Abstract
Medication recommendation based on electronic health records (EHRs) is a significant research direction in the biomedical field, which aims to provide a reasonable prescription for patients according to their historical and current health conditions. However, the existing recommended methods have many limitations in dealing with the structural and temporal characteristics of EHRs. These methods either only consider the current state while ignoring the historical situation, or fail to adequately assess the structural correlations among various medical events. These factors result in poor recommendation quality. To solve this problem, we propose an augmented graph structural–temporal convolutional network (A-GSTCN). Firstly, an augmented graph attention network is used to model the structural features among medical events of patients’ EHRs. Next, the dilated convolution combined with residual connection is applied in the proposed model, which can improve the temporal prediction capability and further reduce the complexity. Moreover, the cache memory module further enhances the model’s learning of the history of EHRs. Finally, the A-GSTCN model is compared with the baselines through experiments, and the efficiency of the A-GSTCN model is verified by Jaccard, F1 and PRAUC. Not only that, the proposed model also reduces the training parameters by an order of magnitude.
1. Introduction
Electronic health records (EHRs) are the primary data carrier for personalized medical research and help accelerate the care process and ensure medical quality. With the increasing potential of EHRs for medical applications, a great deal of research has been applied in this field, which includes diagnosis prediction and medication recommendation [1,2,3,4]. As shown in Figure 1, medication recommendation is of great importance because it can simplify the medical process and assist doctors in making accurate prescriptions. The target of medication recommendation is to recommend personalized and precise drugs for patients based on their current diagnosis and their historical health condition, whereas previous medication recommendation research was based on the rules and facts derived from specialists with abundant clinic experience [5,6,7,8]. With the deepening of medical informatization, deep learning models significantly improve the accuracy of medication recommendation tasks and the feasibility for practical application [9,10,11]. Nevertheless, because of the following characteristics, EHRs bring difficulties to medication recommendation tasks:
Figure 1.
The application of medication recommendation system in a medical scenario. The medication recommendation system learns the collected EHRs in advance and establishes the model to facilitate follow-up patients’ medical treatment and discharge with drugs.
- 1.
- Structural correlation: A patient’s EHRs can be seen as a combination of a set of diagnoses, procedures and medications, where the diagnoses, procedures and medications can be collectively referred to the medical events. Therefore, the EHRs can be expressed as a combination of multiple medical events, and the occurrences of medical events simultaneously in a medical record are referred to as structural correlations. For example, chemical ulcers are often accompanied by gastric perforation, and chickenpox can cause erysipelas. These phenomena can be considered as structural correlations between diagnostic events and diagnostic events themselves. Similarly, the combination of statins with cardiovascular drugs is more beneficial for recovery from coronary heart disease, and this phenomenon is thought to be structurally correlated with diagnostic events and medication combinations.
- 2.
- Temporal dependency: Chronic diseases, such as stroke, diabetes and high blood pressure, do not recover as quickly as common diseases. On the contrary, chronic diseases are often incurable and require multiple visits. Meanwhile, during the patient’s medical treatment process, different treatments and drugs can be used at different times. The connection of these medical events on a temporal level is referred to as temporal dependency. For the same patient, the EHRs at multiple admissions can be regarded as multiple continuous medical processes, which may have rich temporal characteristics. In addition, different medical events (diagnoses, procedures and medications) may show different temporal dependencies in different patients.
To capture the structural correlation and temporal dependency of the EHRs, a lot of work has been performed in the early research [12,13,14,15]. However, these methods are rule-based or based on simple classifications, resulting in poor learning ability of EHRs. With the gradual popularization of neural-network-based methods, the graph structure is introduced to capture the structural correlation. Some studies [16,17,18] introduce the graph convolutional network (GCN) for structural modeling, which learns the internal correlation between medical events adequately. However, they ignore the temporal dependency of patients’ records, so that the change of EHRs is not restricted and the models cannot recommend medications accurately. Moreover, some models [19,20,21] consider the temporal change of EHRs, but they cannot cope with the medical events with a complex topological structure, which leads to their inability to describe the structural correlation of EHRs.
Therefore, to simultaneously learn the structural correlation and temporal dependency of EHRs, we propose a novel medication recommendation model called augmented graph structural-temporal convolutional network (A-GSTCN). As shown in Figure 2, we use ICD-9 encoding and ATC encoding to standardize the datasets. Moreover, we use an augmented graph attention network (GAT) to learn the structural correlations of EHRs and further utilize dilated convolution combined with residual connection to capture the temporal features.
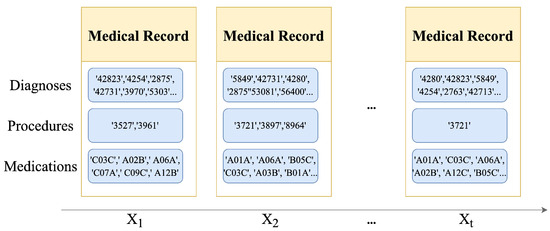
Figure 2.
A standardized sample of EHRs. ICD-9 encoding and ATC encoding are used to standardize the EHRs.
Our contributions can be summarized as follows:
- 1.
- We treat EHRs as time-series records with structural correlation and use ICD-9 encoding and ATC encoding to standardize the records in pretraining. Meanwhile, the A-GSTCN model is proposed to realize personalized medication recommendation based on the standardized records, and the model has excellent performance and can be used in specific medical environments.
- 2.
- In the A-GSTCN model, we construct global structural correlation diagrams for diagnoses and procedures, capturing the structural correlation of EHRs based on these diagrams and augmented GAT. In addition, we learn the temporal dependency of EHRs by dilated convolution combined with residual connection. Furthermore, we employ a cache mechanism to enhance the medication recommendation accuracy of the proposed model.
- 3.
- The proposed model outperforms the baselines in all evaluation metrics (Jaccard, F1, PRAUC) for the MIMIC-III datasets and ZJ-CVD datasets. Compared to the baselines, the A-GSTCN model has more accurate drug recommendation ability and requires far fewer parameters, which greatly reduces the training time and significantly improves the inference speed.
