Efficient Cytotoxicity of Recombinant Azurin in Escherichia coli Nissle 1917-Derived Minicells against Colon Cancer Cells
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Methods
2.2.1. Cell Cultures and Plasmid Constructs
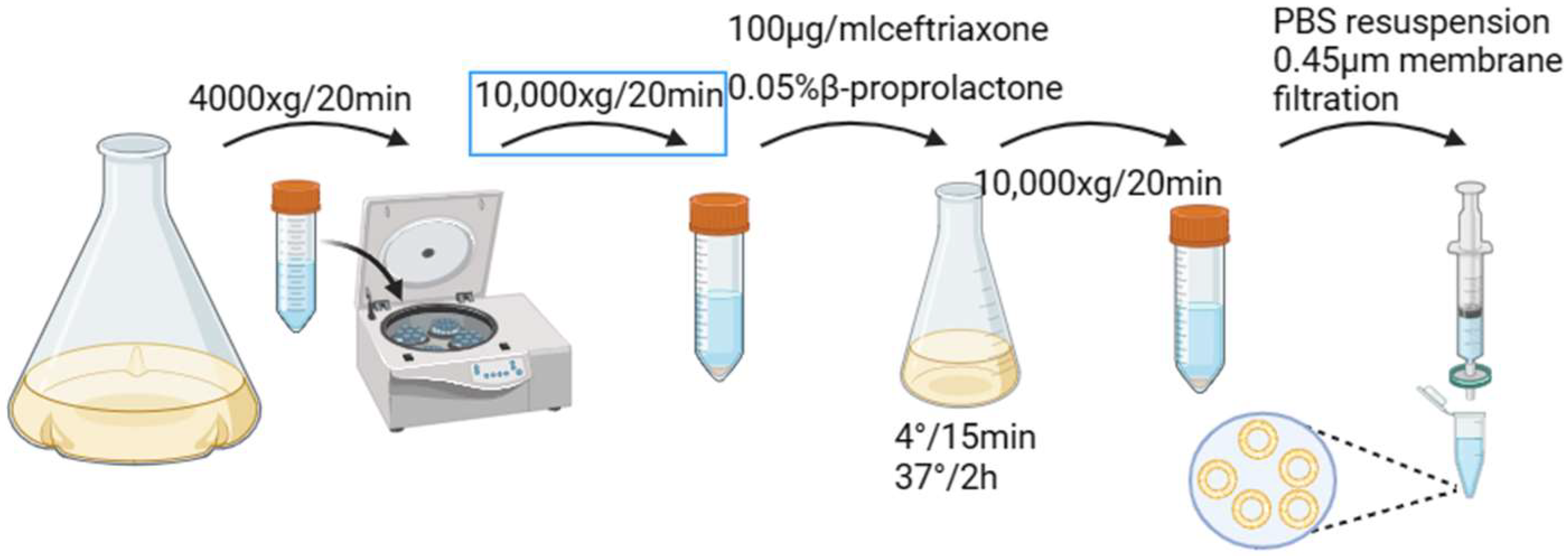
2.2.2. Preparation and Characterization of Minicells
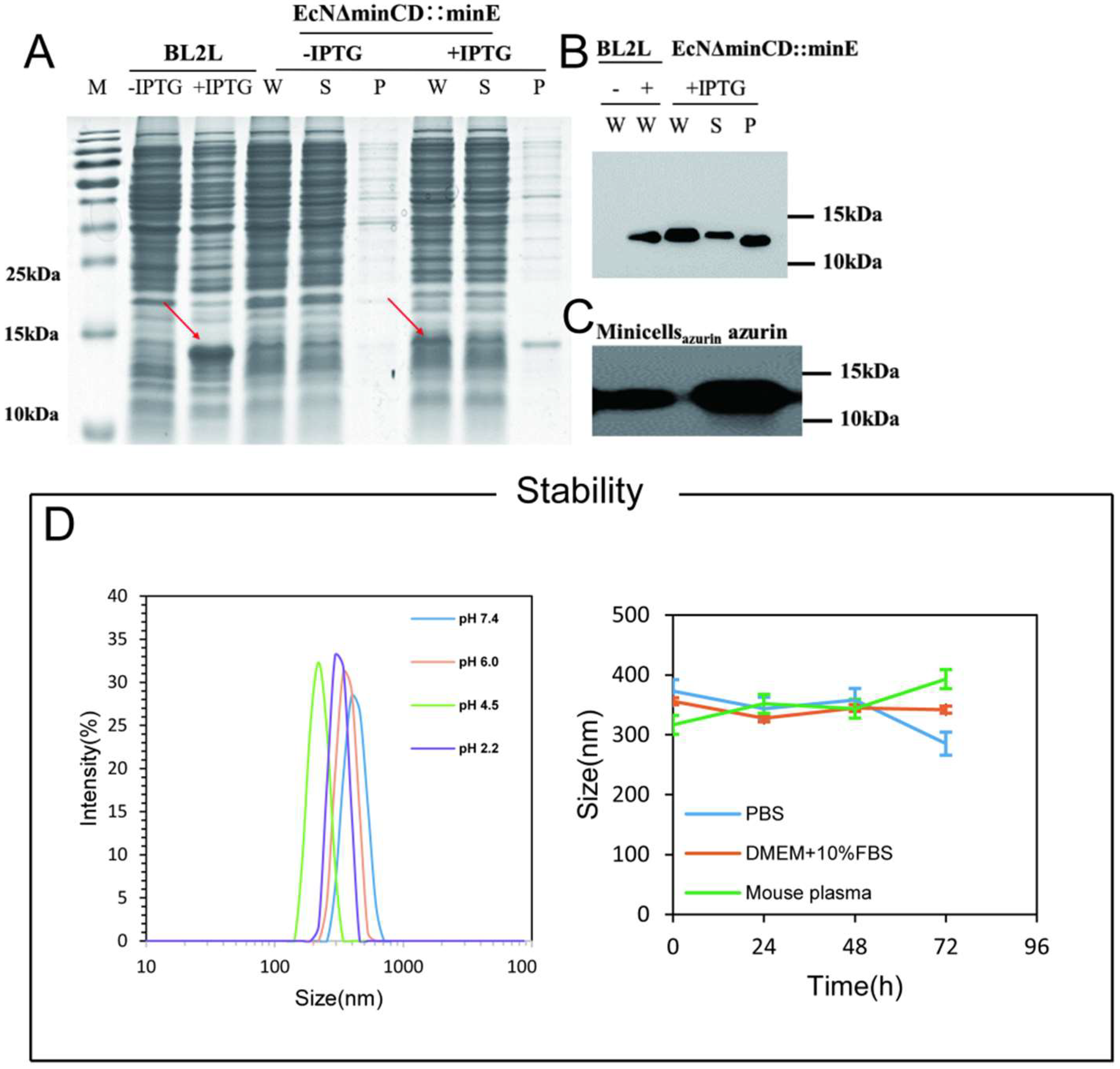
2.2.3. SDS-PAGE and Western Blot
2.2.4. Extracellular Stability of Minicellsazurin
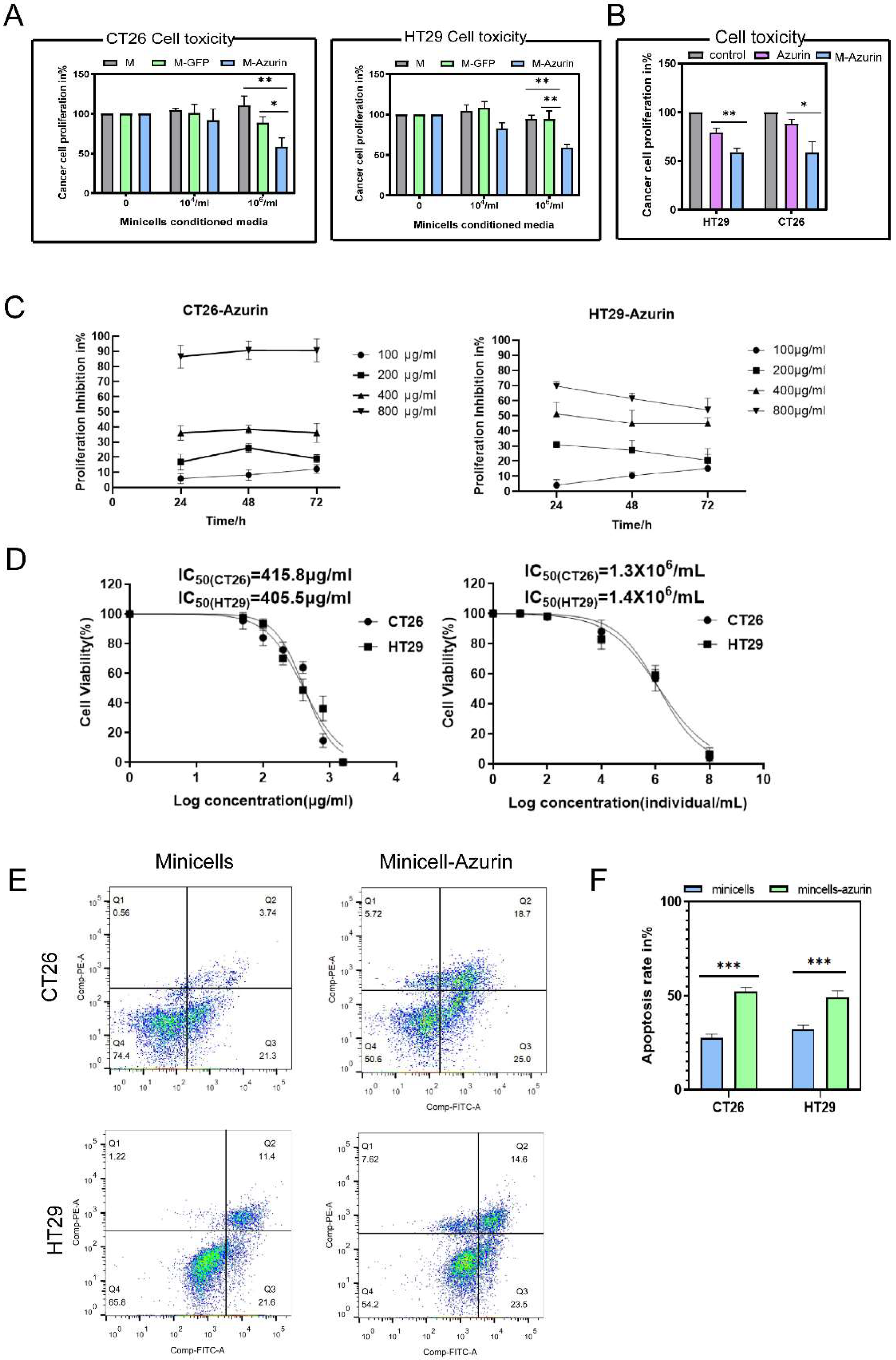
2.2.5. Cytotoxicity of Minicellsazurin against Colon Cancer Cells and IC50
2.2.6. Assessment of Cancer Cell Apoptosis
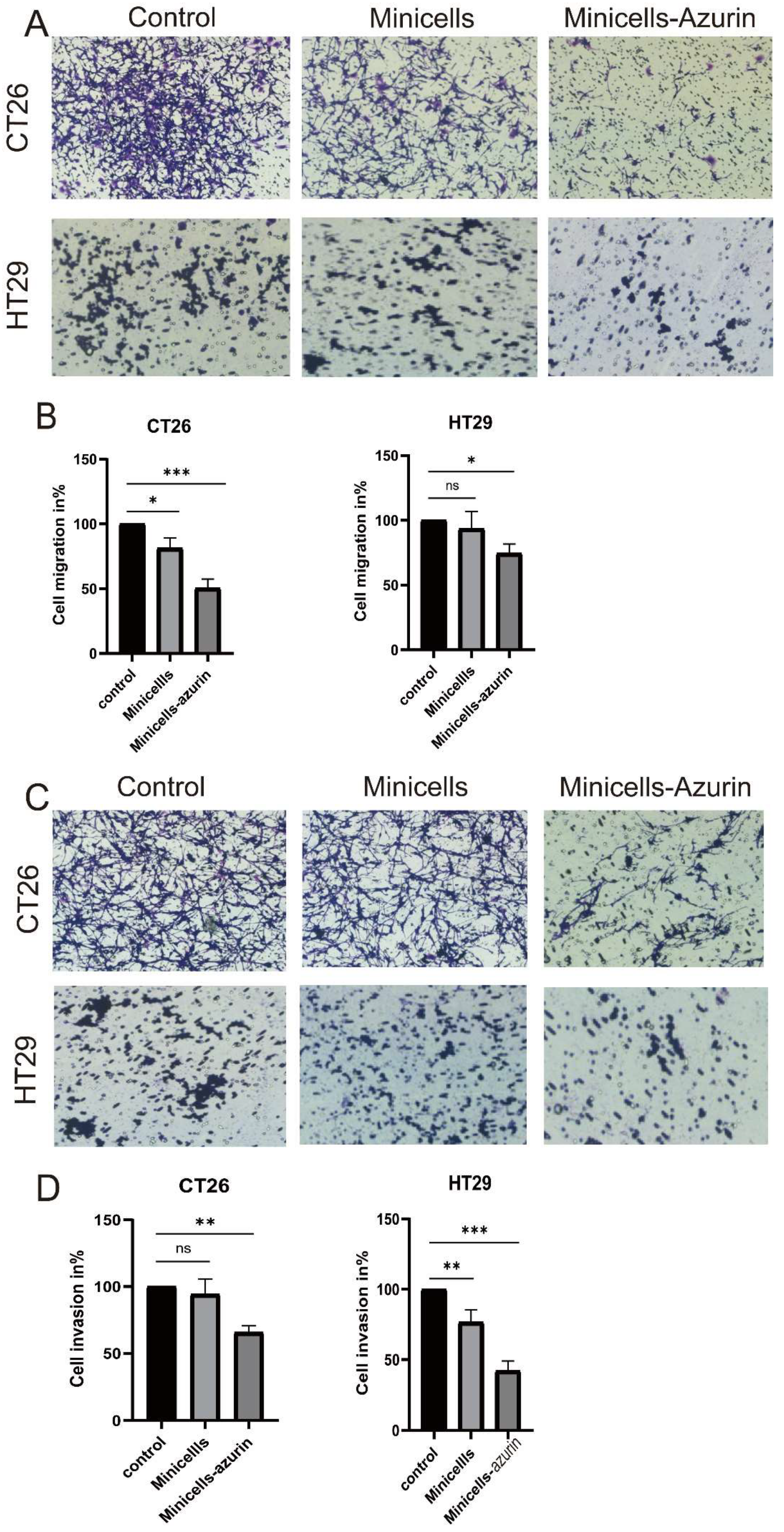
2.2.7. Inhibition of Migration and Invasion of Colon Cancer Cells by Minicellsazurin
2.2.8. Cancer-Cell-Specific Uptake of Minicells
2.2.9. The Expressions of Apoptosis-Related Genes and Tumor-Related Proteins Were Detected by Q-PCR
3. Results and Discussion
3.1. Construction of the Minicell for Delivery of Self-Generated Azurin to Cancer Cell
3.2. Cancer Cell Proliferation Decreases and Cell Death Increases upon Treatment with Azurin and Minicellazurin
3.3. Cancer Cell Migration and Invasion Decrease upon Treatment with Minicellazurin
3.4. Minicells with Tumor Tropism Promote Internalization
3.5. Secretion of Cytokines Involved in Different Kinds of Colon Cancers by Engineered Minicellsazurin Quantified by Q-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El-Shenawy, A.A.; Elsayed, M.M.; Atwa, G.M.; Abourehab, M.A.; Mohamed, M.S.; Ghoneim, M.M.; Mahmoud, R.A.; Sabry, S.A.; Anwar, W.; El-Sherbiny, M.; et al. Anti-Tumor Activity of Orally Administered Gefitinib-Loaded Nanosized Cubosomes against Colon Cancer. Pharmaceutics 2023, 15, 680. