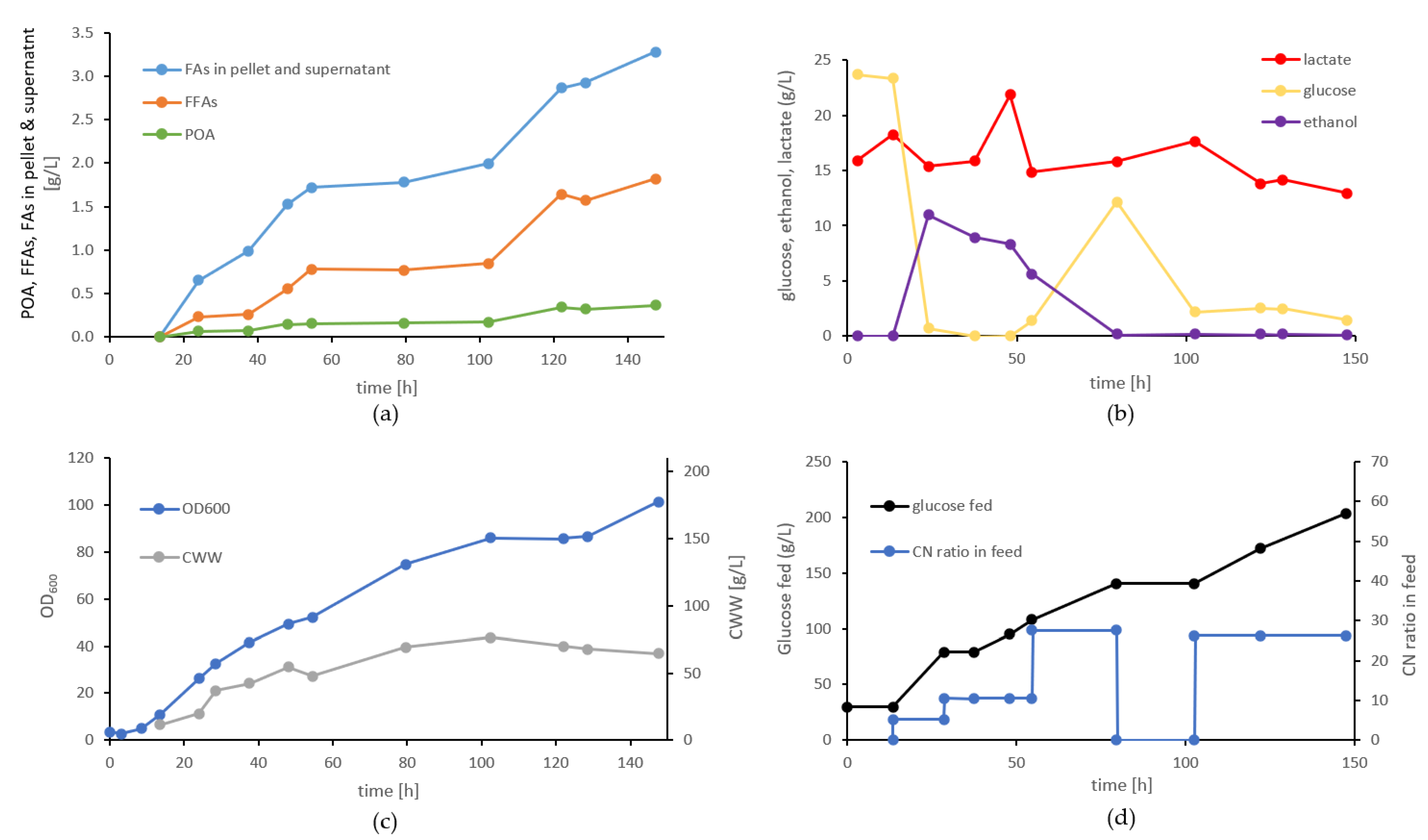
3.9. Evaluation of FA Production of Strain Pp#85 in Bioreactor Cultivations
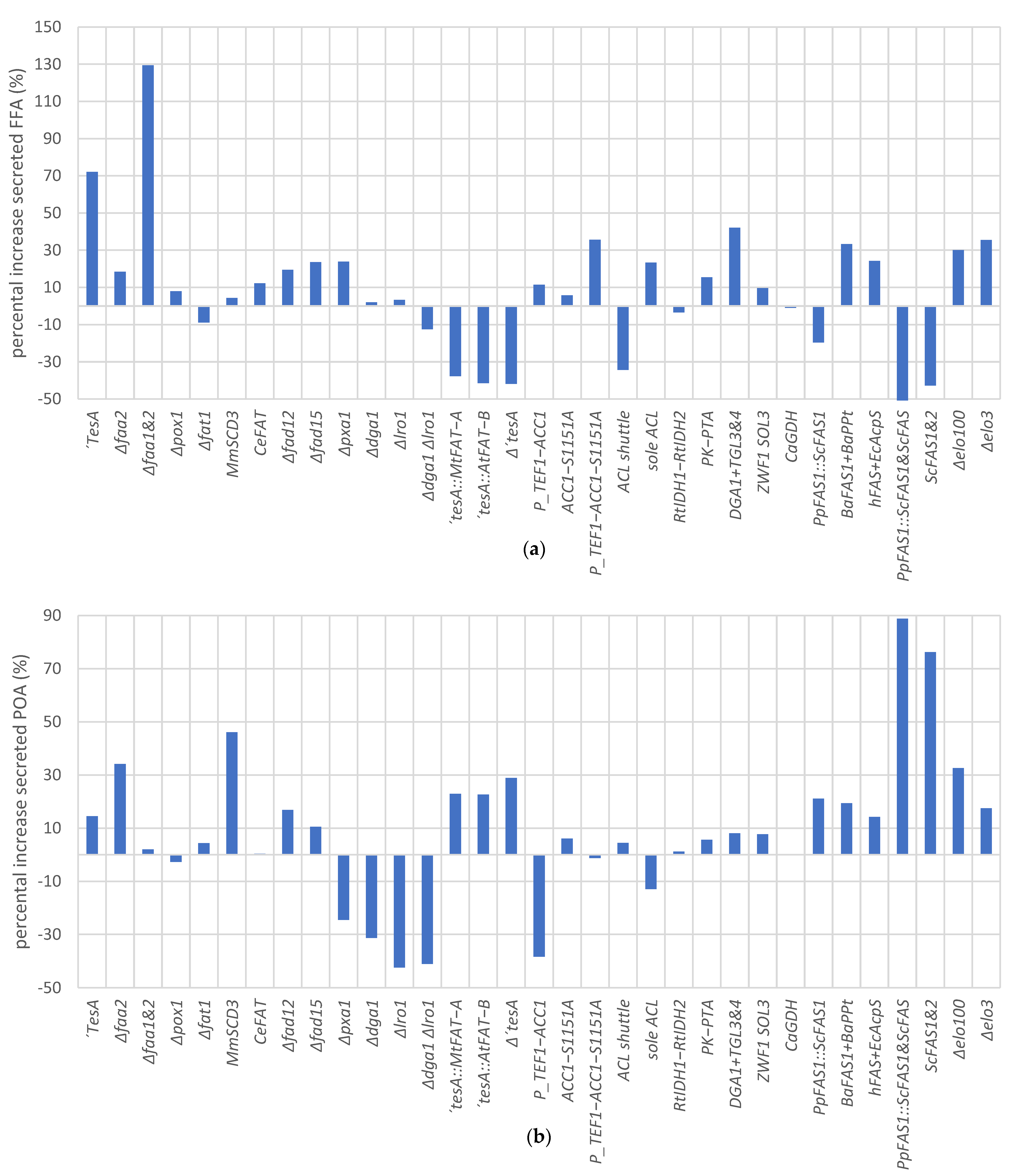
3.10. Evaluation of Metabolic Engineering Strategies for POA Production and Secretion
| Strains (Wild-Type) | Conditions | Fatty Acids (%) | POA (g/L) | FA (g/L) | Ref. | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C16:1 POA | C16:0 PA | C18:3 αLEA | C18:2 LA | C18:1 OA | C18:0 SA | other FA | |||||
| P. pastoris CBS7435 | YPD, SF, (72 h) | 5.5 | 5.1 | 10.9 | 29.5 | 45.7 | 0.8 | 2.5 | 0.05 | 1 (i) | this study |
| Kluyveromyces polysporus DBM 2171 | N-lim. MM, SF (n/d~48–96 h est.) | 74.5 | 7.2 | 0 | 0.9 | 17.1 | 0.3 | 0 | 0.16 g/g DW ** | n/d | [74] |
| S. cerevisiae DBM 2115 | N-lim. MM, SF (n/d~48–96 h est.) | 59.8 | 11.5 | 0 | 2 | 24 | 2.7 | 0 | 0.08 g/g DW ** | n/d | [74] |
| Strains (engineered) | Conditions | C16:1 POA | C16:0 PA | C18:3 αLEA | C18:2 LA | C18:1 OA | C18:0 SA | other FA | POA (g/L) | FA (g/L) | Ref. |
| P. pastoris Pp#85 | N-lim. MM, FB (150 h) | 18.7 | 27.9 | * | 6.3 | 40.7 | 3.4 | 3.0 | 0.37(s) 0.61 (t) | 1.8 (s) 3.3 (t) | this study |
| P. pastoris PC124H | N-lim. MM, FB (220 h) | 9 | 42 | n/d | 27 | 17 | 4 | n/d | 2.1 | 23.4 (t) | [62] |
| S. cerevisiae MK | N-lim. MM, FB (144 h) | 57.5 | 9.4 | 0 | 0 | 23 | 2 | 8.1 | 6.56 | 11.4 (i) | [103] |
| S. cerevisiae Y & Z055E (CEN.PK113-5D) | N-lim. MM, FB (~250 h) | 36.5 | 32.9 | 0.0 | 0.0 | 18.7 | 6.3 | 5.4 | 9.1 | 25 (t) | [101] |
| Scheffersomyces segobiensis SS-12 | MM, FB (~185 h) | 24.9 | 9.6 | 0.0 | 4.0 | 58.1 | 0.0 | 3.4 | 7.3 | 29.6 (i) | [104] |
| E. coli SBF50 | N-lim. MM, FB (~55 h) | 30.3 | 14.0 | 0.0 | 0.0 | 16.1 | 0.0 | 39.5 | 10 | 33.6 (s) | [18] |
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cerone, M.; Smith, T.K. A Brief Journey into the History of and Future Sources and Uses of Fatty Acids. Front. Nutr. 2021, 8, 570401. [Google Scholar] [CrossRef] [PubMed]
- Dartiailh, C.; Cicek, N.; Sorensen, J.L.; Levin, D.B. Production and Modification of PHA Polymers Produced from Long-Chain Fatty Acids. In The Handbook of Polyhydroxyalkanoates; CRC Press: Boca Raton, FL, USA, 2020; ISBN 978-0-429-29661-1. [Google Scholar]
- De Carvalho, C.C.C.R.; Caramujo, M.J. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef]
- Koch, B.; Schmidt, C.; Daum, G. Storage Lipids of Yeasts: A Survey of Nonpolar Lipid Metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol. Rev. 2014, 38, 892–915. [Google Scholar] [CrossRef]
- Yan, Q.; Pfleger, B.F. Revisiting Metabolic Engineering Strategies for Microbial Synthesis of Oleochemicals. Metab. Eng. 2020, 58, 35–46. [Google Scholar] [CrossRef]
- Yu, A.-Q.; Pratomo Juwono, N.K.; Leong, S.S.J.; Chang, M.W. Production of Fatty Acid-Derived Valuable Chemicals in Synthetic Microbes. Front. Bioeng. Biotechnol. 2014, 2, 78. [Google Scholar] [CrossRef]
- Kim, J.Y.; Ahn, Y.-J.; Lee, J.A.; Lee, S.Y. Recent Advances in the Production of Platform Chemicals Using Metabolically Engineered Microorganisms. Curr. Opin. Green Sustain. Chem. 2023, 40, 100777. [Google Scholar] [CrossRef]
- Ko, Y.-S.; Woong Kim, J.; An Lee, J.; Han, T.; Bae Kim, G.; Eum Park, J.; Yup Lee, S. Tools and Strategies of Systems Metabolic Engineering for the Development of Microbial Cell Factories for Chemical Production. Chem. Soc. Rev. 2020, 49, 4615–4636. [Google Scholar] [CrossRef] [PubMed]