The subsequent contents are arranged as follows: Section 2 introduces some related work used in the paper, and Section 3 reviews the framework of the A-GSTCN. In Section 4, the A-GSTCN model and the baselines are compared for the MIMIC-III datasets and ZJ-CVD datasets from several angles, and meanwhile, the high efficiency of the proposed model is proved by experiments. Finally, the conclusion and future work are described in Section 5.
2. Related Work
Medication recommendation is a significant research direction in the field of medicine, and it can assist doctors to formulate safe and effective prescriptions quickly. Moreover, the existing medication recommendation approaches can be divided into two categories, i.e., model-driven approaches and data-driven approaches.
Early medication recommendation approaches are mainly based on the model-driven approach, which focuses on the rules and the causal relationship among diagnoses, procedures and medication combinations. These model-driven methods require experts in the field of medicine to model medical events in detail based on prior knowledge. Specifically, Chen et al. [22] developed the reasoning templates based on the knowledge patterns to encode the clinical guidelines for chronic heart failure (CHF) management. Ajmi et al. [23] proposed a backward rule-based expert system, which could be used for a headache diagnosis and medication recommendation system. In addition, a backward rule-based expert system [24] is presented, which can be used for a headache diagnosis and medication recommendation system. In addition, medication recommendation can be influenced by many factors, such as different areas of the hospital, different medical habits of doctors and different disease characteristics of patients [12]. Furthermore, medication recommendation rules that rely on experts’ prior knowledge produce a huge amount of work and affect the efficiency of the recommendations [14,15].
With the continuous accumulation of medical records, the data-driven approach has gradually become an important application for medication recommendation. Specifically, Choi et al. [20,21] employed a traditional recursive neural network (RNN) and an attention-based RNN to learn the multiple admission sequence of patients, thereby obtaining the temporal characteristic of EHRs. Pang et al. [25] added medical records to the pretraining module of BERT by using artificial time tokens. In fact, these approaches learn the temporal characteristics of EHRs and further improve the accuracy of medication recommendation. Nevertheless, early data-driven approaches ignore the structural correlation between medical events.
With the continuous deepening of research on medication recommendation, many comprehensive approaches to learn EHR characteristics have appeared. To be specific, Wang et al. [26] proposed an adversarially regularized model for medication recommendation, which could model the temporal information of EHRs and built a key value memory network based on information from historical admissions. Shang et al. [27] proposed a graph augmented memory network named GAMENet, which could integrate the drug–drug interactions and model longitudinal patient records as a query. Methods [28,29] could model the correlation between medical events and learn the structural correlation of EHRs by constructing medical ontology trees. Mao et al. [16] proposed an intelligent medical system that can accurately estimate the lab values and automatically recommend medication combinations based on patients’ incomplete lab tests. Furthermore, the COGNet model [30] introduces a novel copy-or-predict mechanism to generate the set of medicines. While these models have improved the accuracy of medication recommendation compared to previous models, they also have certain limitations, such as difficulty in applying to real environments, high complexity and so on.
For the above reasons, we propose a novel model named A-GSTCN, which can simultaneously model the structural and temporal characteristics of EHRs. Meanwhile, the proposed model can be also used for medication recommendation tasks in practical applications.
3. The A-GSTCN Model
The A-GSTCN model is described in three parts. Firstly, the structure of the proposed model and the goal of the medication recommendation tasks are described. Next, the A-GSTCN’ framework is presented. Last but not least, the optimizer and the training algorithm of the proposed model are introduced. For ease of description, the notations used in the A-GSTCN model are shown in Table 1.

Table 1.
Notations used in the A-GSTCN model.
3.1. Problem Formulation
An efficient medication recommendation model requires high precision of datasets. To improve the availability of the datasets, the EHRs need to be cleaned and standardized. To be specific, the definition of standardized EHRs, the medical event correlation diagram constructed in pretraining and the goal of the medication recommendation tasks are presented as follows.
3.1.1. Standardized EHRs
The pretrained EHRs can be represented as a collection of temporal records as follows: =, where represent the total number of patients and T represents the maximum number of one’s visits. To describe the algorithm more clearly, we omit the superscript n and introduce the proposed model only by unit patient. Each visit = of a patient contains diagnosis codes, , procedure codes, , and medication codes,
3.1.2. Medical Events Correlation Diagrams
To obtain the structural correlation between the medical events, we construct a diagnosis graph matrix and a procedure graph matrix for all the diagnosis events and procedure events, where and respectively represent the total number of diagnosis events and procedure events in the data set. Moreover, since and are built in the same way, we use to express them. Finally, the positive point-wise mutual information (PPMI) [31] is used to calculate the correlation between medical event i and medical event j of . The formula of is defined as follows:
where represents the probability of simultaneous occurrence of the event i and event j, and and represent the probability of event i and event j, respectively.
3.1.3. Medication Recommendation Tasks
Given a patient’s historical visits =, diagnosis events and procedure events at the tth visit, the goal of medication recommendation tasks is to generate a personalized medication combination = at the tth visit based on the patient’s current clinical events , and historical visits , where represents the total number of the medications.
3.2. The Framework of A-GSTCN
The A-GSTCN model includes four components: medical entity embedding module, structural correlation enhancement module, temporal dependency progressive module and cache memory enhancement module. Next, the modules presented in Figure 3 and the algorithm processes of the A-GSTCN model will be described as follows.
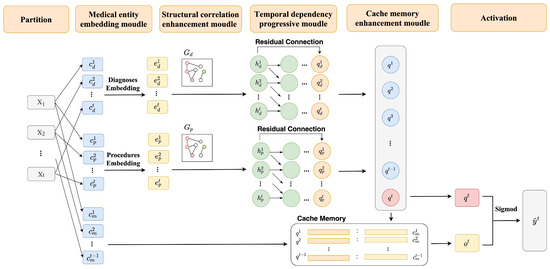
Figure 3.
The training process of A-GSTCN model. Each visit = of a patient contains diagnosis codes, , procedure codes, , and medication codes, . Among them, are used in the medical entity embedding module to output the hidden embedding with Equation (1). Then, structural correlation enhancement module generates by accepting and described in Equations (1) and (3)–(6). Next, are input into the temporal dependency progressive module to output using the dilated convolution combined with residual connection by Equations (7) and (8). After that, the output is generated by integrating the key-value pairs stored in cache memory using Equations (9)–(11). In the end, query and output are activated by Equation (12) for medication recommendation.
