Genetic Alterations in Renal Cancers: Identification of The Mechanisms Underlying Cancer Initiation and Progression and of Therapeutic Targets
Abstract
1. Introduction
2. Hereditary RCCs
3. Von Hippel-Lindau Disease
4. Hereditary Papillary Renal Carcinoma Type I
5. Germline PTEN Mutation Cowden Syndrome
6. Hereditary BAP1 Tumor Syndrome
7. Succinate Dehydrogenase (SDH) and Fumarate Hydratase (FH)-Deficient Renal Cell Carcinoma
8. Birt-Hogg-Dubé (BHD) Syndrome
9. Familial MITF Microphtalmia-Associated Transcription Factor
10. Chromophobe Renal Cancer
11. Papillary Renal Carcinoma
12. Genetic Alterations of CCRCC
13. Genetic Abnormalities of Renal Medullary Carcinoma (RMC)
14. Genetic Alterations of Tubulocystic Renal Carcinoma (TCRCC)
15. Wilms Nephroblastoma
16. RCCs with Sarcomatoid (sRCC) Features
17. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2019, 69, 7. [Google Scholar] [CrossRef]
- Ferlay, J.; Steliarova-Foucher, E.; Leortet-Tieulent, J. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer 2013, 49, 1374. [Google Scholar] [CrossRef] [PubMed]
- Saad, A.M.; Gad, M.M.; Al-Husseine, M.J.; Ruhban, M.I.; Sonbol, M.B.; Ho, T.H. Trends in renal cell carcinoma incidence and mortality in the United States in the last wo decades: A SEER study. Clin. Genitourin. Cancer 2019, 17, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.S.; Goggins, W.B.; Yip, B.H.K.; Fung, F.D.H.; Leung, C.; Fang, Y.; Wong, S.Y.S.; Ng, C.F. Incidence and mortality of kidney cancer: Temporal patterns and global trends in 39 countries. Sci. Rep. 2017, 7, 15698. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO classification of tumours of the urinary system and male genital organs—Part A: Renal, penile and testicular tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef]
- Carlo, M.I.; Hakimi, A.A.; Stewart, G.D.; Bratslavsky, G.; Brugoroles, J.; Chen, Y.B.; Linehan, W.M.; Maher, E.R.; Merino, M.J.; Offit, K.; et al. Familial kidney cancer: Implications of new syndromes and molecular insights. Eur. Urol. 2019, 76, 754–764. [Google Scholar] [CrossRef]
- Shuch, B.; Vourganti, S.; Ricketts, C.J.; Middletoh, L.; Petrson, J.; Merino, M.J.; Metwalli, A.R.; Svrinivasan, R.; Linehan, W.M. Defining early-onset kidney cancer: Implications for germline and somatic mutation testing and clinical management. J. Clin. Oncol. 2013, 32, 431–437. [Google Scholar] [CrossRef]
- Carlo, M.I.; Mukherjee, S.; Mandelker, D.; Vijai, J.; Kemel, Y.; Zhang, L.; Krezecic, A.; Patil, S.; Cayan-birsoy, O.; Huang, K.C.; et al. Prevalence of germline mutations in cancer susceptibility genes in patients with advanced red cell carcinoma. JAMA Oncol. 2018, 4, 1228–1235. [Google Scholar] [CrossRef]
- Varshney, N.; Kebede, A.A.; Owusu-Dapaah, H.; Lather, J.; Kaushik, M.; Bullar, J.S. A review of Von Hippel-Lindau syndrome. J. Kidney Cancer VHL 2017, 4, 20–29. [Google Scholar] [CrossRef]
- Gossage, L.; Essen, T.; Maher, E.R. VHL, the story of a tumor suppressor gene. Nat. Rev. Cancer 2015, 15, 55–64. [Google Scholar] [CrossRef]
- Liu, Q.; Yuan, G.; Tong, D.; Liu, G.; Yi, Y.; Zhang, J.; Zhang, Y.; Wang, L.; Wang, L.; Zhang, D.; et al. Novel genotype-phenotype correlations in five Chinese families with von Hippel-Lindau disease. Endocr. Connect. 2018, 7, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Wang, J.Y.; Peng, S.H.; Li, T.; Ning, X.H.; Hong, B.A.; Liu, J.Y.; Wu, P.J.; Zhou, B.W.; Zhou, J.C.; et al. Genotype and phenotype correlation in von Hippel-Lindau disease based on alteration of the HIF-α binding site in VHL protein. Genet. Med. 2018, 20, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Maher, E.R. Hereditary renal cell carcinoma syndromes: Diagnosis, surveillance and management. World J. Urol. 2018, 36, 1891–1898. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Brunet, J.P.; Di Napoli, A.; Mertz, K.D.; Seeley, A.; Pires, M.M.; Linhart, D.; Warrell, R.A.; Moch, H.; Rubin, M.A.; et al. Patterns of gene expression and copy-number alterations in von-Hippel Lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009, 69, 4674–4681. [Google Scholar] [CrossRef]