- Lamers, D.; van Biezen, N.; Martens, D.; Peters, L.; van de Zilver, E.; Jacobs-van Dreumel, N.; Wijffels, R.H.; Lokman, C. Selection of Oleaginous Yeasts for Fatty Acid Production. BMC Biotechnol. 2016, 16, 45. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Benitez, M.G.; Chen, J.; Harrison, E.; Khusnutdinova, A.N.; Mahadevan, R. Opportunities and Challenges for Microbial Synthesis of Fatty Acid-Derived Chemicals (FACs). Front. Bioeng. Biotechnol. 2021, 9, 613322. [Google Scholar] [CrossRef]
- Fujita, Y.; Matsuoka, H.; Hirooka, K. Regulation of Fatty Acid Metabolism in Bacteria. Mol. Microbiol. 2007, 66, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, K.; Jackowski, S.; Rock, C.O.; Cronan, J.E. Regulation of Fatty Acid Biosynthesis in Escherichia coli. Microbiol. Rev. 1993, 57, 522–542. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.J.; Choi, K.R.; Lee, S.Y. Microbial Production of Fatty Acids and Derivative Chemicals. Curr. Opin. Biotechnol. 2020, 65, 129–141. [Google Scholar] [CrossRef]
- Janßen, H.J.; Steinbüchel, A. Fatty Acid Synthesis in Escherichia coli and Its Applications towards the Production of Fatty Acid Based Biofuels. Biotechnol. Biofuels 2014, 7, 7. [Google Scholar] [CrossRef]
- Kim, S.-K.; Park, Y.-C. Biosynthesis of ω-Hydroxy Fatty Acids and Related Chemicals from Natural Fatty Acids by Recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 2019, 103, 191–199. [Google Scholar] [CrossRef]
- Lennen, R.M.; Pfleger, B.F. Engineering Escherichia coli to Synthesize Free Fatty Acids. Trends Biotechnol. 2012, 30, 659–667. [Google Scholar] [CrossRef]
- Shin, K.S.; Lee, S.K. Introduction of an Acetyl-CoA Carboxylation Bypass into Escherichia coli for Enhanced Free Fatty Acid Production. Bioresour. Technol. 2017, 245, 1627–1633. [Google Scholar] [CrossRef]
- Park, W.S.; Shin, K.S.; Jung, H.W.; Lee, Y.; Sathesh-Prabu, C.; Lee, S.K. Combinatorial Metabolic Engineering Strategies for the Enhanced Production of Free Fatty Acids in Escherichia coli. J. Agric. Food Chem. 2022, 70, 13913–13921. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Duan, X.; Zhou, P.; Liu, P.; Pang, Z.; Wang, Y.; Wang, X.; Li, W.; Dong, M. Construction of Artificial Micro-Aerobic Metabolism for Energy- and Carbon-Efficient Synthesis of Medium Chain Fatty Acids in Escherichia coli. Metab. Eng. 2019, 53, 1–13. [Google Scholar] [CrossRef]
- Jiang, W.; Li, C.; Li, Y.; Peng, H. Metabolic Engineering Strategies for Improved Lipid Production and Cellular Physiological Responses in Yeast Saccharomyces cerevisiae. J. Fungi 2022, 8, 427. [Google Scholar] [CrossRef]
- Ratledge, C. Fatty Acid Biosynthesis in Microorganisms Being Used for Single Cell Oil Production. Biochimie 2004, 86, 807–815. [Google Scholar] [CrossRef]
- Patel, A.; Karageorgou, D.; Rova, E.; Katapodis, P.; Rova, U.; Christakopoulos, P.; Matsakas, L. An Overview of Potential Oleaginous Microorganisms and Their Role in Biodiesel and Omega-3 Fatty Acid-Based Industries. Microorganisms 2020, 8, 434. [Google Scholar] [CrossRef]
- Spagnuolo, M.; Yaguchi, A.; Blenner, M. Oleaginous Yeast for Biofuel and Oleochemical Production. Curr. Opin. Biotechnol. 2019, 57, 73–81. [Google Scholar] [CrossRef]
- Szczepańska, P.; Hapeta, P.; Lazar, Z. Advances in Production of High-Value Lipids by Oleaginous Yeasts. Crit. Rev. Biotechnol. 2022, 42, 1–22. [Google Scholar] [CrossRef]
- Wang, K.; Shi, T.-Q.; Lin, L.; Wei, P.; Ledesma-Amaro, R.; Ji, X.-J. Engineering Yarrowia lipolytica to Produce Tailored Chain-Length Fatty Acids and Their Derivatives. ACS Synth. Biol. 2022, 11, 2564–2577. [Google Scholar] [CrossRef]
- Wu, C.-C.; Honda, K.; Kazuhito, F. Current Advances in Alteration of Fatty Acid Profile in Rhodotorula toruloides: A Mini-Review. World J. Microbiol. Biotechnol. 2023, 39, 234. [Google Scholar] [CrossRef]