3.2.1. Medical Entity Embedding Module
The patient’s tth visit consists of , where both are multi-hot vectors, so is used to indicate the unified definition. The medical embeddings for are derived separately, and the embedding matrixes and are obtained by embedding entities, where and represent the total number of diagnosis events and procedure events at the tth visit, and l represents the characteristic dimensions. Specifically, the embedding formula of ( is used for and ) is shown as follows:
Here, presents the embedding matrix, and is the total number of medical events. Through the medical entity embedded module, the input = is transformed into =.
3.2.2. Structural Correlation Enhancement Module
The function of the structural correlation enhancement module is to make the embedding matrix contain information about other related medical events and obtain a more comprehensive matrix representation. For this reason, we propose an enhanced multi-head graph attention network. Specifically, the medical events correlation diagram constructed in pretraining is used as the global weight matrix. For the value =, graph transformation is performed for each of its sub-events and the hidden layer = is obtained with more structural information. The specific calculation formula [32] of can be written as follows:
where ‖ is the concatenation operation; represents the sub-event graph transformation; K is interpreted as the number of multiple attention; represents a nonlinear function; can be interpreted as the collection of other sub-events related to the event i; and represent the weight matrix and bias, respectively; represents the weight coefficient of attention at the tth visit. To be specific, the calculation formula of [33] is illustrated as follows:
where is the feedforward neural network training vector; represents the weight matrix; can be interpreted as the corresponding eigenvector for events ∗. Inspired by previous research [34], instead of complex pretraining, the medical events correlation diagram is applied to calculate the weight of medical events in each visit. Therefore, there is no need to train the specific training parameters, such as and , and the calculation of can be simplified as:
Here, and are the correlation between event i and event j, event i and event k in the graph matrix , respectively. The graph matrix is derived from the medical events correlation diagram as follows:
3.2.3. Temporal Dependency Progressive Module
GRU and LSTM are firstly considered to capture the temporal dynamic changes of EHRs, but these models have high memory usage. Thanks to the prior research [35], it is more appropriate to use the method of dilated convolution combined with residual connection to learn the temporal characteristics of EHRs. Specifically, simple convolutional networks can only deal with sequential tasks with relatively small sequence length and perform poorly in long sequential tasks, so they cannot be applied to EHRs with an uncertain number of visits. Therefore, the method of combining dilated convolution with residual connection is considered, and we propose a new approach to capture medical events’ temporal dependency for medication recommendation inspired by references [36,37]. As shown in Figure 4, the dilated convolution contains two more significant parameters: filter and factor. The size of filter is set to 7 and the factor is set to 1. As the hidden layer deepens, the receptive field can cover all values from the length of patients’ visits, and the output results are obtained through the residual connection layer. Specifically, and are trained separately, and the specific inputs of the network are : and :, which could be expressed by . After the dilated convolution and residual connection, the output : contained temporal characteristics can be obtained as follows:
where is a residual mapping and represents the set of parameter matrix. represents the hidden layer results obtained through dilated convolution, and it can be expressed as :. The in can be derived as follows:
where is the dilation factor and k represents the filter size; t − ·i accounts for the direction of the past; represents the filter function in the dilated convolution process.
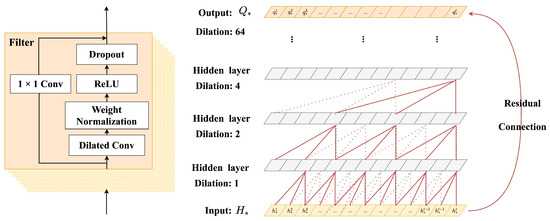
Figure 4.
The structure of the temporal dependency progressive module. Both residual and parameterized skip connections are used throughout this module.
In the temporal dependency progressive module, diagnosis representations : and procedure representations : are obtained, and they capture rich temporal features by the method of combining dilated convolution with the residual connection. Therefore, = is transformed into =.
3.2.4. Cache Memory Enhancement Module
The cache memory enhancement module pre-stores the historical records of patients in a dynamic bank with key-value pairs, and it can optimize the current recommendation by comparing the similarity between the current recommendation and the historical records. In addition, the conclusions can be drawn from the research [38] that an effective cache memory enhancement module can improve the model’s learning rate of historical conditions, so the cache memory enhancement module is applied and further divided into four steps:
- 1.
- Create a query vector of the tth visit. To be specific, from the set can be generated a query as follows:where represents a transformation function, and this function can connect the diagnosis representation and the procedure representation .
- 2.
- Use the and medication representation as dependent variables, and generate the cache records before the tth visit in the form of key-value pairs as follows:where is empty when t = 1, and represents the historical visit before the tth visit. : is denoted as the key vector, and : is denoted as the value vector to represent the history cache of the tth visit.
- 3.
- Based on the similarity between the representation vector and its historical cache, the attention strategy is applied as follows:where the similarity between the key vector matrix and the representation vector is first considered. Furthermore, the similarity relationship is obtained by matrix multiplication and activation, and the transposed vector matrix is further multiplied to obtain .
- 4.
- Activate and , obtain the multi-label recommended medication combination . The formula can be expressed as follows:where is the activation function.
3.3. Optimization
The quality of the medication recommendation model can be explained by the gap between the drug recommendation combination generated by the model and the real drug recommendation combination . Meanwhile, whether a single drug is recommended can be likened to binary classification, so the task of drug combination recommendation can be further classified into multiple classification problems. In this case, the multi-label margin loss and the binary cross-entropy loss are applied as optimizations, which are combined as model’ optimizer as follows:
Here, is the mixture weights; and represent the medication i in the tth visit; is the jth label indexed by predicted label set .