- Fisher, R.; Horsewell, S.; Rowan, A.; Salm, M.P.; De Bruin, E.C.; Gulati, S.; Mc Granahan, N.; Stares, M.; Gerlinger, M.; Varela, I.; et al. Deverlopment of synchronous VHL syndrome tumors reveals contingencies and constraints ti tumor evolution. Genome Biol. 2014, 15, 433. [Google Scholar] [CrossRef]
- Fei, S.S.; Mitchell, A.D.; Heskett, M.B.; Vocke, C.D.; Ricketts, C.J.; Peto, M.; Whang, N.J.; Sonmez, K.; Linehan, W.M.; Spellman, P.T. Patient-specific factors influence somatic variation patterns in von Hippel-Lindau disease renal tumors. Nat. Commun. 2016, 7, 11588. [Google Scholar] [CrossRef]
- Mandriota, S.J.; Turner, K.J.; Davies, D.R.; Murray, P.G.; Morgan, N.V.; Sowter, H.M.; Wykoff, C.C.; Maher, E.R.; Harris, A.L.; Ratcliffe, P.J.; et al. HIG identifies early lesions in VHL kidneys: Evidence for site-specific tumor suppressor function in the nephron. Cancer Cell 2002, 1, 459–468. [Google Scholar] [CrossRef]
- Mitchell, T.J.; Turajilic, S.; Rowan, A.; Nicol, D.; Farmery, J.; O’Brien, T.; Martincorena, I.; Tarpey, P.; Angelopoulos, N.; Yates, L.R.; et al. Timing and landmark events in the evolution of clear cell cancer: TRACERx renal. Cell 2018, 173, 611–623. [Google Scholar] [CrossRef]
- Peng, X.; Chen, J.; Wang, J.; Peng, S.; Liu, S.; Ma, K.; Zhou, J.; Hong, B.; Zhou, B.; Zhang, J.; et al. Natural history of tumors in von Hippel-Lindau disease: A large retrospective study of Chinese patients. J. Med. Genet. 2019, 56, 380–387. [Google Scholar] [CrossRef]
- Dharmawardama, P.G.; Giubellino, A.; Bottaro, D.P. Hereditary papillary renal carcinoma type I. Curr. Mol. Med. 2004, 4, 855–868. [Google Scholar] [CrossRef]
- Schmidt, L.; Duh, F.M.; Chen, F.; Kishida, T.; Glenn, G.; Choyke, P.; Scherer, S.W.; Zhuang, Z.; Lubensky, I.; Dean, M.; et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat. Genet. 1997, 16, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, L.; Junker, K.; Nakaigawa, N.; Kinjerski, T.; Wierich, G.; Miller, M.; Lubensky, I.; Neumann, H.P.H.; Brauch, H.; Decker, J.; et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene 1999, 18, 2343–2350. [Google Scholar] [CrossRef]
- Lubensky, I.A.; Schmidt, L.; Zhuang, Z.; Meirich, G.; Pack, S.; Zambreno, N.; Walther, M.C.M.; Choyke, P.; Linehan, W.M.; Zbar, B. Hereditary and sporadic papillary renal carcinomas with c-met mutations share a distinct morphological phenotype. Am. J. Pathol. 1999, 155, 517–526. [Google Scholar] [CrossRef]
- Bentz, M.; Bergerheim, U.S.R.; Li, C.; Joos, S.; Werner, C.A.; Baudis, M.; Gnarra, T.; Merino, M.; Zbar, B.; Linehan, W.M.; et al. Chromosome imbalances in papillary renal cell carcinoma and first cytogenetic data of familial cases analyzed by comparative genomic hybridization. Cytogenet. Cell Genet. 1996, 75, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Park, W.S.; Pack, S.; Schmidt, L.; Vortmeyer, A.O.; Pak, E.; Pham, T.; Weil, R.J.; Candidus, S.; Lubensky, I.A.; et al. Trisomy 7-harbouring non-random duplication of the mutant MET allele in hereditary papillary renal carcinomas. Nat. Genet. 1998, 17, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Palmedo, G.; Von Knobloch, R.; Bugert, P.; Prayer-Galetti, T.; Pagano, F.; Kovacs, G. Duplication and overexpression of the mutant allele of the MET proto-oncogene in multiple hereditary papillary renal cell tumors. Oncogene 1998, 17, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Wadt, K.A.; Gerdes, A.M.; Hansen, T.V.; Toft, B.G.; Fziis-Hausen, L.; Andersen, M.K. Novel germline c-MET mutation in a family with hereditary papillary renal carcinoma. Fam. Cancer 2012, 11, 535–537. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Vaishimpayan, U.; Rosenberg, J.E.; Logan, T.F.; Harszatark, A.L.; Bukowski, R.M.; Rini, B.I.; Srinival, S.; Skin, M.N.; Adams, L.M.; et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor Foretinib in patients with papillary renal cell carcinoma. J. Clin. Oncol. 2013, 31, 181–186. [Google Scholar] [CrossRef]