- Bergenholm, D.; Gossing, M.; Wei, Y.; Siewers, V.; Nielsen, J. Modulation of Saturation and Chain Length of Fatty Acids in Saccharomyces cerevisiae for Production of Cocoa Butter-like Lipids. Biotechnol. Bioeng. 2018, 115, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Knothe, G. Improving Biodiesel Fuel Properties by Modifying Fatty Ester Composition. Energy Environ. Sci. 2009, 2, 759–766. [Google Scholar] [CrossRef]
- Konzock, O.; Matsushita, Y.; Zaghen, S.; Sako, A.; Norbeck, J. Altering the Fatty Acid Profile of Yarrowia lipolytica to Mimic Cocoa Butter by Genetic Engineering of Desaturases. Microb. Cell Factories 2022, 21, 25. [Google Scholar] [CrossRef]
- Botella-Martínez, C.; Pérez-Álvarez, J.Á.; Sayas-Barberá, E.; Navarro Rodríguez de Vera, C.; Fernández-López, J.; Viuda-Martos, M. Healthier Oils: A New Scope in the Development of Functional Meat and Dairy Products: A Review. Biomolecules 2023, 13, 778. [Google Scholar] [CrossRef] [PubMed]
- Sales-Campos, H.; de Souza, P.R.; Peghini, B.C.; da Silva, J.S.; Cardoso, C.R. An Overview of the Modulatory Effects of Oleic Acid in Health and Disease. Mini Rev. Med. Chem. 2013, 13, 201–210. [Google Scholar]
- Porokhovinova, E.A.; Matveeva, T.V.; Khafizova, G.V.; Bemova, V.D.; Doubovskaya, A.G.; Kishlyan, N.V.; Podolnaya, L.P.; Gavrilova, V.A. Fatty Acid Composition of Oil Crops: Genetics and Genetic Engineering. Genet. Resour. Crop Evol. 2022, 69, 2029–2045. [Google Scholar] [CrossRef]
- Bermúdez, M.A.; Pereira, L.; Fraile, C.; Valerio, L.; Balboa, M.A.; Balsinde, J. Roles of Palmitoleic Acid and Its Positional Isomers, Hypogeic and Sapienic Acids, in Inflammation, Metabolic Diseases and Cancer. Cells 2022, 11, 2146. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Fitzgerald, M.; Topp, B.; Alam, M.; O’Hare, T.J. A Review of Biological Functions, Health Benefits, and Possible de Novo Biosynthetic Pathway of Palmitoleic Acid in Macadamia Nuts. J. Funct. Foods 2019, 62, 103520. [Google Scholar] [CrossRef]
- Araujo Nunes, E.; Rafacho, A. Implications of Palmitoleic Acid (Palmitoleate) On Glucose Homeostasis, Insulin Resistance and Diabetes. Curr. Drug Targets 2017, 18, 619–628. [Google Scholar] [CrossRef]
- Wu, Y.; Li, R.; Hildebrand, D.F. Biosynthesis and Metabolic Engineering of Palmitoleate Production, an Important Contributor to Human Health and Sustainable Industry. Prog. Lipid Res. 2012, 51, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Yamamoto, Y.; Miura, M.; Konno, H.; Yano, S.; Nonomura, Y. Systematic Analysis of Selective Bactericidal Activity of Fatty Acids against Staphylococcus aureus with Minimum Inhibitory Concentration and Minimum Bactericidal Concentration. J. Oleo Sci. 2019, 68, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Solà Marsiñach, M.; Cuenca, A.P. The Impact of Sea Buckthorn Oil Fatty Acids on Human Health. Lipids Health Dis. 2019, 18, 145. [Google Scholar] [CrossRef]
- Barone, G.D.; Emmerstorfer-Augustin, A.; Biundo, A.; Pisano, I.; Coccetti, P.; Mapelli, V.; Camattari, A. Industrial Production of Proteins with Pichia pastoris—Komagataella phaffii. Biomolecules 2023, 13, 441. [Google Scholar] [CrossRef]
- Moser, S.; Strohmeier, G.A.; Leitner, E.; Plocek, T.J.; Vanhessche, K.; Pichler, H. Whole-Cell (+)-Ambrein Production in the Yeast Pichia pastoris. Metab. Eng. Commun. 2018, 7, e00077. [Google Scholar] [CrossRef]
