Improving OCT Image Segmentation of Retinal Layers by Utilizing a Machine Learning Based Multistage System of Stacked Multiscale Encoders and Decoders
Abstract
:1. Introduction and Motivation
1.1. Related Work
1.2. Contribution of This Work and Future Prospects
1.3. Section Overview
2. Materials, Methods, and Implementation
2.1. Data Sets
2.1.1. Peripapillary Data Set
2.1.2. Duke SD-OCT Data Set
2.1.3. UMN Data Set
2.1.4. Heidelberg Data Set
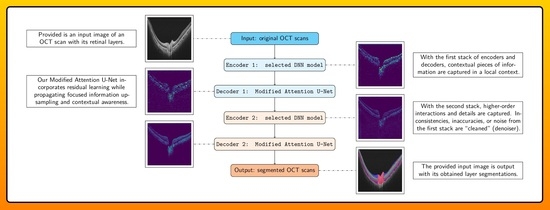
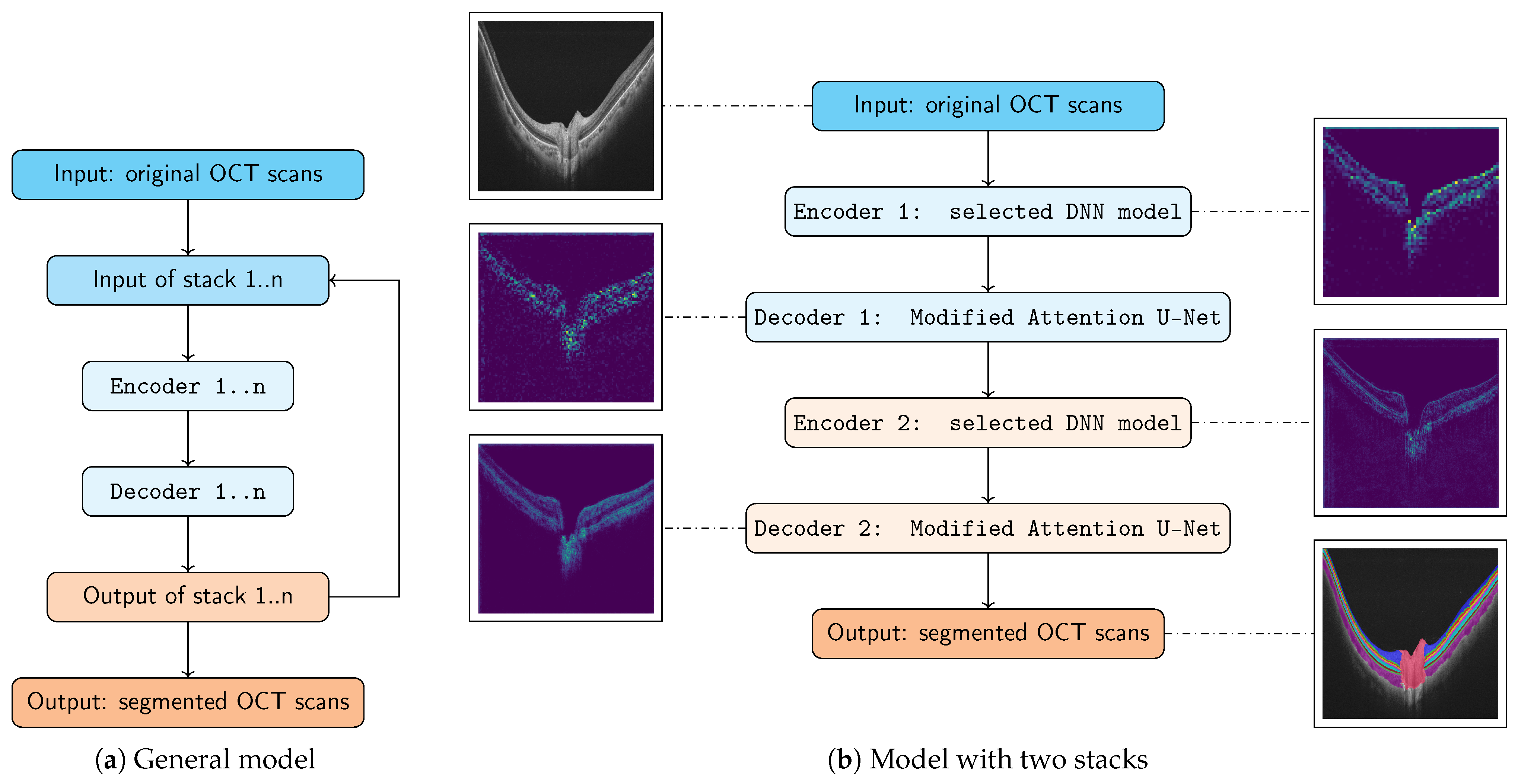
2.2. Model Architecture
2.2.1. Stack 1: Encoder 1 + Decoder 1 (Local Context)
2.2.2. Stack 2: Encoder 2 + Decoder 2 (Denoiser)
2.2.3. Modified Attention U-Net
2.3. Training Setup
2.4. Loss Functions
2.4.1. Dice Loss
2.4.2. Lovász Loss
- = predicted probability sorted in decreasing order
- = the corresponding ground truth label
- = the cumulative sum of ground truth labels up to index i
- =
- = denotes the positive part of x, defined as .