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhou, J.; Hu, Y.; Lin, Z.; Ma, Y.; Richardson, J.J.; Caruso, F. Polyphenol-Based Nanoparticles for Intracellular Protein Delivery via Competing Supramolecular Interactions. ACS Nano 2020, 14, 12972–12981. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Jain, A.; Chakraborty, M.; Sahni, J.K.; Ali, J.; Dang, S. Oral delivery of therapeutic proteins and peptides: A review on recent developments. Drug Deliv. 2013, 20, 237–246. [Google Scholar] [CrossRef] [PubMed]
- de Almeida Pachioni-Vasconcelos, J.; Lopes, A.M.; Apolinário, A.C.; Valenzuela-Oses, J.K.; Costa, J.S.R.; de Oliveira Nascimento, L.; Pessoa, A.; Barbosa, L.R.S.; de Oliveira Rangel-Yagui, C. Nanostructures for protein drug delivery. Biomater. Sci. 2016, 4, 205–218. [Google Scholar] [CrossRef]
- Bak, A.; Leung, D.; Barrett, S.E.; Forster, S.; Minnihan, E.C.; Leithead, A.W.; Cunningham, J.; Toussaint, N.; Crocker, L.S. Physicochemical and formulation developability assessment for therapeutic peptide delivery—A primer. AAPS J. 2015, 17, 144–155. [Google Scholar] [CrossRef]
- Kafka, A.P.; McLeod, B.J.; Rades, T.; McDowell, A. Release and bioactivity of PACA nanoparticles containing D-Lys⁶-GnRH for brushtail possum fertility control. J. Control. Release. 2011, 149, 307–313. [Google Scholar] [CrossRef]
- Kv, S.; Devi, G.S.; Mathew, S.T. Liposomal formulations of serratiopeptidase: In vitro studies using PAMPA and Caco-2 models. Mol. Pharm. 2008, 5, 92–97. [Google Scholar] [CrossRef]
- Koppelman, C.M.; Blaauwen, T.D.; Duursma, M.C.; Heeren, R.M.; Nanninga, N. Escherichia coli minicell membranes are enriched in cardiolipin. J. Bacteriol. 2001, 183, 6144–6147. [Google Scholar] [CrossRef]