- Vijayakumar, V.E.; Venkataraman, K. A Systematic Review of the Potential of Pichia pastoris (Komagataella phaffii) as an Alternative Host for Biologics Production. Mol. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Wriessnegger, T.; Augustin, P.; Engleder, M.; Leitner, E.; Müller, M.; Kaluzna, I.; Schürmann, M.; Mink, D.; Zellnig, G.; Schwab, H.; et al. Production of the Sesquiterpenoid (+)-Nootkatone by Metabolic Engineering of Pichia pastoris. Metab. Eng. 2014, 24, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Ata, Ö.; Ergün, B.G.; Fickers, P.; Heistinger, L.; Mattanovich, D.; Rebnegger, C.; Gasser, B. What Makes Komagataella phaffii Non-Conventional? FEMS Yeast Res. 2021, 21, foab059. [Google Scholar] [CrossRef] [PubMed]
- García-Ortega, X.; Cámara, E.; Ferrer, P.; Albiol, J.; Montesinos-Seguí, J.L.; Valero, F. Rational Development of Bioprocess Engineering Strategies for Recombinant Protein Production in Pichia pastoris (Komagataella phaffii) Using the Methanol-Free GAP Promoter. Where Do We Stand? New Biotechnol. 2019, 53, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P. Biotechnological Strains of Komagataella (Pichia) pastoris Are Komagataella phaffii as Determined from Multigene Sequence Analysis. J. Ind. Microbiol. Biotechnol. 2009, 36, 1435. [Google Scholar] [CrossRef] [PubMed]
- Sturmberger, L.; Chappell, T.; Geier, M.; Krainer, F.; Day, K.J.; Vide, U.; Trstenjak, S.; Schiefer, A.; Richardson, T.; Soriaga, L.; et al. Refined Pichia pastoris Reference Genome Sequence. J. Biotechnol. 2016, 235, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Küberl, A.; Schneider, J.; Thallinger, G.G.; Anderl, I.; Wibberg, D.; Hajek, T.; Jaenicke, S.; Brinkrolf, K.; Goesmann, A.; Szczepanowski, R.; et al. High-Quality Genome Sequence of Pichia pastoris CBS7435. J. Biotechnol. 2011, 154, 312–320. [Google Scholar] [CrossRef]
- Weninger, A.; Fischer, J.E.; Raschmanová, H.; Kniely, C.; Vogl, T.; Glieder, A. Expanding the CRISPR/Cas9 Toolkit for Pichia pastoris with Efficient Donor Integration and Alternative Resistance Markers. J. Cell. Biochem. 2018, 119, 3183–3198. [Google Scholar] [CrossRef]
- Liu, Q.; Shi, X.; Song, L.; Liu, H.; Zhou, X.; Wang, Q.; Zhang, Y.; Cai, M. CRISPR-Cas9-Mediated Genomic Multiloci Integration in Pichia pastoris. Microb. Cell Factories 2019, 18, 144. [Google Scholar] [CrossRef]
- Cai, P.; Duan, X.; Wu, X.; Gao, L.; Ye, M.; Zhou, Y.J. Recombination Machinery Engineering Facilitates Metabolic Engineering of the Industrial Yeast Pichia pastoris. Nucleic Acids Res. 2021, 49, 7791–7805. [Google Scholar] [CrossRef]
- Lin-Cereghino, J.; Wong, W.W.; Xiong, S.; Giang, W.; Luong, L.T.; Vu, J.; Johnson, S.D.; Lin-Cereghino, G.P. Condensed Protocol for Competent Cell Preparation and Transformation of the Methylotrophic Yeast Pichia pastoris. BioTechniques 2005, 38, 44–48. [Google Scholar] [CrossRef]
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of Genomic DNA from Yeasts for PCR-Based Applications. BioTechniques 2011, 50, 325–328. [Google Scholar] [CrossRef]
- Straathof, A. The Proportion of Downstream Costs in Fermentative Production Processes. In Comprehensive Biotechnology; Moo-Young, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 811–814. ISBN 978-0-08-088504-9. [Google Scholar]
- d’Espaux, L.; Ghosh, A.; Runguphan, W.; Wehrs, M.; Xu, F.; Konzock, O.; Dev, I.; Nhan, M.; Gin, J.; Reider Apel, A.; et al. Engineering High-Level Production of Fatty Alcohols by Saccharomyces cerevisiae from Lignocellulosic Feedstocks. Metab. Eng. 2017, 42, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Tehlivets, O.; Scheuringer, K.; Kohlwein, S.D. Fatty Acid Synthesis and Elongation in Yeast. Biochim. Biophys. Acta 2007, 1771, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Færgeman, N.J.; Knudsen, J. Role of Long-Chain Fatty Acyl-CoA Esters in the Regulation of Metabolism and in Cell Signalling. Biochem. J. 1997, 323, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Steen, E.J.; Kang, Y.; Bokinsky, G.; Hu, Z.; Schirmer, A.; McClure, A.; del Cardayre, S.B.; Keasling, J.D. Microbial Production of Fatty-Acid-Derived Fuels and Chemicals from Plant Biomass. Nature 2010, 463, 559–562. [Google Scholar] [CrossRef]