2.4.3. Tversky Loss
2.4.4. Combined Loss
2.5. Evaluation Principles
3. Test Results, Evaluation, and Discussion
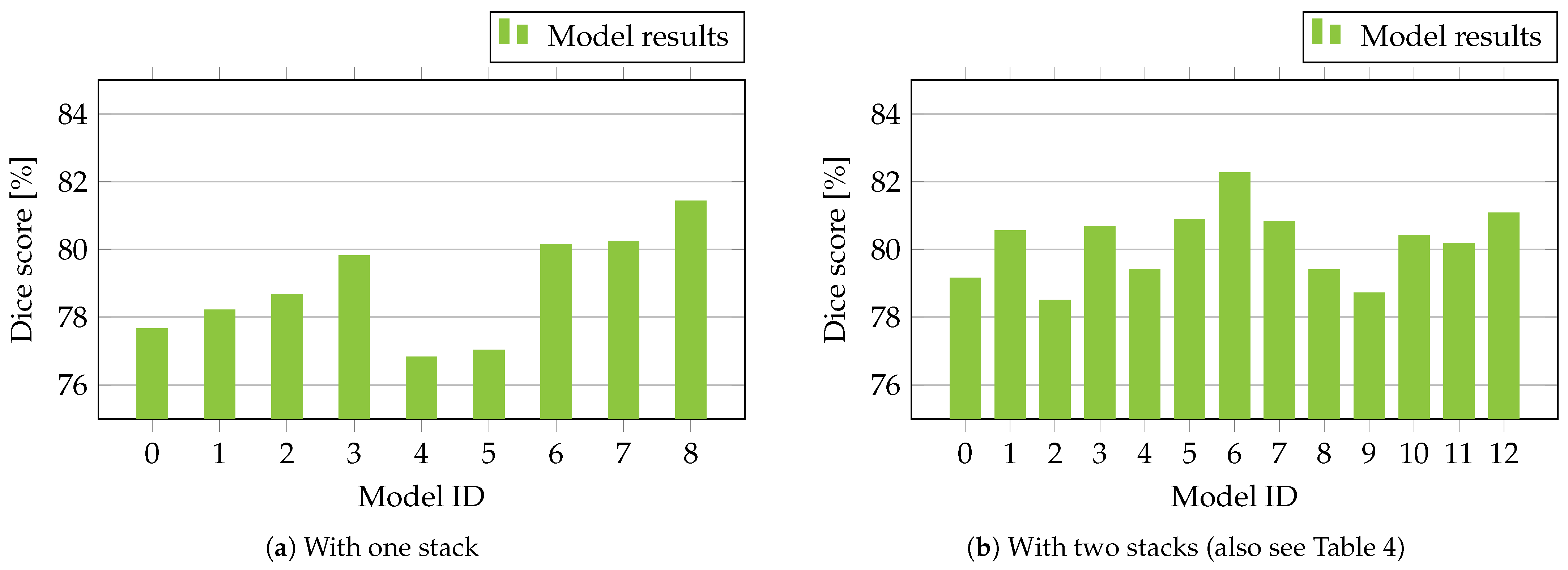
3.1. Quantitative Results
3.2. Qualitative Results
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMD | Age-related macula degeneration |
| DNN | Deep neural network |
| GCL | Ganglion cell layer |
| INL | Inner nuclear layer |
| IPL | Inner plexiform layer |
| IRF | Intraretinal fluid |
| IS/OS | Inner/outer photoreceptor segment |
| ONL | Outer nuclear layer |
| OPL | Outer plexiform layer |
| PED | Pigment epithelial detachment |
| RNFL | Retinal nerve fiber layer |
| RNN | Recurrent neural network |
| RPE | Retinal pigment epithelium |
| SRF | Subretinal fluid |
References
- Anders, J.; Dapp, U.; Laub, S.; Renteln-Kruse, W. Impact of fall risk and fear of falling on mobility of independently living senior citizens transitioning to frailty: Screening results concerning fall prevention in the community. Z. Gerontol. Geriatr. 2007, 40, 255–267. [Google Scholar] [CrossRef]
- E, J.Y.; Li, T.; Mclnally, L.; Thomson, K.; Shahani, U.; Gray, L.; Howe, T.; Skelton, D. Environmental and behavioural interventions for reducing physical activity limitation and preventing falls in older people with visual impairment. Cochrane Database Syst. Rev. 2020, 9, CD009233. [Google Scholar] [CrossRef]
- Pascolini, D.; Mariotti, S. Global Estimates of Visual Impairment: 2010. Br. J. Ophthalmol. 2011, 96, 614–618. [Google Scholar] [CrossRef]
- Reitmeir, P.; Linkohr, B.; Heier, M.; Molnos, S.; Strobl, R.; Schulz, H.; Breier, M.; Faus, T.; Küster, D.M.; Wulff, A.; et al. Common eye diseases in older adults of southern Germany: Results from the KORA-Age study. Age Ageing 2017, 46, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Finger, R.P.; Fimmers, R.; Holz, F.G.; Scholl, H.P. Incidence of blindness and severe visual impairment in Germany: Projections for 2030. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4381–4389. [Google Scholar] [CrossRef] [PubMed]
- Kansal, V.; Armstrong, J.J.; Pintwala, R.; Hutnik, C. Optical coherence tomography for glaucoma diagnosis: An evidence based meta-analysis. PLoS ONE 2018, 13, e0190621. [Google Scholar] [CrossRef]
- Aumann, S.; Donner, S.; Fischer, J.; Müller, F. Optical coherence tomography (OCT): Principle and technical realization. In High Resolution Imaging in Microscopy and Ophthalmology: New Frontiers in Biomedical Optics; Springer: Berlin, Germany, 2019; pp. 59–85. [Google Scholar]
- Shu, X.; Beckmann, L.; Zhang, H.F. Visible-light optical coherence tomography: A review. J. Biomed. Opt. 2017, 22, 121707. [Google Scholar] [CrossRef]
- Maldonado, R.S.; Toth, C.A. Optical coherence tomography in retinopathy of prematurity: Looking beyond the vessels. Clin. Perinatol. 2013, 40, 271–296. [Google Scholar] [CrossRef]