In summary, Algorithm 1 describes the training algorithm of the A-GSTCN.
| Algorithm 1: Training algorithm of the A-GSTCN |
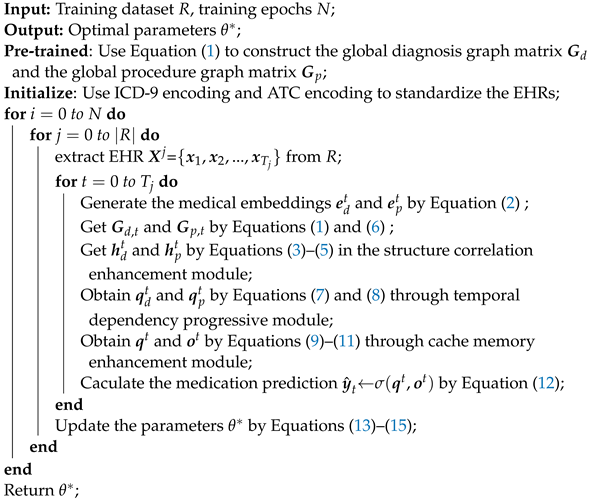 |
4. Experiments
The experiments are divided into three parts. Firstly, preparations of the experimental environment are presented, such as the datasets and the baselines. Secondly, the performance of the A-GSTCN model and baselines is compared in four experiments. Next, a case study is applied for proving the feasibility of the A-GSTCN model in specific medical environments. Finally, through the engineering applications, the A-GSTCN is well applied in the medication recommendation process of a digital hospital.
4.1. Experimental Setup
4.1.1. Datasets
The proposed model and the baselines are performed on MIMIC-III and ZJ-CVD datasets, and the relevances of the two datasets are presented as follows:
- MIMIC-III is a sizable single-center database, which includes more than 50,000 cases admitted to intensive care units from 2001 to 2012 and 7870 newborns admitted from 2001 to 2008. To be specific, the MIMIC-III dataset includes medical orders, medications, procedures, diagnoses, and so on. Meanwhile, to improve the dataset availability, the records are generated into a temporal list of diagnosis, procedure and medication codes.
- ZJ-CVD is a Chinese medical dataset collected by our laboratory, which contains the medical records of more than 8000 patients with cerebrovascular disease from the First Hospital of Zhejiang Province, the Fourth Affiliated Hospital Zhejiang University of Medicine and Taizhou Municipal Hospital. Each patient may have multiple hospitalizations, so the number of EHRs in ZJ-CVD datasets exceeds 10,000. To be specific, ZJ-CVD datasets are cleaned and augmented in pretraining and consist of admission diagnosis, hospitalization, discharge medication and some other medical information.
Furthermore, the medical events of the datasets are converted into vector representations according to the ATC and the ICD-9 medical standards. The characteristics of MIMIC-III datasets and ZJ-CVD datasets can be seen in Table 2.

Table 2.
The characteristics of MIMIC-III and ZJ-CVD datasets.
4.1.2. Baselines
The baselines are introduced as follows:
- Leap [39] can predict target event through an attention mechanism by establishing mappings between medical events and tensors.
- RETAIN [21] generates a medication recommendation through building a two-layer RNN with attention model, and this model can consider the influence of temporal factors.
- DMNC [38] strengthens the capturing of temporal characteristics for medical events by establishing a memory enhancement networks.
- GAMENet [27] integrates the drug–drug interactions and model longitudinal patient records as the query, which can capture the temporal dependency of EHRs.
- G-Bert [28] uses the BERT to pretrain the correlations between medical events in EHRs and constructs an ontological tree for medication recommendation.
4.1.3. Metrics
Jaccard Similarity Score (Jaccard), Precision–Recall AUC (PRAUC) and Average F1 (F1) are used as the scoring functions in the experiments. Next, the scoring functions are explained separately.
The caculation formula of Jaccard can be described as follows:
where N is the total number of patients, and represents the max visits of the kth patient.
PRAUC is calculated by the trapezoidal integral for the area under the PR curve, and this scoring function is used for the datasets with imbalanced positive and negative sample numbers.
The F1 score can transform the multi classification problem into n bipartitions. Meanwhile, it calculates the average score of the bipartition to obtain the final evaluation index, whose caculation formula can be written below:
where t represents tth visit, and k can be interpreted as the kth patient in the test set.
4.2. Experimental Results
The effectiveness of the A-GSTCN model is demonstrated by four comparative experiments. Specifically, the A-GSTCN model is compared with the baselines on Jaccard, F1 and PRAUC in the first experiment. In the second part, the validity of each module of A-GSTCN is verified. Next, the third part compares the drug recommendation performance of the model on different recommended frequency drugs. Finally, the last experiment compares the drug recommendation performance of the model for patients with different visits.
4.2.1. Recommendation Performance
Table 3 indicates the comparisons of Jaccard, PRAUC and F1 between the proposed model and the baselines on MIMIC-III and ZJ-CVD datasets. Among them, it is obviously observed that the A-GSTCN model obtains the best recommendation performance under all evaluation metrics, which can prove the effectiveness of the A-GSTCN in medication recommendation. To be specific, compared with the previous best method (G-Bert), the A-GSTCN model improves 1.78%, 1.24% and 1.86% in Jaccard, PRAUC and F1 score, respectively, for the MIMIC-III dataset. In a similar way, the A-GSTCN model increases 2.76%, 8.37% and 2.67% in Jaccard, PRAUC and F1 score, respectively, for the ZJ-CVD dataset. Moreover, the average recommended number of medications for A-GSTCN for the MIMIC-III datasets and ZJ-CVD datasets are 15.34 and 13.22, which have the smallest gap with the real value of 14.61 and 12.89. Futhermore, compared with the baseline methods, the most significant feature of the A-GSTCN model is the correlation diagrams for pretrained medical events and the dilated convolution applied in the temporal dependency progressive module. These features lead to fewer parameters in the A-GSTCN model, which effectively decreases the memory occupancy rate and cache training pressure.

Table 3.
Medication recommendation performance between the A-GSTCN model and baselines on MIMIC-III and ZJ-CVD datasets. In addition, the gold average number of medicines on the test set is 14.61 and 12.89 for the MIMIC-III datasets and ZJ-CVD datasets, respectively.
4.2.2. Module Validity
To further prove the effectiveness of the structure correlation enhancement module, the temporal dependency progressive module and the cache memory enhancement module, the A-GSTCN model is compared with its variants.