- Runguphan, W.; Keasling, J.D. Metabolic Engineering of Saccharomyces cerevisiae for Production of Fatty Acid-Derived Biofuels and Chemicals. Metab. Eng. 2014, 21, 103–113. [Google Scholar] [CrossRef]
- Scharnewski, M.; Pongdontri, P.; Mora, G.; Hoppert, M.; Fulda, M. Mutants of Saccharomyces cerevisiae Deficient in Acyl-CoA Synthetases Secrete Fatty Acids Due to Interrupted Fatty Acid Recycling. FEBS J. 2008, 275, 2765–2778. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Cronan, J.E. Defective Export of a Periplasmic Enzyme Disrupts Regulation of Fatty Acid Synthesis (*). J. Biol. Chem. 1995, 270, 4216–4219. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Buijs, N.A.; Zhu, Z.; Qin, J.; Siewers, V.; Nielsen, J. Production of Fatty Acid-Derived Oleochemicals and Biofuels by Synthetic Yeast Cell Factories. Nat. Commun. 2016, 7, 11709. [Google Scholar] [CrossRef]
- Cai, P.; Wu, X.; Deng, J.; Gao, L.; Shen, Y.; Yao, L.; Zhou, Y.J. Methanol Biotransformation toward High-Level Production of Fatty Acid Derivatives by Engineering the Industrial Yeast Pichia pastoris. Proc. Natl. Acad. Sci. USA 2022, 119, e2201711119. [Google Scholar] [CrossRef]
- Dmochowska, A.; Dignard, D.; Maleszka, R.; Thomas, D.Y. Structure and Transcriptional Control of the Saccharomyces cerevisiae POX1 Gene Encoding Acylcoenzyme A Oxidase. Gene 1990, 88, 247–252. [Google Scholar] [CrossRef]
- Zou, Z.; Tong, F.; Færgeman, N.J.; Børsting, C.; Black, P.N.; DiRusso, C.C. Vectorial Acylation in Saccharomyces cerevisiae: Fat1p and fatty ACYL-CoA Synthetase Are Interacting Components of a Fatty Acid Import Complex. J. Biol. Chem. 2003, 278, 16414–16422. [Google Scholar] [CrossRef]
- Martin, C.E.; Oh, C.-S.; Jiang, Y. Regulation of Long Chain Unsaturated Fatty Acid Synthesis in Yeast. Biochim. Biophys. Acta 2007, 1771, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L.; Browse, J. A Palmitoyl-CoA-Specific Delta9 Fatty Acid Desaturase from Caenorhabditis Elegans. Biochem. Biophys. Res. Commun. 2000, 272, 263–269. [Google Scholar] [CrossRef]
- Qiao, K.; Imam Abidi, S.H.; Liu, H.; Zhang, H.; Chakraborty, S.; Watson, N.; Kumaran Ajikumar, P.; Stephanopoulos, G. Engineering Lipid Overproduction in the Oleaginous Yeast Yarrowia lipolytica. Metab. Eng. 2015, 29, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; Bruggink, S.M.; Ntambi, J.M. Identification of Mouse Palmitoyl-Coenzyme A Delta9-Desaturase. J. Lipid Res. 2006, 47, 700–704. [Google Scholar] [CrossRef]
- Geier, M.; Fauland, P.; Vogl, T.; Glieder, A. Compact Multi-Enzyme Pathways in P. Pastoris. Chem. Commun. 2015, 51, 1643–1646. [Google Scholar] [CrossRef]
- Yazawa, H.; Iwahashi, H.; Kamisaka, Y.; Kimura, K.; Uemura, H. Improvement of Polyunsaturated Fatty Acids Synthesis by the Coexpression of CYB5 with Desaturase Genes in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2010, 87, 2185–2193. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.S.; Li, M.C.; Zhang, X.X.; Zhou, H.; Xing, L.J. A Novel Delta12-Fatty Acid Desaturase Gene from Methylotrophic Yeast Pichia pastoris GS115. Acta Biochim. Pol. 2006, 53, 753–759. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.; Wei, D.; Xing, L. Identification and Characterization of a Novel Yeast Omega3-Fatty Acid Desaturase Acting on Long-Chain n-6 Fatty Acid Substrates from Pichia pastoris. Yeast 2008, 25, 21–27. [Google Scholar] [CrossRef]