- Iqbal, S.; Qureshi, A.; Li, J.; Mahmood, T. On the Analyses of Medical Images Using Traditional Machine Learning Techniques and Convolutional Neural Networks. Arch. Comput. Methods Eng. 2023, 30, 3173–3233. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, C.E.; Yang, J.; Anderson, B.M.; Court, L.E.; Brock, K.B. Advances in Auto-Segmentation. Semin. Radiat. Oncol. 2019, 29, 185–197. [Google Scholar] [CrossRef]
- Schlosser, T.; Beuth, F.; Meyer, T.; Kumar, A.S.; Stolze, G.; Furashova, O.; Engelmann, K.; Kowerko, D. Visual Acuity Prediction on Real-Life Patient Data Using a Machine Learning Based Multistage System. arXiv 2022, arXiv:2204.11970. [Google Scholar]
- Garvin, M.K.; Abramoff, M.D.; Wu, X.; Russell, S.R.; Burns, T.L.; Sonka, M. Automated 3-D Intraretinal Layer Segmentation of Macular Spectral-Domain Optical Coherence Tomography Images. IEEE Trans. Med. Imaging 2009, 28, 1436–1447. [Google Scholar] [CrossRef]
- Chiu, S.J.; Allingham, M.J.; Mettu, P.S.; Cousins, S.W.; Izatt, J.A.; Farsiu, S. Kernel regression based segmentation of optical coherence tomography images with diabetic macular edema. Biomed. Opt. Express 2015, 6, 1172–1194. [Google Scholar] [CrossRef]
- Fang, L.; Cunefare, D.; Wang, C.; Guymer, R.H.; Li, S.; Farsiu, S. Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search. Biomed. Opt. Express 2017, 8, 2732–2744. [Google Scholar] [CrossRef] [PubMed]
- Elgafi, M.; Sharafeldeen, A.; Elnakib, A.; Elgarayhi, A.; Alghamdi, N.S.; Sallah, M.; El-Baz, A. Detection of Diabetic Retinopathy Using Extracted 3D Features from OCT Images. Sensors 2022, 22, 7833. [Google Scholar] [CrossRef]
- Li, J.; Jin, P.; Zhu, J.; Zou, H.; Xu, X.; Tang, M.; Zhou, M.; Gan, Y.; He, J.; Ling, Y.; et al. Multi-scale GCN-assisted two-stage network for joint segmentation of retinal layers and discs in peripapillary OCT images. Biomed. Opt. Express 2021, 12, 2204–2220. [Google Scholar] [CrossRef]
- Everingham, M.; Eslami, S.A.; Van Gool, L.; Williams, C.K.; Winn, J.; Zisserman, A. The pascal visual object classes challenge: A retrospective. Int. J. Comput. Vis. 2015, 111, 98–136. [Google Scholar] [CrossRef]
- Brostow, G.J.; Fauqueur, J.; Cipolla, R. Semantic object classes in video: A high-definition ground truth database. Pattern Recognit. Lett. 2009, 30, 88–97. [Google Scholar] [CrossRef]
- Song, S.; Lichtenberg, S.P.; Xiao, J. SUN RGB-D: A RGB-D Scene Understanding Benchmark Suite. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Shotton, J.; Johnson, M.; Cipolla, R. Semantic texton forests for image categorization and segmentation. In Proceedings of the 2008 IEEE Conference on Computer Vision and Pattern Recognition, Anchorage, AK, USA, 23–28 June 2008; pp. 1–8. [Google Scholar] [CrossRef]
- Leon, F.; Floria, S.A.; Badica, C. Evaluating the effect of voting methods on ensemble-based classification. In Proceedings of the 2017 IEEE International Conference on INnovations in Intelligent Systems and Applications (INISTA), Gdynia, Poland, 3–5 July 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Sturgess, P.; Alahari, K.; Ladicky, L.; Torr, P. Combining Appearance and Structure from Motion Features for Road Scene Understanding. In Proceedings of the BMVC-British Machine Vision Conference, London, UK, 7–10 September 2009. [Google Scholar] [CrossRef]