- Churchward, G.; Bremer, H.; Young, R. Transcription in bacteria at different DNA concentrations. J. Bacteriol. 1982, 150, 572–581. [Google Scholar] [CrossRef]
- Cook, D.I.; Revzin, A. Intracellular location of catabolite activator protein of Escherichia coli. J. Bacteriol. 1980, 141, 1279–1283. [Google Scholar] [CrossRef]
- Fralick, J.A.; Fisher, W.D.; Adler, H.I. Polyuridylic acid-directed phenylalanine incorporation in minicell extracts. J. Bacteriol. 1969, 99, 621–622. [Google Scholar] [CrossRef] [PubMed]
- Reeve, J. Macromolecular synthesis in bacteriophage infected minicells of Bacillus subtilis. In Microbiology-1976; American Society for Microbiology: Washington, DC, USA, 1976; pp. 332–339. [Google Scholar]
- Adler, H.; Fisher, W.; Cohen, A.; Hardigree, A.A. Miniature Escherichia coli cells deficient in DNA. Proc. Natl. Acad. Sci. USA 1967, 57, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Black, J.W. Growth Characteristics of Miniature Escherichia coli Cells Deficient in DNA. Ph.D. Thesis, University of Tennessee, Knoxville, TN, USA, 1967. [Google Scholar]
- MacDiarmid, J.A.; Langova, V.; Bailey, D.; Pattison, S.T.; Pattison, S.L.; Christensen, N.; Armstrong, L.R.; Brahmbhatt, V.N.; Smolarczyk, K.; Harrison, M.T.; et al. Targeted Doxorubicin Delivery to Brain Tumors via Minicells: Proof of Principle Using Dogs with Spontaneously Occurring Tumors as a Model. PLoS ONE. 2016, 11, e0151832. [Google Scholar] [CrossRef]