- Beopoulos, A.; Cescut, J.; Haddouche, R.; Uribelarrea, J.-L.; Molina-Jouve, C.; Nicaud, J.-M. Yarrowia lipolytica as a Model for Bio-Oil Production. Prog. Lipid Res. 2009, 48, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Kolouchová, I.; Sigler, K.; Schreiberová, O.; Masák, J.; Řezanka, T. New Yeast-Based Approaches in Production of Palmitoleic Acid. Bioresour. Technol. 2015, 192, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Pádrová, K.; Kolouchová, I.; Řezanka, T.; Čejková, A. Using Nutritional and Oxidative Stress to Increase Content of Healthbeneficial Fatty Acids in Oleaginous and Non-Oleaginous Yeasts. Chem. Pap. 2016, 70, 1351–1359. [Google Scholar] [CrossRef]
- Hynes, M.J.; Murray, S.L. ATP-Citrate Lyase Is Required for Production of Cytosolic Acetyl Coenzyme A and Development in Aspergillus Nidulans. Eukaryot. Cell 2010, 9, 1039–1048. [Google Scholar] [CrossRef]
- Beopoulos, A.; Chardot, T.; Nicaud, J.-M. Yarrowia Lipolytica: A Model and a Tool to Understand the Mechanisms Implicated in Lipid Accumulation. Biochimie 2009, 91, 692–696. [Google Scholar] [CrossRef]
- Leber, C.; Polson, B.; Fernandez-Moya, R.; Da Silva, N.A. Overproduction and Secretion of Free Fatty Acids through Disrupted Neutral Lipid Recycle in Saccharomyces cerevisiae. Metab. Eng. 2015, 28, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Salvador López, J.M.; Van Bogaert, I.N.A. Microbial Fatty Acid Transport Proteins and Their Biotechnological Potential. Biotechnol. Bioeng. 2021, 118, 2184–2201. [Google Scholar] [CrossRef]
- Jing, F.; Cantu, D.C.; Tvaruzkova, J.; Chipman, J.P.; Nikolau, B.J.; Yandeau-Nelson, M.D.; Reilly, P.J. Phylogenetic and Experimental Characterization of an Acyl-ACP Thioesterase Family Reveals Significant Diversity in Enzymatic Specificity and Activity. BMC Biochem. 2011, 12, 44. [Google Scholar] [CrossRef]
- Mayer, K.M.; Shanklin, J. Identification of Amino Acid Residues Involved in Substrate Specificity of Plant Acyl-ACP Thioesterases Using a Bioinformatics-Guided Approach. BMC Plant Biol. 2007, 7, 1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moreno-Pérez, A.J.; Sánchez-García, A.; Salas, J.J.; Garcés, R.; Martínez-Force, E. Acyl-ACP Thioesterases from Macadamia (Macadamia tetraphylla) Nuts: Cloning, Characterization and Their Impact on Oil Composition. Plant Physiol. Biochem. PPB 2011, 49, 82–87. [Google Scholar] [CrossRef]
- McKeon, T.A.; Stumpf, P.K. Purification and Characterization of the Stearoyl-Acyl Carrier Protein Desaturase and the Acyl-Acyl Carrier Protein Thioesterase from Maturing Seeds of Safflower. J. Biol. Chem. 1982, 257, 12141–12147. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.M.; Nau, K.; Geraghty, M.T.; Erdmann, R.; Gould, S.J. Identification of Peroxisomal Acyl-CoA Thioesterases in Yeast and Humans*. J. Biol. Chem. 1999, 274, 9216–9223. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Chen, Y.; Siewers, V.; Nielsen, J. Improving Production of Malonyl Coenzyme A-Derived Metabolites by Abolishing Snf1-Dependent Regulation of Acc1. mBio 2014, 5, e01130-14. [Google Scholar] [CrossRef] [PubMed]
- Sumper, M.; Riepertinger, C.; Lynen, F.; Oesterhelt, D. Die Synthese Verschiedener Carbonsäuren Durch Den Multienzymkomplex der Fettsäuresynthese Aus Hefe Und Die Erklärung Ihrer Bildung. Eur. J. Biochem. 1969, 10, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.-J.; Jiang, J.-G. Characterization of Malic Enzyme and the Regulation of Its Activity and Metabolic Engineering on Lipid Production. RSC Adv. 2015, 5, 45558–45570. [Google Scholar] [CrossRef]