- Roy, A.G.; Conjeti, S.; Karri, S.P.K.; Sheet, D.; Katouzian, A.; Wachinger, C.; Navab, N. ReLayNet: Retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks. Biomed. Opt. Express 2017, 8, 3627–3642. [Google Scholar] [CrossRef] [PubMed]
- Kiaee, F.; Fahimi, H.; Rabbani, H. Intra-Retinal Layer Segmentation of Optical Coherence Tomography Using 3D Fully Convolutional Networks. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 2795–2799. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R.B. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Lin, T.; Maire, M.; Belongie, S.J.; Bourdev, L.D.; Girshick, R.B.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014. [Google Scholar]
- Pérez-Nicolás, M.; Colinas-León, T.; Alia-Tejacal, I.; Peña-Ortega, G.; González-Andrés, F.; Beltrán-Rodríguez, L. Morphological Variation in Scarlet Plume (Euphorbia fulgens Karw ex Klotzsch, Euphorbiaceae), an Underutilized Ornamental Resource of Mexico with Global Importance. Plants 2021, 10, 2020. [Google Scholar] [CrossRef]
- Liu, W.; Sun, Y.; Ji, Q. MDAN-UNet: Multi-Scale and Dual Attention Enhanced Nested U-Net Architecture for Segmentation of Optical Coherence Tomography Images. Algorithms 2020, 13, 60. [Google Scholar] [CrossRef]
- Lin, T.; Dollár, P.; Girshick, R.B.; He, K.; Hariharan, B.; Belongie, S.J. Feature Pyramid Networks for Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Orlando, J.I.; Seeböck, P.; Bogunović, H.; Klimscha, S.; Grechenig, C.; Waldstein, S.; Gerendas, B.S.; Schmidt-Erfurth, U. U2-net: A bayesian u-net model with epistemic uncertainty feedback for photoreceptor layer segmentation in pathological oct scans. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; IEEE: Piscatway, NJ, USA, 2019; pp. 1441–1445. [Google Scholar]
- Farshad, A.; Yeganeh, Y.; Gehlbach, P.; Navab, N. Y-Net: A Spatiospectral Dual-Encoder Networkfor Medical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022. [Google Scholar] [CrossRef]
- Li, Y.; Qi, H.; Dai, J.; Ji, X.; Wei, Y. Fully Convolutional Instance-aware Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Tran, A.; Weiss, J.; Albarqouni, S.; Faghi Roohi, S.; Navab, N. Retinal Layer Segmentation Reformulated as OCT Language Processing. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part V. Springer: Berlin/Heidelberg, Germany, 2020; pp. 694–703. [Google Scholar] [CrossRef]
- Kugelman, J.; Alonso-Caneiro, D.; Read, S.; Vincent, S.; Collins, M. Automatic segmentation of OCT retinal boundaries using recurrent neural networks and graph search. Biomed. Opt. Express 2018, 9, 5759. [Google Scholar] [CrossRef]
- Meester, R.; Doubeni, C.; Zauber, A.; Goede, S.; Levin, T.; Corley, D.; Jemal, A.; Lansdorp-Vogelaar, I. 969 Public Health Impact of Achieving 80% Colorectal Cancer Screening RATES in the United States by 2018. Cancer 2015, 81, AB181–AB182. [Google Scholar] [CrossRef]