- Lynch, E.D.; Ostermayer, E.A.; Lee, M.K. Inherited mutations in PTEN that are associated with breast cancer, Cowden disease, and juvenile polyposis. Am. J. Hum. Genet. 1997, 61, 1254. [Google Scholar] [CrossRef]
- Mester, J.L.; Zhou, M.; Prescott, N. Papillary renal cell carcinoma is associated with PTEN hamartoma tumor syndrome. Urology 2012, 79, 1187. [Google Scholar] [CrossRef]
- Shugh, B.; Ricketts, C.J.; Vocke, C.D.; Komiya, T.; Middleton, L.A.; Kauffman, E.C.; Merino, M.J.; Metwalli, A.R.; Dennis, P.; Linehan, W.M. Germline PTEN mutation Cowden syndrome: An under-appreciated form of hereditary kidney cancer. J. Urol. 2013, 190, 1990–1998. [Google Scholar] [CrossRef]
- Cavaillé, M.; Ponelle-Chachuat, F.; Urhammer, N.; Viala, S.; Gay-Balille, M.; Privat, M.; Bidet, Y.; Bignon, J.Y. Early onset multiple primary tumors in atypical presentation of Cowden syndrome identified by whole-exome-sequencing. Front. Genet. 2018, 9, 353. [Google Scholar] [CrossRef]
- Walpole, S.; Pritchard, A.L.; Cebulla, C.M.; Pilorski, R.; Stautberg, M.; Davidorf, F.H.; De la Fouchardiere, A.; Cabaret, O.; Golamrd, L.; Stoppa-Lyonnet, D.; et al. Comprehensive study of the clinical phenotype of germline BAP1 variant-carrying families worldwide. J. Natl. Cancer Inst. 2018, 110, 1328–1341. [Google Scholar] [CrossRef]
- Testa, J.R.; Cheung, M.; Pei, J.; Below, J.E.; Tan, Y.; Sementino, E.; Cox, N.J.; Dogan, A.U.; Pass, H.I.; Testa, J.R.; et al. Germline BAP1 mutations predispose to malignant mesotheliomas. Nat. Genet. 2011, 43, 1022–1025. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, T.; Obenauf, A.C.; Murali, R.; Fried, I.; Griewank, K.G.; Ulz, P.; Windpassinger, C.; Wackernagel, W.; Loy, S.; Wolf, I.; et al. Germline mutations in BAP1 predipose to melanocytic tumors. Nat. Genet. 2011, 43, 1018–1021. [Google Scholar] [CrossRef] [PubMed]
- Popova, T.; Hebert, L.; Jaquemin, V.; Gad, S.; Caux-Moncoutier, V.; Dubois-d’Enghien, C.; Richadeau, B.; Renaudin, X.; Sellers, J.; Nicolas, A.; et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am. J. Hum. Genet. 2013, 92, 974–980. [Google Scholar] [CrossRef]
- Farley, M.N.; Schmidt, L.S.; Mester, J.L.; Pena-Llopis, S.; Pavia-Jimenez, A.; Christie, A.; Vocke, C.D.; Ricketts, C.J.; Peterson, J.; Middleton, L. A novel germline mutation in Bap1 predisposes to familial clear-cell renal cell carcinoma. Mol. Cancer Res. 2013, 11, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- Ball, M.W.; An, J.Y.; Gomella, P.T.; Gautam, R.; Ricketts, C.J.; Vocke, C.D.; Schmidt, L.S.; Merino, M.J.; Srinivisan, R.; Malayeri, A.A.; et al. Growth rates of genetically defined renal tumors: Implications for active surveillance and intervention. J. Clin. Oncol. 2020, 38, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- AL-Rasheed, M.R.H.; Tarjan, G. Succinate dehydrogenase complex: An updated review. Arch. Pathol. Lab. Med. 2018, 142, 1564–1570. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.; Barnocoat, A.; Mumtaz, F.; Aitchinson, M.; Side, L.; Brittain, H.; Bates, A.; Gale, D.P. Cascade fumarate hydratase mutation screening allows early detection of kidney tumour: A case report. BMC Med. Genet. 2017, 18, 79. [Google Scholar] [CrossRef]