Variant types of the A-GSTCN model in Figure 5a,b are shown below:
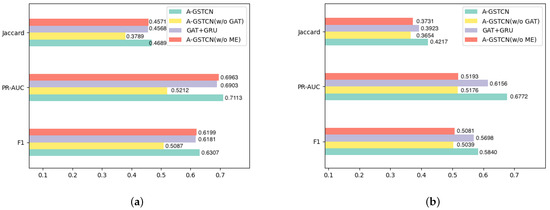
Figure 5.
(a,b) are the performance comparisons (Jaccard, PRAUC and F1 score) between different variants of proposed methods on MIMIC-III and ZJ-CVD datasets.
- A-GSTCN: the proposed model.
- A-GSTCN (w/o GAT): removes the structure correlation enhancement module of the A-GSTCN model.
- GAT + GRU: changes the temporal dependency progressive module into the GRU model for the A-GSTCN model.
- A-GSTCN (w/o ME): removes the cache memory enhancement module of the A-GSTCN model.
By comparing the performance of the A-GSTCN and the A-GSTCN (w/o GAT) in Figure 5a,b, it indicates that the performance of each metric has a significant decrease when the structural correlation enhancement module is removed. Specifically, Jaccard and F1 score decrease by nearly 8% and 6%, and PRAUC decreases by nearly 16% for the ZJ-CVD datasets. The reductions in Jaccard, F1 score and PRAUC for the MIMIC-III datasets are more prominent. Therefore, it can be concluded that the structural correlation enhancement module behaves excellently in structural modeling and can adequately capture the structural characteristics of medical entities of EHRs.
Through the comparative experiments of the A-GSTCN and the GAT + GRU in Figure 5a,b, it apparently shows that Jaccard, F1 Score and PRAUC for the GAT + GRU decrease by nearly 2% compared with the A-GSTCN for the MIMIC-III datasets, and these metrics decline by nearly 2%, 2%, 6.16% for the GAT + GRU compared with the A-GSTCN model for the ZJ-CVD datasets. Therefore, the conclusions can be drawn from the significant reduction in metrics: use dilated convolution instead of GRU can reduce the amount of parameters used while maintaining model performance in A-GSTCN.
Compared with the proposed model, the Jaccard, PRAUC and F1 score for the A-GSTCN (w/o ME) decline by 1.17%, 1.5% and 1.08%, respectively, for the MIMIC-III datasets. These metrics decline by nearly 4.86%, 15.79% and 7.59% for the ZJ-CVD datasets also. Meanwhile, it is obviously observed that the performance gap between the A-GSTCN and the A-GSTCN (w/o ME) for the ZJ-CVD datasets is larger than that for the MIMIC-III datasets because of the relatively short number of patient visits in the ZJ-CVD datasets. In summary, the cache memory enhancement module can cooperate with the temporal dependency progressive module to fully preserve the temporal features of EHRs, thus improving the accuracy of medication recommendation.
4.2.3. Comparison for Different Recommended Frequency Drugs
Some drugs have a high recommended frequency, and others may be used less often. The A-GSTCN model can decrease the impact of data imbalance by applying the global structural correlation diagrams for diagnoses and procedures and adding a caching mechanism. Specifically, Figure 6a,b count the number of medications in different recommended frequencies in the MIMIC-III and ZJ-CVD datasets, and it can be seen that 58 of the 145 medication types appear less than 100 times, while nearly 40 types are recommended more than 1000 times in the MIMIC-III datasets. In the ZJ-CVD datasets, 133 of the 453 medication types are recommended less than 100 times, while nearly 40 types occur more than 1000 times. Figure 6c,d calculate the average F1 score of medication recommendation results in different recommended frequencies, and it indicate that the A-GSTCN model significantly improves the recommended accuracy of less frequent medications based on its global structural correlation diagrams and caching mechanism.
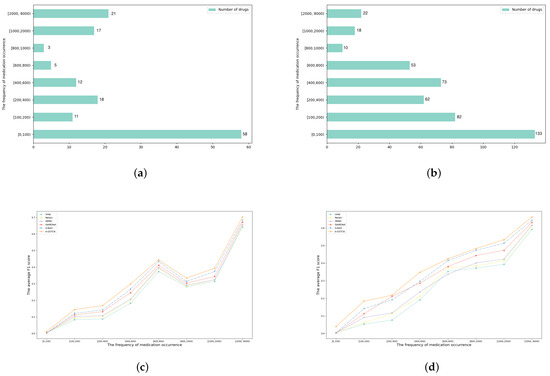
Figure 6.
(a,b) are the total number of medications in different frequency ranges in MIMIC-III and ZJ-CVD datasets; (c,d) are the comparisons of average F1 score between the A-GSTCN model and baselines in different frequency ranges in MIMIC-III and ZJ-CVD datasets.
4.2.4. Comparison for Patients with Different Visits
As shown in Table 2, the max visits of patients in the MIMIC-III and ZJ-CVD datasets are 29 and 4, respectively. Logically speaking, different numbers of admissions of patients also affect the accuracy of medication recommendation. To be specific, Figure 7a,b indicate the comparisons of average F1 score between the A-GSTCN model and baselines with different temporal lengths of EHRs in the MIMIC-III and ZJ-CVD datasets, and it can be found that the A-GSTCN model is superior to the baselines over most of the temporal horizon, especially for long sequences. Meanwhile, it can be apparently observed that the A-GSTCN model also has a significant learning ability in short visit sequences and recommends more precise medication combination for patients than the baseline models. These results prove that the A-GSTCN model has efficient modeling ability for long temporal dependency.
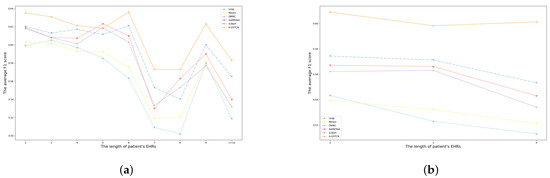
Figure 7.
(a,b) are the comparisons of average F1 score between the A-GSTCN model and baselines with different temporal length of EHRs in MIMIC-III and ZJ-CVD datasets.
4.3. Case Study
To clearly clarify the effectiveness of the A-GSTCN model in the task of drug recommendation, we further compare the drug recommendation results of the model through two specific cases.