- Mohammed, A.; Yildirim, S.; Farup, I.; Pedersen, M.; Hovde, Ø. Y-net: A deep convolutional neural network for polyp detection. arXiv 2018, arXiv:1806.01907. [Google Scholar] [CrossRef]
- Chen, S.; Guo, W. Auto-Encoders in Deep Learning—A Review with New Perspectives. Mathematics 2023, 11, 1777. [Google Scholar] [CrossRef]
- Lin, G.; Shen, C.; Reid, I.D.; van den Hengel, A. Efficient piecewise training of deep structured models for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Noh, H.; Hong, S.; Han, B. Learning Deconvolution Network for Semantic Segmentation. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A. Semantic Image Segmentation with Deep Convolutional Nets and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2014, 40, 834–848. [Google Scholar] [CrossRef]
- Martí, M.; Maki, A. A multitask deep learning model for real-time deployment in embedded systems. arXiv 2017, arXiv:1711.00146. [Google Scholar]
- Hariharan, B.; Arbeláez, P.A.; Girshick, R.B.; Malik, J. Hypercolumns for Object Segmentation and Fine-grained Localization. arXiv 2014, arXiv:1411.5752. [Google Scholar]
- Mostajabi, M.; Yadollahpour, P.; Shakhnarovich, G. Feedforward semantic segmentation with zoom-out features. arXiv 2014, arXiv:1412.0774. [Google Scholar]
- Zang, P.; Wang, J.; Hormel, T.T.; Liu, L.; Huang, D.; Jia, Y. Automated segmentation of peripapillary retinal boundaries in OCT combining a convolutional neural network and a multi-weights graph search. Biomed. Opt. Express 2019, 10, 4340–4352. [Google Scholar] [CrossRef]
- Devalla, S.K.; Renukanand, P.K.; Sreedhar, B.K.; Subramanian, G.; Zhang, L.; Perera, S.; Mari, J.M.; Chin, K.S.; Tun, T.A.; Strouthidis, N.G.; et al. DRUNET: A dilated-residual U-Net deep learning network to segment optic nerve head tissues in optical coherence tomography images. Biomed. Opt. Express 2018, 9, 3244–3265. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Advances in Neural Information Processing Systems; MIT Press: Boston, MA, USA, 2014; Volume 27. [Google Scholar]
- You, A.; Kim, J.K.; Ryu, I.H.; Yoo, T.K. Application of generative adversarial networks (GAN) for ophthalmology image domains: A survey. Eye Vis. 2022, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Burlina, P.M.; Joshi, N.; Pacheco, K.D.; Liu, T.A.; Bressler, N.M. Assessment of deep generative models for high-resolution synthetic retinal image generation of age-related macular degeneration. JAMA Ophthalmol. 2019, 137, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Rashno, A.; Nazari, B.; Koozekanani, D.D.; Drayna, P.M.; Sadri, S.; Rabbani, H.; Parhi, K.K. Fully-automated segmentation of fluid regions in exudative age-related macular degeneration subjects: Kernel graph cut in neutrosophic domain. PLoS ONE 2017, 12, e0186949. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; pp. 630–645. [Google Scholar] [CrossRef]
- Iakubovskii, P. Segmentation Models Pytorch. 2019. Available online: https://github.com/qubvel/segmentation_models.pytorch (accessed on 30 August 2023).