- MacDiarmid, J.A.; Amaro-Mugridge, N.B.; Madrid-Weiss, J.; Sedliarou, I.; Wetzel, S.; Kochar, K.; Brahmbhatt, V.N.; Phillips, L.; Pattison, S.T.; Petti, C.; et al. Sequential treatment of drug-resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nat. Biotechnol. 2009, 27, 643–651. [Google Scholar] [CrossRef]
- Taylor, B.N.; Mehta, R.R.; Yamada, T.; Lekmine, F.; Christov, K.; Chakrabarty, A.M.; Green, A.; Bratescu, L.; Shilkaitis, A.; Beattie, C.W.; et al. Noncationic peptides obtained from azurin preferentially enter cancer cells. Cancer Res. 2009, 69, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.S.; Fattah, S.A.; Mostafa, H.M. Azurin as antitumor protein and its effect on the cancer cell lines. Curr. Res. J. Biol. Sci. 2010, 2, 396–401. [Google Scholar]
- Paydarnia, N.; Nikkhoi, S.K.; Fakhravar, A.; Mehdiabdol, M.; Heydarzadeh, H.; Ranjbar, S. Synergistic effect of granzyme B-azurin fusion protein on breast cancer cells. Mol. Biol. Rep. 2019, 46, 3129–3140. [Google Scholar] [CrossRef]
- Yamada, T.; Christov, K.; Shilkaitis, A.; Bratescu, L.; Green, A.; Santini, S.; Bizzarri, A.R.; Cannistraro, S.; Gupta, T.K.D.; Beattie, C.W. p28, a first in class peptide inhibitor of cop1 binding to p53. Br. J. Cancer Suppl. 2013, 108, 2495–2504. [Google Scholar] [CrossRef]
- Bizzarri, A.R.; Santini, S.; Coppari, E.; Bucciantini, M.; Di Agostino, S.; Yamada, T.; Beattie, C.W.; Cannistraro, S. Interaction of an anticancer peptide fragment of azurin with p53 and its isolated domains studied by atomic force spectroscopy. Int. J. Nanomed. 2011, 6, 3011–3019. [Google Scholar] [CrossRef]
- Chaudhari, A.; Mahfouz, M.; Fialho, A.M.; Yamada, T.; Granja, A.T.; Zhu, Y.; Hashimoto, W.; Schlarb-Ridley, B.; Cho, W.; Gupta, T.K.D.; et al. Cupredoxin–cancer interrelationship: Azurin binding with EphB2, interference in EphB2 tyrosine phosphorylation, and inhibition of cancer growth. Biochemistry 2007, 46, 1799–1810. [Google Scholar] [CrossRef]