The first special case is tested for the MIMIC-III dataset. This case selects a patient’s EHRs of four temporal admissions in the test set, and the patient has various symptoms, such as gout, depression and heart disease. As can be seen in Table 4, the correct recommended combination of drugs for the patient is 15 drugs, and the A-GSTCN model performed best in this case, recommending the right 14 drugs. In contrast, the model with the best recommendations in the baselines is G-Bert, which recommends 13 drugs correctly and misses 2. Other models in baselines are less effective. Moreover, it can be seen that none of the models successfully hit the drug “Anxiolytics”, and this is where subsequent models need to improve.

Table 4.
A specific case selects a patient’s EHRs of four temporal admissions from the MIMIC-III datasets; “unseen” indicates the drugs that do not appear in the actual recommendation results, and “missed” refers to the drugs that should be recommended in the actual situation but are not recommended.
Similar to Table 4, Table 5 represents a recommended result of a patient who accesses a total of three visits from the ZJ-CVD datasets, and this patient suffered from stroke, diabetes and high blood pressure. In addition, compared with the MIMIC-III datasets, this typical case evidently reflects the recommendation ability of the A-GSTCN model in medication recommendation. Specifically, it obviously shows that the actual number of recommended drugs in the patient’s last visit is eight. Meanwhile, the DMNC model, GAMENet model and G-Bert model perform best among all baselines, but they only recommend five drugs correctly. In contrast, the A-GSTCN model correctly recommends seven drugs and misses only one drug. Furthermore, the missed drug “Rabeprazole Sodium Enteric-coated Capsules” from the A-GSTCN model is also lost in all baseline models due to the low utilization rate of this drug.

Table 5.
A specific case of a patient who accesses a total of three visits from ZJ-CVD datasets, and this patient suffered from stroke, diabetes and high blood pressure. Missing drugs include Rabeprazole Sodium Enteric-coated Capsules (RSEC), Betahistine mesilate Tablets (BMT), Trimetazidine Hydrochloride Tablets (THT), Perindopril And Indapamide Tablets (PAIT) and Aspirin Enteric-Coated Sustained Release Tablets (AESRT). For convenience, corresponding abbreviations are used below.
Compared with other baseline models, the A-GSTCN model achieves the best medication recommendation effect in both cases, which fully proves that A-GSTCN model can better learn the structural correlation and temporal dependency of EHRs.
4.4. Engineering Applications
Medical service informatization is the development trend of Internet medical treatment in the digital age. With the rapid development of information technology, more and more hospitals are accelerating the overall construction of hospital information systems (HISs) to improve the service level and core competitiveness of hospitals. As a new application of the Internet in the medical industry, the digital hospital is an important form of medical service informatization [40]. Since the requirements to ensure the universality and accuracy of medical services, most of the current research focuses on applying deep learning models to learn the structural–temporal characteristics of medical data and then apply these models to medical services, such as medication recommendation, diagnostic prediction, treatment guidance, etc. [41]. Among them, medication recommendation is one of the key issues in the research on the digital hospital. Figure 8 presents the link of medication recommendation in Internet medical treatment.
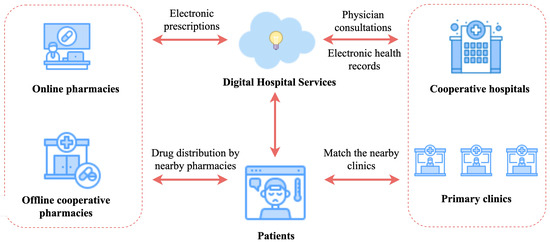
Figure 8.
Medication recommendation process in Internet medical treatment.
However, the structural–temporal characteristics of medical records have a great influence on the accuracy of medication recommendation, which directly affects the applicability of the final recommended prescriptions. In this regard, the priority is to produce more accurate deep learning models that can intelligently generate recommended medications. Therefore, as shown in Figure 9, the data-driven approach can be used to collect medical data from patients in cooperative hospitals and clinics for integration into the A-GSTCN model. To be specific, firstly, real medical records are imported into the A-GSTCN model. Then, the structural correlation enhancement module and the temporal dependency progressive module are employed to learn the structural–temporal characteristics of the data, respectively, so as to optimize the recommendation performance of the model and recommend more accurate prescriptions.
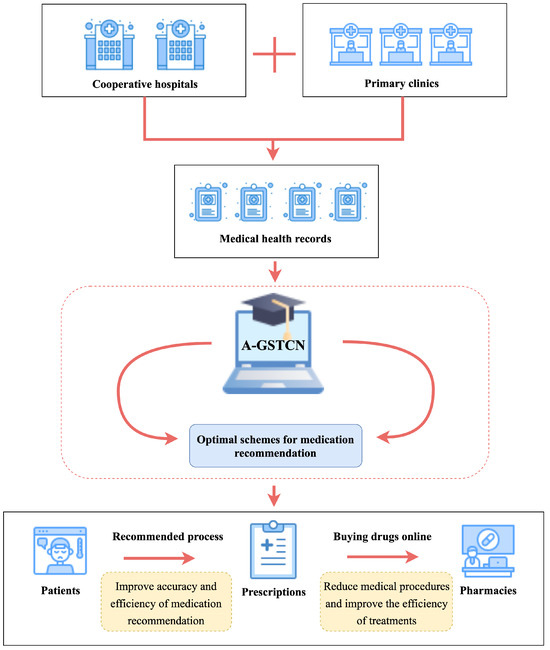
Figure 9.
An application diagram of the A-GSTCN in medication recommendation.
5. Conclusions and Future Work
In this article, we propose a novel medication recommendation model that can effectively learn the structural correlation and temporal dependency of EHRs. To be specific, we establish the global correlation diagrams for medical events and apply an augmented GAT to capture the structural correlation. Next, dilated convolution combined with residual connection are used to capture temporal features on the premise of greatly reducing training parameters. Meanwhile, the caching mechanism is introduced to improve the medication recommendation accuracy. Finally, through comparative experiments, case studies and engineering applications, it proves that the proposed model has higher medication recommendation accuracy and better landing possibility compared to the previous models.