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Schlosser, T.; Friedrich, M.; Beuth, F.; Kowerko, D. Improving automated visual fault inspection for semiconductor manufacturing using a hybrid multistage system of deep neural networks. J. Intell. Manuf. 2022, 33, 1099–1123. [Google Scholar] [CrossRef]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Cardoso, M.J. Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations. arXiv 2017, arXiv:1707.03237. [Google Scholar]
- Berman, M.; Blaschko, M.B. Optimization of the Jaccard index for image segmentation with the Lovász hinge. arXiv 2017, arXiv:1705.08790. [Google Scholar]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. arXiv 2017, arXiv:1706.05721. [Google Scholar]
- Liu, Q.; Tang, X.; Guo, D.; Qin, Y.; Jia, P.; Zhan, Y.; Zhou, X.; Wu, D. Multi-class gradient harmonized dice loss with application to knee MR image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part VI 22. Springer: Berlin, Germany, 2019; pp. 86–94. [Google Scholar]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Sorensen, T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skr. 1948, 5, 1–34. [Google Scholar]
- Van Rijsbergen, C.J. The Geometry of Information Retrieval; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- He, T.; Zhang, Z.; Zhang, H.; Zhang, Z.; Xie, J.; Li, M. Bag of tricks for image classification with convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 558–567. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Wightman, R. PyTorch Image Models. 2019. Available online: https://github.com/rwightman/pytorch-image-models (accessed on 30 August 2023).
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Proceedings 4. Springer: Berlin, Germany, 2018; pp. 3–11. [Google Scholar]
- Xia, K.j.; Yin, H.s.; Zhang, Y.d. Deep semantic segmentation of kidney and space-occupying lesion area based on SCNN and ResNet models combined with SIFT-flow algorithm. J. Med. Syst. 2019, 43, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, H.; Zhao, T.; Guo, Y.; Xu, Z.; Liu, Z.; Liu, S.; Lan, X.; Sun, X.; Feng, M. Classification of cardiac abnormalities from ECG signals using SE-ResNet. In Proceedings of the 2020 Computing in Cardiology, Rimini, Italy, 13–16 September 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Abedalla, A.; Abdullah, M.; Al-Ayyoub, M.; Benkhelifa, E. Chest X-ray pneumothorax segmentation using U-Net with EfficientNet and ResNet architectures. PeerJ Comput. Sci. 2021, 7, e607. [Google Scholar] [CrossRef] [PubMed]