- Saha, N.; Robev, D.; Mason, E.O.; Himanen, J.P.; Nikolov, D.B. Therapeutic potential of targeting the Eph/ephrin signaling complex. Int. J. Biochem. Cell Biol. 2018, 105, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.R.; Yamada, T.; Taylor, B.N.; Christov, K.; King, M.L.; Majumdar, D.; Lekmine, F.; Tiruppathi, C.; Shilkaitis, A.; Bratescu, L.; et al. A cell penetrating peptide derived from azurin inhibits angiogenesis and tumor growth by inhibiting phosphorylation of VEGFR-2, FAK and Akt. Angiogenesis 2011, 14, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, A.M. Bacterial azurin in potential cancer therapy. Cell Cycle. 2016, 15, 1665–1666. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Mehta, R.; Majumdar, D.; Chakrabarty, A.M.; Das Gupta, T.K. Azurin, highly potent and selective anticancer agent for breast cancer. J. Clin. Oncol. 2006, 24 (Suppl. S18), 13106. [Google Scholar] [CrossRef]
- Sereena, M.C.; Sebastian, D. Evaluation of Anticancer and Anti-hemolytic Activity of Azurin, a Novel Bacterial Protein from Pseudomonas aeruginosa SSj. Int. J. Pept. Res. Ther. 2020, 26, 459–466. [Google Scholar] [CrossRef]
- Sanders, M.E. Probiotics: Considerations for human health. Nutr. Rev. 2003, 61, 91–99. [Google Scholar] [CrossRef]
- Thamm, D.H.; Kurzman, I.D.; King, I.; Li, Z.; Sznol, M.; Dubielzig, R.R.; Vail, D.M.; MacEwen, E.G. Systemic administration of an attenuated, tumor-targeting Salmonella typhimurium to dogs with spontaneous neoplasia: Phase I evaluation. Clin. Cancer Res. 2005, 11, 4827–4834. [Google Scholar] [CrossRef]