- Gill, A.J.; Pachter, N.S.; Chou, A.; Young, B.; Clarkson, A.S.; Tucker, K.M.; Winship, I.M.; Early, P.; Benn, D.E.; Robinson, B.G.; et al. Renal tumors associated with with germline SDHB mutation show distinctive morphology. Am. J. Surg. Pathol. 2011, 35, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Gill, A.J.; Hes, O.; Papathomas, T.; Sedivcova, M.; Tan, P.H.; Agaimy, A.; Andresen, P.A.; Kedziora, A.; Clarkson, A.; Toon, C.W.; et al. Succinate dehydrogenase (SDH)-deficient renal carcinoma: A morphologically distinct entity. A clinicopathologic series of 36 tumors from 27 patients. Am. J. Surg. Pathol. 2014, 38, 1588–1602. [Google Scholar] [CrossRef] [PubMed]
- Williamson, S.R.; Eble, J.N.; Amin, M.B.; Gupta, N.S.; Smith, S.G.; Sholl, L.M.; Montironi, R.; Hirsch, M.S.; Hornick, J.L. Succinate dehydrogenase-deficient renal cell carcinoma: Detailed characterization of 11 tumors defining a unique subtype of renal cell carcinoma. Mod. Pathol. 2015, 28, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Caliò, A.; Grignon, D.J.; Stohr, B.A.; Williamson, S.R.; Eble, J.N.; Cheng, L. Renal cell carcinoma with TFE3 translocation and succinate dehydrogenase B mutation. Mod. Pathol. 2017, 30, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Swanson, A.A.; Chen, Y.B.; Lopez, T.; Milosevic, D.; Kipp, B.R.; Leibovich, B.C.; Thompson, R.H.; Herrera-Hernandez, L.; Cheville, J.C.; et al. Incidence of succinate dehydrogenase and fumarate hydratase-deficient renal cell carcinoma based on immunohistochemical screening with SDHA/SDHB and FH/2SC. Hum. Pathol. 2019, 91, 114–122. [Google Scholar] [CrossRef]
- Ajhamir, S.M.K.; Haeshmat, R.; Ebrahimi, M.; Katabchi, S.E.; Dizaji, S.P.; Khatami, F. The impact of succinate dehydrogenase gene (SDH) mutations in renal cell carcinoma (RCC): A systematic review. OncoTarg. Ther. 2019, 12, 7929–7940. [Google Scholar] [CrossRef]
- Hakirevich, E.; Ali, S.M.; Mega, A.; McMahon, C.; Brodsky, A.S.; Ross, J.S.; Allen, J.; Elvin, J.A.; Resnick, M.B. A novel SDHA-deficient renal cell carcinoma revealed by comprehensive genomic profiling. Am. J. Surg. Pathol. 2015, 39, 858–863. [Google Scholar] [CrossRef]
- Saxena, N.; Maio, N.; Crooks, D.R.; Ricketts, C.J.; Yang, Y.; Wei, M.H.; Fan, T.; Lane, A.N.; Soubier, C.; Singh, A.; et al. SDHB-deficient cancers: The role of mutations that impair iron sulfur cluster delivery. J. Natl. Cancer Inst. 2016, 108, djv287. [Google Scholar] [CrossRef]
- Maio, N.; Singh, A.; Uhrigishardt, H.; Saxena, N.; Tong, W.H.; Rouault, T.A. Chaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014, 19, 445–457. [Google Scholar] [CrossRef]
- Cardaci, S.; Zheng, L.; MacKay, G.; Van den Broek, N.; MacKenzie, E.D.; Nixon, C.; Stevenson, D.; Tumanov, S.; Bulusu, V.; Kamphorst, J.J.; et al. Pyruvate carboxylation enables growth of SDH-deficinet cells by supporting aspartate biosynthesis. Nat. Cell Biol. 2015, 17, 1317–1326. [Google Scholar] [CrossRef]
- Lussey-Lepoutre, C.; Bellucci, A.; Morin, A.; Buffet, A.; Amar, L.; Janin, M.; Ottolenghi, C.; Zinzindohoué, F.; Autret, G.; Burnichon, N.; et al. In vivo detection of succinate by magnetic resonance spectroscopy as a hallmark of SDHx mutations in paraganglioma. Clin. Cancer Res. 2016, 22, 1120–1129. [Google Scholar] [CrossRef] [PubMed]
- Casey, R.T.; McLean, M.A.; Madhu, B.; Challis, R.G.; Ten Hoopen, R.; Roberts, T.; Clark, G.R.; Pittfield, D.; Simpson, H.L.; Bulusu, V.R.; et al. Translating in vivo metabolomic analysis of succinate dehydrogenase deficient tumors into clinical utility. JCO Precis. Oncol. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Yang, M.; Soga, T.; Pollard, P.J. Oncometabolites: Linking altered metabolism with cancer. J. Clin. Investig. 2013, 123, 3652–3658. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.; Stewart, G.D.; Frezza, C. Oncometabolites in renal cancer. Nat. Rev. Nephrol. 2019, in press. [Google Scholar] [CrossRef]