- Midena, E.; Torresin, T.; Schiavon, S.; Danieli, L.; Polo, C.; Pilotto, E.; Midena, G.; Frizziero, L. The Disorganization of Retinal Inner Layers Is Correlated to Müller Cells Impairment in Diabetic Macular Edema: An Imaging and Omics Study. Int. J. Mol. Sci. 2023, 24, 9607. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Touvron, H.; Cord, M.; Sablayrolles, A.; Synnaeve, G.; Jégou, H. Going deeper with image transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 32–42. [Google Scholar]
- Han, K.; Xiao, A.; Wu, E.; Guo, J.; Xu, C.; Wang, Y. Transformer in transformer. In Advances in Neural Information Processing Systems; MIT Press: Boston, MA, USA, 2021; Volume 34, pp. 15908–15919. [Google Scholar]






| Data Set Name | Number of Images | Image Resolution | Annotations |
|---|---|---|---|
| Duke SD-OCT [14] | 88 | ✓ | |
| UMN [54] | 125 | ✓ | |
| Heidelberg [55] | 125 | - |
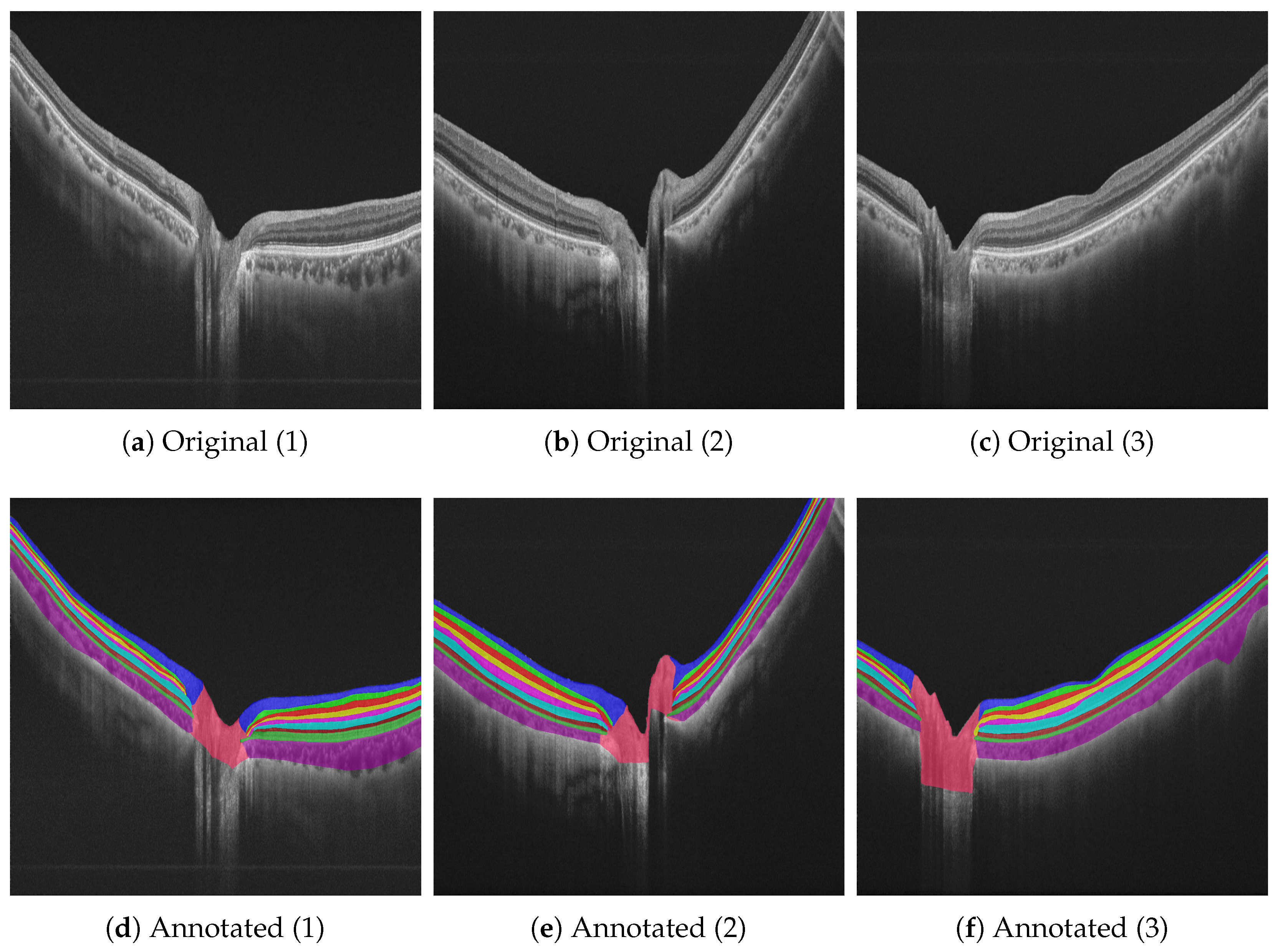
| Layer | Color Scheme |
|---|---|
| Retinal nerve fiber layer (RNFL) |  |
| Ganglion cell layer (GCL) |  |
| Inner plexiform layer (IPL) |  |
| Inner nuclear layer (INL) |  |
| Outer plexiform layer (OPL) |  |
| Outer nuclear layer (ONL) |  |
| Inner/outer photoreceptor segment (IS/OS) |  |
| Retinal pigment epithelium (RPE) |  |
| Choroid |  |
| Optic disc |  |
| Model ID | Encoder | Decoder | Dice Score [%] |
|---|---|---|---|
| 0 | EfficientNet B0 | Modified Attention U-Net | |
| 1 | EfficientNet B1 | Modified Attention U-Net | |
| 2 | EfficientNet B2 | Modified Attention U-Net | |
| 3 | EfficientNet B3 | Modified Attention U-Net | |
| 4 | EfficientNet B4 | Modified Attention U-Net | |
| 5 | EfficientNet B5 | Modified Attention U-Net | |
| 6 | ResNet34D | Modified Attention U-Net | |
| 7 | ResNet50D | Modified Attention U-Net | |
| 8 | SEResNeXt50-32x4D | Modified Attention U-Net |
| Model ID | Encoder 1 | Encoder 2 | Decoders 1 and 2 | Dice Score [%] |
|---|---|---|---|---|
| 0 | EfficientNet B0 | EfficientNet B1 | Modified Attention U-Net | |
| 1 | EfficientNet B2 | EfficientNet B3 | Modified Attention U-Net | |
| 2 | EfficientNet B4 | EfficientNet B5 | Modified Attention U-Net | |
| 3 | EfficientNet B0 | ResNet34D | Modified Attention U-Net | |
| 4 | EfficientNet B2 | ResNet34D | Modified Attention U-Net | |
| 5 | ResNet50D | ResNet34D | Modified Attention U-Net | |
| 6 | SEResNeXt50-32x4D | ResNet34D | Modified Attention U-Net | |