In light of the current situation, the EHRs introduce a significant amount of uncertainty into medication recommendations due to the lack of information, imprecise information and contradictory nature. Therefore, it is essential to explore the characteristics of other important influencing factors in EHRs, such as inspection indicators and operation status. Meanwhile, as we continuously collect and integrate the EHRs, it is important to consider the introduction of pretrained models like BERT, GPT and other large language models to enhance the performance of the recommendation model. Furthermore, the application of EHRs needs to be expanded; in addition to medication recommendation, it also can be further applied to disease prediction, disease prevention and other issues. Finally, in the process of medication recommendation, it is significant to consider the safety of medication recommendation, and we need to further consider adding drug–drug interactions (DDIs) to ensure the safety of recommended drugs.
Author Contributions
Conceptualization, L.Z. (Lei Zhang) and J.W.; methodology, W.Y. and L.Z. (Lijuan Zhang); software, W.Y.; validation, W.Y., M.W. and L.Z. (Lijuan Zhang); formal analysis, W.Y. and L.Z. (Lijuan Zhang); investigation, J.H.; resources, L.Z. (Lei Zhang); data curation, W.Y.; writing—original draft preparation, W.Y.; writing—review and editing, N.X. and A.V.V.; project administration, J.W.; funding acquisition, L.Z. (Lei Zhang) and L.Z. (Lijuan Zhang). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Key Research and Development Program of Zhejiang Province (Grant No. 2020C03071), the National Natural Science Youth Science Foundation Project (Grant No. 62201508) and Zhejiang Provincial Natural Science Foundation Youth Fund Project (Grant No. LQ23F010004).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- de Moraes, B.A.F.; Miraglia, J.; Donato, T.; Filho, A. COVID-19 Diagnosis Prediction in Emergency Care Patients: A Machine Learning Approach. 2020. Available online: https://www.medrxiv.org/content/medrxiv/early/2020/04/07/2020.04.04.20052092.full.pdf (accessed on 1 August 2023).
- Wagner, T.; Shweta, F.; Murugadoss, K.; Awasthi, S.; Venkatakrishnan, A.; Bade, S.; Puranik, A.; Kang, M.; Pickering, B.W.; O’Horo, J.C.; et al. Augmented curation of clinical notes from a massive EHR system reveals symptoms of impending COVID-19 diagnosis. eLife 2020, 9, e58227. [Google Scholar] [CrossRef]
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.; Dahly, D.L.; Damen, J.A.; Debray, T.P.; et al. Prediction models for diagnosis and prognosis of COVID-19: Systematic review and critical appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef]
- Arndt, B.G.; Beasley, J.W.; Watkinson, M.D.; Temte, J.L.; Tuan, W.J.; Sinsky, C.A.; Gilchrist, V.J. Tethered to the EHR: Primary care physician workload assessment using EHR event log data and time-motion observations. Ann. Fam. Med. 2017, 15, 419–426. [Google Scholar] [CrossRef]
- Boussadi, A.; Caruba, T.; Karras, A.; Berdot, S.; Degoulet, P.; Durieux, P.; Sabatier, B. Validity of a clinical decision rule-based alert system for drug dose adjustment in patients with renal failure intended to improve pharmacists’ analysis of medication orders in hospitals. Int. J. Med. Inform. 2013, 82, 964–972. [Google Scholar] [CrossRef]
- Kropf, M.; Modre-Osprian, R.; Gruber, K.; Fruhwald, F.; Schreier, G. Evaluation of a clinical decision support rule-set for medication adjustments in mHealth-based heart failure management. In eHealth; IOS Press: Amsterdam, The Netherlands, 2015; pp. 81–87. [Google Scholar]
- Mahmoud, N.; Elbeh, H. IRS-T2D: Individualize recommendation system for type2 diabetes medication based on ontology and SWRL. In Proceedings of the 10th International Conference on Informatics and Systems; Giza, Egypt, 9–11 May 2016, pp. 203–209.
- Farzi, S.; Farzi, S.; Alimohammadi, N.; Moladoost, A. Medication errors by the intensive care units’ nurses and the Preventive Strategies. Anesthesiol. Pain 2016, 6, 33–45. [Google Scholar]
- Elhoseny, M.; Shankar, K.; Uthayakumar, J. Intelligent diagnostic prediction and classification system for chronic kidney disease. Sci. Rep. 2019, 9, 9583. [Google Scholar] [CrossRef]
- Shickel, B.; Tighe, P.J.; Bihorac, A.; Rashidi, P. Deep EHR: A survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J. Biomed. Health Inform. 2017, 22, 1589–1604. [Google Scholar] [CrossRef] [PubMed]
- Erraguntla, M.; Zapletal, J.; Lawley, M. Framework for Infectious Disease Analysis: A comprehensive and integrative multi-modeling approach to disease prediction and management. Health Inform. J. 2019, 25, 1170–1187. [Google Scholar] [CrossRef]
- John, A.; Vasudevan, V. Medication recommendation system based on clinical documents. In Proceedings of the 2016 International Conference on Information Science (ICIS), Kochi, India, 12–13 August 2016; pp. 180–184. [Google Scholar]
- Zhang, Y.; Zhang, D.; Hassan, M.M.; Alamri, A.; Peng, L. CADRE: Cloud-assisted drug recommendation service for online pharmacies. Mob. Netw. Appl. 2015, 20, 348–355. [Google Scholar] [CrossRef]
- Syed-Abdul, S.; Nguyen, A.; Huang, F.; Jian, W.S.; Iqbal, U.; Yang, V.; Hsu, M.H.; Li, Y.C. A smart medication recommendation model for the electronic prescription. Comput. Methods Programs Biomed. 2014, 117, 218–224. [Google Scholar] [CrossRef]
- Liu, H.; Xie, G.; Mei, J.; Shen, W.; Sun, W.; Li, X. An efficacy driven approach for medication recommendation in type 2 diabetes treatment using data mining techniques. Stud. Health Technol. Inform. 2013, 192, 1071. [Google Scholar] [PubMed]