- Toso, J.F.; Gill, V.J.; Hwu, P.; Marincola, F.M.; Restifo, N.P.; Schwartzentruber, D.J.; Sherry, R.M.; Topalian, S.L.; Yang, J.C.; Stock, F.; et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J. Clin. Oncol. 2002, 20, 142–152. [Google Scholar] [CrossRef]
- Grozdanov, L.; Zähringer, U.; Blum-Oehler, G.; Brade, L.; Henne, A.; Knirel, Y.A.; Schombel, U.; Schulze, J.; Sonnenborn, U.; Gottschalk, G.; et al. A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype serum sensitivity of Escherichia coli strain Nissle 1917. J. Bacteriol. 2002, 184, 5912–5925. [Google Scholar] [CrossRef]
- De Vrese, M.; Schrezenmeir, A.J. Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. Biotechnol. 2008, 111, 1–66. [Google Scholar]
- Zhang, Y.; Ji, W.; He, L.; Chen, Y.; Ding, X.; Sun, Y.; Hu, S.; Yang, H.; Huang, W.; Zhang, Y.; et al. E. coli Nissle 1917-Derived Minicells for Targeted Delivery of Chemotherapeutic Drug to Hypoxic Regions for Cancer Therapy. Theranostics. 2018, 8, 1690–1705. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Khokhlatchev, A.V.; Chew, C.; Illendula, A.; Conaway, M.; Dryden, K.; Maeda, D.L.N.F.; Rajasekaran, V.; Kester, M.; Zeichner, S.L. Minicells from Highly Genome Reduced Escherichia coli: Cytoplasmic and Surface Expression of Recombinant Proteins and Incorporation in the Minicells. ACS Synth. Biol. 2021, 10, 2465–2477. [Google Scholar] [CrossRef] [PubMed]
- Jivrajani, M.; Shrivastava, N.; Nivsarkar, M. A combination approach for rapid and high yielding purification of bacterial minicells. J. Microbiol. Methods 2013, 92, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B. Resistance of minicells to penicillin lysis: A method of obtaining large quantities of purified minicells. J. Bacteriol. 1970, 103, 836–839. [Google Scholar] [CrossRef] [PubMed]