| 7 | ResNet34D | SEResNeXt50-32x4D | Modified Attention U-Net | |
| 8 | SEResNeXt50-32x4D | EfficientNet B2 | Modified Attention U-Net | |
| 9 | SEResNeXt50-32x4D | EfficientNet B3 | Modified Attention U-Net | |
| 10 | ResNet34D | ResNet34D | Modified Attention U-Net | |
| 11 | ResNet50D | ResNet50D | Modified Attention U-Net | |
| 12 | SEResNeXt50-32x4D | SEResNeXt50-32x4D | Modified Attention U-Net |
| Model | Dice Score [%] |
|---|---|
| State-of-the-art model with a single stage [17] | |
| One stack with one encoder and one decoder with attention pooling (SEResNeXt50-32x4D) | |
| Two stacks of encoders and decoders with attention pooling (best model from Table 4, model ID 6: SEResNeXt50-32x4D and ResNet34D) |
| Test Sample(s) | RNFL | GCL | IPL | INL | OPL | ONL | IS/OS | RPE | Choroid | Optic Disc | Overall Dice Score [%] |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Figure 5c | 85.05 | 75.21 | 79.12 | 81.73 | 84.13 | 83.63 | 51.89 | 67.84 | 92.91 | 63.65 | 76.52 |
| Figure 5f | 92.72 | 79.64 | 80.78 | 84.17 | 88.73 | 95.03 | 92.50 | 90.00 | 96.67 | 90.23 | 89.05 |
| Overall | 83.25 | 68.49 | 73.93 | 80.65 | 81.39 | 91.73 | 85.84 | 84.55 | 86.92 | 85.56 | 82.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sampath Kumar, A.; Schlosser, T.; Langner, H.; Ritter, M.; Kowerko, D. Improving OCT Image Segmentation of Retinal Layers by Utilizing a Machine Learning Based Multistage System of Stacked Multiscale Encoders and Decoders. Bioengineering 2023, 10, 1177. https://doi.org/10.3390/bioengineering10101177
Sampath Kumar A, Schlosser T, Langner H, Ritter M, Kowerko D. Improving OCT Image Segmentation of Retinal Layers by Utilizing a Machine Learning Based Multistage System of Stacked Multiscale Encoders and Decoders. Bioengineering. 2023; 10(10):1177. https://doi.org/10.3390/bioengineering10101177
Chicago/Turabian StyleSampath Kumar, Arunodhayan, Tobias Schlosser, Holger Langner, Marc Ritter, and Danny Kowerko. 2023. "Improving OCT Image Segmentation of Retinal Layers by Utilizing a Machine Learning Based Multistage System of Stacked Multiscale Encoders and Decoders" Bioengineering 10, no. 10: 1177. https://doi.org/10.3390/bioengineering10101177
APA StyleSampath Kumar, A., Schlosser, T., Langner, H., Ritter, M., & Kowerko, D. (2023). Improving OCT Image Segmentation of Retinal Layers by Utilizing a Machine Learning Based Multistage System of Stacked Multiscale Encoders and Decoders. Bioengineering, 10(10), 1177. https://doi.org/10.3390/bioengineering10101177