- Martinez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Selak, M.A.; Amour, B.M.; MacKenzie, E.D.; Boulahbel, H.; Watson, D.G.; Mansfield, K.D.; Pan, Y.; Simon, M.C.; Thompson, C.B.; Gottlieb, E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 2005, 7, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Pollard, P.J.; Briere, J.J.; Alam, N.A.; Barwell, J.; Barclay, E.; Wortham, N.C.; Hunt, T.; Mitchell, M.; Olpin, S.; Moat, S.J.; et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet. 2005, 14, 2231–2239. [Google Scholar] [CrossRef]
- Pollard, P.; Wotham, N.; Barclay, E.; Alam, A.; Elia, G.; Manek, S.; Poulson, R.; Tomlinson, I. Evidence of increased microvessel density and activation of the hypoxia pathway in tumours from the hereditary leiomyomatosis and renal cell cancer syndrome. J. Pathol. 2005, 205, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Letouzé, E.; Martinelli, C.; Loriot, C.; Burnichon, N.; Abemili, N.; Ottolenghi, C.; Janin, M.; Menara, M.; Nguyen, A.T.; Benit, P.; et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell 2013, 23, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Sulkowski, P.L.; Sundaram, R.K.; Oeck, S.; Corso, C.D.; Liu, Y.; Noorbakhsh, S.; Niger, M.; Boeke, M.; Ueno, D.; Kalathil, A.N.; et al. Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat. Genet. 2018, 50, 1086–1092. [Google Scholar] [CrossRef]
- Tomlinson, I.P.; Alam, N.A.; Rowan, A.J.; Barclay, E.; Jaeger, E.E.; Kelsell, D.; Laigh, I.; Groman, P.; Lamlum, H.; Rahman, S.; et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cancer. Nat. Genet. 2002, 30, 406–410. [Google Scholar] [PubMed]
- Pan, X.; Zhang, M.; Yao, J.; Zeng, H.; Nie, L.; Gong, J.; Chen, X.; Xu, M.; Zhou, Q.; Chen, N. Fumarate hydratase-deficient renal cell carcinoma: A clinicopathological and molecular study of 13 cases. J. Clin. Pathol. 2019, 72, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Linehan, W.M.; Spellman, P.T.; Ricketts, C.J.; Creighton, C.J.; Fei, S.S.; Davis, C.; Wheeler, D.A.; Murray, B.A.; Schmidt, L.; et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 2016, 374, 135–145. [Google Scholar]
- Chen, Y.B.; Xu, J.; Skanderup, J.; Dong, Y.; Brannon, A.R.; Wang, L.; Wan, H.H.; Wang, P.J.; Nanjangud, G.J.; Jungbluth, A.A.; et al. Molecular analysis of aggressive renal cell carcinoma with unclassified histology reveals distinct subsets. Nat. Commun. 2016, 7, 13131. [Google Scholar] [CrossRef]
- Wei, M.H.; Toure, O.; Glenn, G.M.; Pithukpakorn, M.; Neckers, L.; Stolle, C.; Choyke, P.; Grubb, R.; Middleton, L.; Turner, M.L.; et al. Novel mutations in FH and expansion of the spectrum of phenotypes expressed in families with hereditary leiomyomatosis and renal cell cancer. J. Med. Genet. 2006, 43, 18–27. [Google Scholar] [CrossRef]
- Muller, M.; Ferlicot, S.; Guillaud-Bataille, M.; Le Teuff, G.; Genestie, C.; Deveaux, S.; Slama, A.; Poulahon, N.; Escudier, B.; Albiges, L.; et al. Reassessing the clinical spectrum associated with hereditary leiomyomatosis and renal cell carcinoma syndrome in French FH mutation carriers. Clin. Genet. 2017, 92, 606–615. [Google Scholar] [CrossRef]
- Vocke, C.D.; Ricketts, C.J.; Merino, M.J.; Srinivasan, R.; Metwalli, A.R.; Middleton, L.A.; Peterson, J.; Yang, Y.; Linehan, W.M. Comprehensive genomic and phenotypic characterization of germline FH deletion in hereditary leiomyomatosis and renal cell carcinoma. Genes Chromosom. Cancer 2017, 56, 484–492. [Google Scholar] [CrossRef]
- Lau, H.D.; Chan, E.; Fan, A.C.; Kunder, C.A.; Williamson, C.R.; Zhou, M.; Idrees, M.T.; MacLean, F.M.; Gill, A.J.; Cao, C.S. A clinicopathologic and molecular analysis of fumarate hydratase-deficient renal cell carcinoma in 32 patients. Am. J. Surg. Pathol. 2020, 44, 98–110. [Google Scholar] [CrossRef]
- Forde, C.; Lim, D.; Alwan, Y.; Burghel, G.; Butland, L.; Cleaver, R.; Dixit, A.; Evans, D.G.; Hnason, H.; Lalloo, F.; et al. Hereditary leiomyomatosis and renal cell cancer: Clinical, molecular, and screening features in a cohort of 185 affected individuals. Eur. Urol. Oncol. 2020, in press. [Google Scholar] [CrossRef]