- Ni, B.; Colin, R.; Sourjik, V. Production and Characterization of Motile and Chemotactic Bacterial Minicells. ACS Synth. Biol. 2021, 10, 1284–1291. [Google Scholar] [CrossRef]
- Apiyo, D.; Wittung-Stafshede, P. Unique complex between bacterial azurin and tumor-suppressor protein p53. Biochem. Biophys. Res. Commun. 2005, 332, 965–968. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Zhu, G.; Feng, L.; Jiang, S.; Xiang, Q.; Wang, J. Efficient Cytotoxicity of Recombinant Azurin in Escherichia coli Nissle 1917-Derived Minicells against Colon Cancer Cells. Bioengineering 2023, 10, 1188. https://doi.org/10.3390/bioengineering10101188
Ma Y, Zhu G, Feng L, Jiang S, Xiang Q, Wang J. Efficient Cytotoxicity of Recombinant Azurin in Escherichia coli Nissle 1917-Derived Minicells against Colon Cancer Cells. Bioengineering. 2023; 10(10):1188. https://doi.org/10.3390/bioengineering10101188
Chicago/Turabian StyleMa, Yi, Guanshu Zhu, Lan Feng, Shoujin Jiang, Qi Xiang, and Jufang Wang. 2023. "Efficient Cytotoxicity of Recombinant Azurin in Escherichia coli Nissle 1917-Derived Minicells against Colon Cancer Cells" Bioengineering 10, no. 10: 1188. https://doi.org/10.3390/bioengineering10101188
APA StyleMa, Y., Zhu, G., Feng, L., Jiang, S., Xiang, Q., & Wang, J. (2023). Efficient Cytotoxicity of Recombinant Azurin in Escherichia coli Nissle 1917-Derived Minicells against Colon Cancer Cells. Bioengineering, 10(10), 1188. https://doi.org/10.3390/bioengineering10101188