- Mao, C.; Yao, L.; Luo, Y. MedGCN: Medication recommendation and lab test imputation via graph convolutional networks. J. Biomed. Inform. 2022, 127, 104000. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.; Bahadori, M.T.; Song, L.; Stewart, W.F.; Sun, J. GRAM: Graph-based attention model for healthcare representation learning. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Halifax, NS, Canada, 13–17 August 2017; pp. 787–795. [Google Scholar]
- Gao, C.; Sun, H.; Wang, T.; Tang, M.; Bohnen, N.I.; Müller, M.L.; Herman, T.; Giladi, N.; Kalinin, A.; Spino, C.; et al. Model-based and model-free machine learning techniques for diagnostic prediction and classification of clinical outcomes in Parkinson’s disease. Sci. Rep. 2018, 8, 7129. [Google Scholar] [CrossRef] [PubMed]
- Tutty, M.A.; Carlasare, L.E.; Lloyd, S.; Sinsky, C.A. The complex case of EHRs: Examining the factors impacting the EHR user experience. J. Am. Med. Inform. Assoc. 2019, 26, 673–677. [Google Scholar] [CrossRef]
- Choi, E.; Bahadori, M.T.; Schuetz, A.; Stewart, W.F.; Sun, J. Doctor ai: Predicting clinical events via recurrent neural networks. In Proceedings of the Machine Learning for Healthcare Conference, PMLR, Los Angeles, CA, USA, 19–20 August 2016; pp. 301–318. [Google Scholar]
- Choi, E.; Bahadori, M.T.; Sun, J.; Kulas, J.; Schuetz, A.; Stewart, W. Retain: An interpretable predictive model for healthcare using reverse time attention mechanism. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; Volume 29. [Google Scholar]
- Chen, Z.; Marple, K.; Salazar, E.; Gupta, G.; Tamil, L. A physician advisory system for chronic heart failure management based on knowledge patterns. Theory Pract. Log. Program. 2016, 16, 604–618. [Google Scholar] [CrossRef]
- Al-Ajmi, N.; Almulla, M.A. Rule-Based Expert System for Headache Diagnosis and Medication Recommendation. Int. J. Health Med. Eng. 2020, 14, 388–391. [Google Scholar]
- Almulla, M.A. Location-based Expert System for Diabetes Diagnosis. Kuwait J. Sci. 2021, 48. [Google Scholar] [CrossRef]
- Pang, C.; Jiang, X.; Kalluri, K.S.; Spotnitz, M.; Chen, R.; Perotte, A.; Natarajan, K. CEHR-BERT: Incorporating temporal information from structured EHR data to improve prediction tasks. In Proceedings of the Machine Learning for Health. PMLR, Virtual, 6–7 August 2021; pp. 239–260. [Google Scholar]
- Wang, Y.; Chen, W.; Pi, D.; Yue, L. Adversarially regularized medication recommendation model with multi-hop memory network. Knowl. Inf. Syst. 2021, 63, 125–142. [Google Scholar] [CrossRef]
- Shang, J.; Xiao, C.; Ma, T.; Li, H.; Sun, J. Gamenet: Graph augmented memory networks for recommending medication combination. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 1126–1133. [Google Scholar]
- Shang, J.; Ma, T.; Xiao, C.; Sun, J. Pre-training of graph augmented transformers for medication recommendation. arXiv 2019, arXiv:1906.00346. [Google Scholar]
- Choi, E.; Xu, Z.; Li, Y.; Dusenberry, M.; Flores, G.; Xue, E.; Dai, A. Learning the graphical structure of electronic health records with graph convolutional transformer. In Proceedings of the AAAI conference on artificial intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 606–613. [Google Scholar]
- Wu, R.; Qiu, Z.; Jiang, J.; Qi, G.; Wu, X. Conditional generation net for medication recommendation. In Proceedings of the ACM Web Conference, Lyon, France, 25–29 April 2022; pp. 935–945. [Google Scholar]
- Khan, F.H.; Qamar, U.; Bashir, S. SentiMI: Introducing point-wise mutual information with SentiWordNet to improve sentiment polarity detection. Appl. Soft Comput. 2016, 39, 140–153. [Google Scholar] [CrossRef]
- Li, J.; Tu, Z.; Yang, B.; Lyu, M.R.; Zhang, T. Multi-head attention with disagreement regularization. arXiv 2018, arXiv:1810.10183. [Google Scholar]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph attention networks. arXiv 2017, arXiv:1710.10903. [Google Scholar]
- Su, C.; Gao, S.; Li, S. GATE: Graph-attention augmented temporal neural network for medication recommendation. IEEE Access 2020, 8, 125447–125458. [Google Scholar] [CrossRef]
- Hewage, P.; Behera, A.; Trovati, M.; Pereira, E.; Ghahremani, M.; Palmieri, F.; Liu, Y. Temporal convolutional neural (TCN) network for an effective weather forecasting using time-series data from the local weather station. Soft Comput. 2020, 24, 16453–16482. [Google Scholar] [CrossRef]
- Oord, A.v.d.; Dieleman, S.; Zen, H.; Simonyan, K.; Vinyals, O.; Graves, A.; Kalchbrenner, N.; Senior, A.; Kavukcuoglu, K. Wavenet: A generative model for raw audio. arXiv 2016, arXiv:1609.03499. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Le, H.; Tran, T.; Venkatesh, S. Dual memory neural computer for asynchronous two-view sequential learning. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, London, UK, 19–23 August 2018; pp. 1637–1645. [Google Scholar]
- Esteban, C.; Tresp, V.; Yang, Y.; Baier, S.; Krompaß, D. Predicting the co-evolution of event and knowledge graphs. In Proceedings of the 2016 19th International Conference on Information Fusion (FUSION), Sun City, South Africa, 1–4 November 2016; pp. 98–105. [Google Scholar]
- Kwon, H.; An, S.; Lee, H.Y.; Cha, W.C.; Kim, S.; Cho, M.; Kong, H.J. Review of smart hospital services in real healthcare environments. Healthc. Inform. Res. 2022, 28, 3–15. [Google Scholar] [CrossRef]
- Duncan, R.; Eden, R.; Woods, L.; Wong, I.; Sullivan, C. Synthesizing dimensions of digital maturity in hospitals: Systematic review. J. Med. Internet Res. 2022, 24, e32994. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).