- Furuya, M.; Iribe, Y.; Nagashima, Y.; Kambe, N.; Ohe, C.; Kinoshita, H.; Sato, C.; KishidA, T.; Okubo, Y.; Nakamura, K.; et al. Clinicopathologic and molecular features of hereditary leiomyomatosis and renal cell cancer-associated renal cell carcinomas. J. Clin. Pathol. 2020, in press. [Google Scholar] [CrossRef]
- Hol, J.A.; Jongmans, M.C.J.; Littooij, A.S.; De Krjger, R.R.; Kuiper, R.P.; Van Harssel, J.J.T.; Mensenkamp, A.; Simons, M.; Tytgat, G.A.M.; Van Den Heuvel-Eibrink, M.M.; et al. Renal cell carcinoma in young FH mutation carriers: Case series and review of the literature. Fam. Cancer 2020, 19, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Valera, V.A.; Padilla-Nash, H.M.; Sourbier, C.; Vocke, C.D.; Vira, M.A.; Abu-Asab, M.S.; Bratslavsky, G.; Tsokos, M.; Merino, M.J.; et al. UOK 262 cell line, fumarate hydratase deficient (FH-/FH.) hereditary leiomyomatosis renal cell carcinoma: In vitro and in vivo model of an aberrant energy metabolic pathway in human cancer. Cancer Genet. Cytogenet. 2010, 196, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Mullen, A.R.; Wheaton, W.W.; Jin, E.S.; Chen, P.H.; Sullivan, L.B.; Cheng, T.; Yang, Y.; Linehan, W.M.; Chandel, N.S.; DeBernardinis, R.J.; et al. Reductive carboxylation supports growth in tumor cells with defective mitochondria. Nature 2011, 481, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Valera, V.; Sourbier, C.; Vocke, C.D.; Wei, M.; Pike, L.; Huang, Y.; Merino, M.A.; Bratslavsky, G.; Wu, M.; et al. A novel fumarate hydratase-deficient HLRCC kidney cancer cell line, UOK268: A model of the Warburg effect in cancer. Cancer Genet. 2012, 205, 377–390. [Google Scholar] [CrossRef]
- Yang, Y.; Lane, A.N.; Ricketts, C.J.; Sourbier, C.; Wei, M.H.; Shuch, B.; Pike, L.; Wu, M.; Rouault, T.A.; Boros, L.G.; et al. Metabolic reprogramming for producing energy and reducing power in fumarate hydratase null cells from hereditary leiomyomatosis renal cell carcinoma. PLoS ONE 2013, 8, e72179. [Google Scholar] [CrossRef]
- Tong, W.H.; Sourbier, C.; Kovtunovych, G.; Geong, S.Y.; Vira, M.; Ghosh, M.; Romero, V.V.; Sougrat, R.; Vaulont, S.; Viollet, B.; et al. The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell 2011, 20, 315–327. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Gonçalves, E.; Johnson, T.I.; Zecchini, V.R.; Henriques da Costa, A.S.; Gaude, E.; Drubbel, A.V.; Theobald, S.J.; Abbo, S.; Tran, M.; et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 2016, 537, 544–547. [Google Scholar] [CrossRef]
- Mackenzie, E.D.; Selak, M.A.; Tennant, D.A.; Payne, L.J.; Crosby, S.; Frederiksen, C.M.; Watson, D.G.; Gottlieb, E. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate-dehydrogenase-deficient cells. Mol. Cell Biol. 2007, 27, 3282–3289. [Google Scholar] [CrossRef]
- Alderson, N.L.; Wang, Y.; Blatnik, M.; Frizzell, N.; Walla, M.D.; Lyons, T.J.; Alt, N.; Carson, J.A.; Nagai, R.; Thorpe, S.R.; et al. S-(2-succinyl) cysteine: A novel chemical modification of tissue proteins by a Krebs cycle intermediate. Arch. Biochem. Biophys. 2006, 450, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bardella, C.; El-Bahrawy, M.; Frizzell, N.; Adam, J.; Ternette, N.; Hatipoglu, E.; Howarth, K.; O’Flaherty, L.; Roberts, I.; Turner, G.; et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J. Pathol. 2011, 225, 4–11. [Google Scholar] [CrossRef]
- Tyrakis, P.A.; Yurkovich, M.E.; Sciacovelli, M.; Papachristou, E.K.; Bridges, H.R.; Gaude, E.; Schreiner, A.; D’Santos, C.; Hirst, J.; Hernandez-Fernaud, J.; et al. Fumarate hydratase loss causes combined respiratory chain defects. Cell Rep. 2017, 21, 1036–1047. [Google Scholar] [CrossRef]