- Bergman, A.; Siewers, V.; Nielsen, J.; Chen, Y. Functional Expression and Evaluation of Heterologous Phosphoketolases in Saccharomyces cerevisiae. AMB Express 2016, 6, 115. [Google Scholar] [CrossRef]
- Qiao, K.; Wasylenko, T.M.; Zhou, K.; Xu, P.; Stephanopoulos, G. Lipid Production in Yarrowia lipolytica Is Maximized by Engineering Cytosolic Redox Metabolism. Nat. Biotechnol. 2017, 35, 173–177. [Google Scholar] [CrossRef]
- Nocon, J.; Steiger, M.; Mairinger, T.; Hohlweg, J.; Rußmayer, H.; Hann, S.; Gasser, B.; Mattanovich, D. Increasing Pentose Phosphate Pathway Flux Enhances Recombinant Protein Production in Pichia pastoris. Appl. Microbiol. Biotechnol. 2016, 100, 5955–5963. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Kickenweiz, T.; Pitzer, J.; Sturmberger, L.; Weninger, A.; Biggs, B.W.; Köhler, E.-M.; Baumschlager, A.; Fischer, J.E.; Hyden, P.; et al. Engineered Bidirectional Promoters Enable Rapid Multi-Gene Co-Expression Optimization. Nat. Commun. 2018, 9, 3589. [Google Scholar] [CrossRef] [PubMed]
- Heil, C.S.; Wehrheim, S.S.; Paithankar, K.S.; Grininger, M. Fatty Acid Biosynthesis: Chain-Length Regulation and Control. ChemBioChem 2019, 20, 2298–2321. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, D.T.; HamediRad, M.; Yuan, Y.; Zhao, H. Orthogonal Fatty Acid Biosynthetic Pathway Improves Fatty Acid Ethyl Ester Production in Saccharomyces cerevisiae. ACS Synth. Biol. 2015, 4, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Leber, C.; Da Silva, N.A. Engineering of Saccharomyces cerevisiae for the Synthesis of Short Chain Fatty Acids. Biotechnol. Bioeng. 2014, 111, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Snowden, J.S.; Alzahrani, J.; Sherry, L.; Stacey, M.; Rowlands, D.J.; Ranson, N.A.; Stonehouse, N.J. Structural Insight into Pichia pastoris Fatty Acid Synthase. Sci. Rep. 2021, 11, 9773. [Google Scholar] [CrossRef] [PubMed]
- Toke, D.A.; Martin, C.E. Isolation and Characterization of a Gene Affecting Fatty Acid Elongation in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 18413–18422. [Google Scholar] [CrossRef] [PubMed]
- Schneiter, R.; Tatzer, V.; Gogg, G.; Leitner, E.; Kohlwein, S.D. Elo1p-Dependent Carboxy-Terminal Elongation of C14:1Δ9 to C16:1Δ11 Fatty Acids in Saccharomyces cerevisiae. J. Bacteriol. 2000, 182, 3655–3660. [Google Scholar] [CrossRef]
- Oh, C.-S.; Toke, D.A.; Mandala, S.; Martin, C.E. ELO2 and ELO3, Homologues of the Saccharomyces cerevisiae ELO1 Gene, Function in Fatty Acid Elongation and Are Required for Sphingolipid Formation. J. Biol. Chem. 1997, 272, 17376–17384. [Google Scholar] [CrossRef]
- Zou, Z.; DiRusso, C.C.; Ctrnacta, V.; Black, P.N. Fatty Acid Transport in Saccharomyces cerevisiae: Directed Mutagenesis of Fat1 Distinguishes the Biochemical Activities Associated with Fat1p. J. Biol. Chem. 2002, 277, 31062–31071. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhou, Y.J.; Huang, M.; Liu, Q.; Pereira, R.; David, F.; Nielsen, J. Reprogramming Yeast Metabolism from Alcoholic Fermentation to Lipogenesis. Cell 2018, 174, 1549–1558.e14. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Dulermo, R.; Niehus, X.; Nicaud, J.-M. Combining Metabolic Engineering and Process Optimization to Improve Production and Secretion of Fatty Acids. Metab. Eng. 2016, 38, 38–46. [Google Scholar] [CrossRef]
- Li, S.; Su, C.; Fang, M.; Cai, D.; Deng, L.; Wang, F.; Liu, J. Overproduction of Palmitoleic Acid from Corn Stover Hydrolysate by Engineered Saccharomyces cerevisiae. Bioresour. Technol. 2023, 382, 129211. [Google Scholar] [CrossRef]