- Ooi, A.; Wong, J.C.; Petillo, D.; Roossien, D.; Perrier-Trudova, V.; Whitten, D.; Wong Hui Min, B.; Tan, M.H.; Zhang, Z.; Yang, X.J.; et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 2011, 20, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Adam, J.; Hapitoglu, E.; O’Falherty, L.; Ternette, N.; Sahgal, N.; Lockstone, H.; Baban, D.; Nye, E.; Stamp, G.W.; Wolhuter, K.; et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: Roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 2011, 20, 524–537. [Google Scholar] [CrossRef]
- Ooi, A.; Dykema, K.; Ansari, A.; Petillo, D.; Snider, J.; Kahnoski, R.; Anema, J.; Craig, D.; Carpten, J.; Teh, B.T.; et al. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res. 2013, 73, 2044–2051. [Google Scholar] [CrossRef]
- Sullivan, L.B.; Martinez-Garcia, E.; Nguyen, H.; Mullen, A.R.; Dufour, E.; Sudarshan, S. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol. Cell 2013, 51, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Cardaci, S.; Jerby, L.; MacKenzie, E.D.; Sciacovelli, M.; Johnson, T.I. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat. Commun. 2015, 6, 6001. [Google Scholar] [CrossRef]
- Kulkarni, R.A.; Bak, D.W.; Wei, D.; Bergholtz, S.E.; Briney, C.A.; Shrimps, J.H.; Alpsoy, A.; Thorpe, A.L.; Bavari, A.E.; Crooks, D.R.; et al. A chemoproteomic portrait of the oncometabolite fumarate. Nat. Chem. Biol. 2019, 15, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Sourbier, C.; Ricketts, C.J.; Liao, P.J.; Matsumoto, S.; Wei, D.; Lang, M.; Railkar, R.; Yang, Y.; Wei, M.H.; Agarwal, P.; et al. Proteasome inhibition disrupts the metabolism of fumarate hydratase-deficient tumors by downregulating p62 and c-Myc. Sci. Rep. 2019, 9, 18409. [Google Scholar] [CrossRef] [PubMed]
- Zbar, B.; Alvord, W.G.; Glenn, G.; Turner, M.; Pavlovich, C.P.; Schmidt, L.; Walther, M.C.; Choyke, P.; Weirich, G.; Hewitt, S.M.; et al. Risk of renal and colonic neoplasms and spontaneous pneumothorax in the Birt-Hogg-Dubé syndrome. Cancer Epidemiol. Biomark. Prev. 2002, 11, 393–400. [Google Scholar]
- Pavlovich, C.P.; Walther, M.C.; Eyler, R.A.; Hewitt, S.M.; Zbar, B.; Linehan, M.; Merino, M.J. Renal tumors in the Birt-Hogg-Dubé syndrome. Am. J. Surg. Pathol. 2002, 26, 1542–1552. [Google Scholar] [CrossRef]
- Schmidt, L.S.; Linehan, W.M. Molecular genetics and clinical features of Birt-Hogg-Dubé-syndrome. Nat. Rev. Urol. 2015, 12, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Adami, A.; Lowrance, W.T.; Yee, D.S.; Chong, K.T.; Bernstein, M.; Tickoo, S.K.; Coleman, J.A.; Russo, P. Renal oncocytosis: Management and clinical outcomes. J. Urol. 2011, 185, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Furuya, M.; Hasumi, H.; Yao, M.; Nagashima, Y. Birt-Hogg-Dubé syndrome-associated renal cell carcinoma: Histopathological features and diagnostic conundrum. Cancer Sci. 2019, 111, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, L.S.; Warren, M.B.; Nickerson, M.L.; Weirich, G.; Matrasova, V.; Toro, J.R.; Turner, M.L.; Duray, P.; Merino, M.; Hewitt, S.; et al. Birt-Hogg-Dubé syndrome, a genodermatosis associated with spontaneous pneumothorax and kidney neoplasia, maps to chromosome 17p11.2. Am. J. Hum. Genet. 2001, 69, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, M.L.; Warren, B.; Toro, J.R.; Matrosova, V.; Glenn, G.; Turner, M.L.; Duray, P.; Merino, M.; Choyke, P.; Pavlovich, C.P.; et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dubé syndrome. Cancer Cell 2002, 2, 157–164. [Google Scholar] [CrossRef]
- Schmidt, L.S.; Linehan, W.M. FLCN: The causative gene for Birt-Hogg-Dubé syndrome. Gene 2018, 640, 28–42. [Google Scholar] [CrossRef]
- Vocke, C.D.; Yang, Y.; Pavlovich, C.P.; Schmidt, L.S.; Nicherson, M.L.; Torres-Cabala, C.A.; Merino, M.J.; Walther, M.M.; Zbar, B.; Linehan, W.M.; et al. High frequency of somatic frameshift BHD gene mutations in Birt-Hogg-Dubé-associated renal tumors. J. Natl. Cancer Inst. 2005, 97, 931–935. [Google Scholar] [CrossRef]