- Qian, X.; Lei, H.; Zhou, X.; Zhang, L.; Cui, W.; Zhou, J.; Xin, F.; Dong, W.; Jiang, M.; Ochsenreither, K. Engineering Scheffersomyces segobiensis for Palmitoleic Acid-Rich Lipid Production. Microb. Biotechnol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Ivashov, V.A.; Zellnig, G.; Grillitsch, K.; Daum, G. Identification of triacylglycerol and steryl ester synthases of the methylotrophic yeast Pichia pastoris. Biochim. Biophys. Acta 2013, 1831, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Zweytick, D.; Leitner, E.; Kohlwein, S.D.; Yu, C.; Rothblatt, J.; Daum, G. Contribution of Are1p and Are2p to Steryl Ester Synthesis in the Yeast Saccharomyces Cerevisiae. Eur. J. Biochem. 2000, 267, 1075–1082. [Google Scholar] [CrossRef]


| Strain (Pp#) | Genetic Background | Further Modifications |
|---|---|---|
| Wild-Type (WT) | CBS7435 | |
| 12 | WT | Δfad12 |
| 14 | WT | Δole1-1 |
| 16 | WT | Δole1-2 |
| 28 | Pp#16 | his4::PGAP-‘TesA-TTARG4 |
| 29 | Pp#28 | Δfaa2 |
| 32 | Pp#29 | Δfaa1 |
| 35 | Pp#32 | Δpox1 |
| 37 | Pp#35 | Δfat1 |
| 39 | Pp#35 | FLDup::PTEF1-MmSCD3-TTARG4 |
| 44 | Pp#35 | Δdga1 |
| 45 | Pp#35 | Δlro1 |
| 46 | Pp#35 | Δdga1 Δlro1 |
| 47 | Pp#39 | his4::PGAP-MtFAT-A-TTARG4 |
| 48 | Pp#39 | his4::PGAP-AtFAT-B-TTARG4 |
| 49 | Pp#39 | acc1::PTEF1-ACC1-TTACC1 |
| 50 | Pp#39 | acc1::Acc1S1151A-TTACC1 |
| 51* | Pp#35 | Δdga1 Δlro1 Δare2 |
| 51 | Pp#39 | TEFup::PPGK1-CeFAT-TTARG4 |
| 53 | Pp#39 | his4::PGAP-Δ‘tesA-TTARG4 |
| 54 | Pp#39 | acc1::PTEF1-ACC1S1151A-TTACC1 |
| 67 | Pp#39 | TEFup::PHTX1-BaFAS1-TTARG4-BaPPT-TTAOX1 |
| 85 | Pp#39 | PpFAS1::PHTX1-ScFAS2-TTPpFAS1-ScFAS1-TTPpFAS1 |
| 89 | Pp#39 | TEFup::PHTX1-ScFAS2-TTARG4-ScFAS1-TTAOX1 |
| 91 | Pp#39 | TEFup::PHTX1-hFAS-TTARG4-EcACPS-TTAOX1 |
| 112 | Pp#50 | Chr1_NS6::PHHX1-ZWF1-TTARG4-SOL3 -TTTEF1 |
| 113 | Pp#50 | faa2::PGAP-RAD52-TTAOX1 Chr1_NS10_PGAP-CaGDH-TTAOX1 |
| 114 | Pp#50 | faa2::PGAP-RAD52-TTAOX1 |
| 115 | Pp#85 | PpFAS2::PHIS4-HIS4-TTPpFAS2 |
| 121 | Pp#50 | faa2::PGAP-RAD52 Chr2_NS3::PTPI-RtACL-TTDAS1-PGAP-MmME-TTHTB-PRP-PpCTP1-TTPGK1-PpMDH3-TTTEF1 |
| 127 | Pp#35 | FLDup::PTEF1-MmSCD3-T2A1-PpCypb5-T2A2-PpCyb5R-TTARG4 |
| 129 | Pp#35 | FLDup::PTEF1-MmSCD3-T2A1-MmCypb5-T2A2-MmCyb5R-TTARG4 |
| 135 | Pp#39 | Δpxa1 |
| 136 | Pp#39 | Δelo3 |
| 138 | Pp#39 | Δelo100 |
| 140 | Pp#39 | Δfad15 |
| 141 | Pp#50 | faa2::PGAP-RAD52 Chr4_NS7::PCAT-DGA1-TTTEF1-PHHX2-TGL3-TTHTA-TGL4-TTGAP |
| 142 | Pp#50 | faa2::PGAP-RAD52-TTAOX1 Chr2_NS3::PENO1-RtACL-TTDAS1 |
| 145 | Pp#50 | faa2::PGAP-RAD52 Chr3_NS7::PHHX2-BbPK--TTFDH1-CkPTA-TTFBP1 |
| 148 | Pp#85 | Chr4_NS7::PGAP-ScOLE1-TTDAS1 |
| 150 | Pp#39 | Δfad12 |
| 153 | Pp#50 | faa2::PGAP-RAD52-TTAOX1 Chr2_NS3::PENO1-RtACL PPpIDH1-RtIDH1-TTPpIDH1PPpIDH2-RtIDH2-TTPpIDH2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobalter, S.; Voit, A.; Bekerle-Bogner, M.; Rudalija, H.; Haas, A.; Wriessnegger, T.; Pichler, H. Tuning Fatty Acid Profile and Yield in Pichia pastoris. Bioengineering 2023, 10, 1412. https://doi.org/10.3390/bioengineering10121412
Kobalter S, Voit A, Bekerle-Bogner M, Rudalija H, Haas A, Wriessnegger T, Pichler H. Tuning Fatty Acid Profile and Yield in Pichia pastoris. Bioengineering. 2023; 10(12):1412. https://doi.org/10.3390/bioengineering10121412
Chicago/Turabian StyleKobalter, Simon, Alena Voit, Myria Bekerle-Bogner, Haris Rudalija, Anne Haas, Tamara Wriessnegger, and Harald Pichler. 2023. "Tuning Fatty Acid Profile and Yield in Pichia pastoris" Bioengineering 10, no. 12: 1412. https://doi.org/10.3390/bioengineering10121412
APA StyleKobalter, S., Voit, A., Bekerle-Bogner, M., Rudalija, H., Haas, A., Wriessnegger, T., & Pichler, H. (2023). Tuning Fatty Acid Profile and Yield in Pichia pastoris. Bioengineering, 10(12), 1412. https://doi.org/10.3390/bioengineering10121412