- Hasumi, H.; Hasumi, Y.; Baba, M.; Nishi, H.; Furuya, M.; Vocke, C.D.; Lang, M.; Irie, N.; Esumi, C.; Merino, M.J.; et al. H255Y and K580R missense mutations in tumour suppressor folliculin (FLCN) promote kidney cell proliferation. Hum. Mol. Genet. 2017, 26, 354–366. [Google Scholar]
- Kato, I.; Iribe, Y.; Nagashima, Y.; Kuroda, N.; Tanaka, R.; Nakatani, Y.; Hasumi, H.; Yao, M.; Furuya, M. Fluorescent and chromogenic in situ hybridization of CEN17q as a potent useful diagnostic marker for Birt-Hogg-Dubé syndrome-associated chromophobe renal cell carcinomas. Hum. Pathol. 2016, 52, 74–82. [Google Scholar] [CrossRef]
- Hasumi, H.; Furuya, M.; Tatsuno, K.; Yamamaoto, S.; Baba, M.; Hasumi, Y.; Isono, Y.; Suzuki, K.; Jikuya, R.; Otake, S.; et al. BHD-associated kidney cancer exhibits unique molecular characteristics and a wide variety of variants in chromatin remodeling genes. Hum. Mol. Genet. 2018, 27, 2712–2724. [Google Scholar] [CrossRef]
- Ruiz-Cordero, R.; Rao, P.; Li, L.; Qi, Y.; Atherton, D.; Peng, B.; Singh, R.R.; Kim, T.B.; Kawakami, F.; Routbort, M.J.; et al. Hybrid oncocytic/chromophobe renal tumors are molecularly distinct from oncocytoma and chromophobe renal cell carcinoma. Mod. Pathol. 2019, 32, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.; Hong, S.B.; Sharma, N.; Warren, M.B.; Nickerson, M.L.; Iawamatsu, A. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 15552–15557. [Google Scholar] [CrossRef] [PubMed]
- Hasumi, H.; Baba, M.; Hong, S.B.; Hasumi, Y.; Huang, Y.; Yao, M. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene 2008, 415, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Takagi, Y.; Kobayashi, T.; Shiono, M.; Wang, L.; Piao, X.; Sun, G. Interaction of folliculin (Birt-Hogge-Dubé gene product) with a novel Fnip1-like (PnipL/Fnip2) protein. Oncogene 2008, 27, 5339–5347. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, R.E.; Fromm, S.A.; Fu, Y.; Yokom, A.L.; Kim, D.J.; Thelen, A.M. Structural mechanism of a rag GTPase activation checkpoint by the lysosomal folliculin complex. Science 2019, 366, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Rogala, K.R.; Chou, H.T.; Huang, R.K.; Yu, Z.; Sabatini, D.M. Cryo-EM structure of the human FLCN-FNIP2-Rag-regulator complex. Cell 2019, 179, 1319–1329. [Google Scholar] [CrossRef]
- Hasumi, H.; Baba, M.; Hasumi, Y.; Lang, M.; Huang, Y.; Oh, H.F.; Matsuo, M.; Merino, M.J.; Yao, M.; Ito, Y.; et al. Folliculin-interacting proteins Fnipo1 and Pnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc. Natl. Acad. Sci. USA 2015, 112, E1624–E1631. [Google Scholar] [CrossRef]
- Rabanal-Ruiz, Y.; Korolchuk, V.I. MTORC1 and nutrient homeostasis: The central role of the lysosome. Int. J. Mol. Sci. 2018, 19, 818. [Google Scholar] [CrossRef]
- Baba, M.; Furihata, M.; Hong, S.B.; Tessarollo, L.; Haines, D.C.; Southon, E. Kidney-targeted Birt-Hogg-Dubé, gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J. Natl. Cancer Inst. 2008, 100, 40–54. [Google Scholar] [CrossRef]
- Chen, J.; Futami, K.; Petillo, D.; Peng, J.; Wang, P.; Knol, J. Deficiency of FLCN in mouse kidney led to development of polycystic kidneys and renal neoplasia. PLoS ONE 2008, 3, e3581. [Google Scholar] [CrossRef]
- Chen, J.; Huang, D.; Rubera, I.; Futami, K.; Wang, P.; Zickert, P. Disruption of tubular Flcn expression as a mouse model for renal tumor induction. Kidney Int. 2015, 88, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Hasumi, Y.; Baba, M.; Ajima, R.; Hasumi, H.; Valera, V.A.; Klein, M.E.; Haines, D.C.; Merino, M.J.; Hong, S.B.; Yamaguchi, T.P.; et al. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc. Natl. Acad. Sci. USA 2009, 106, 18722–18727. [Google Scholar] [CrossRef] [PubMed]
- Hartman, T.R.; Klein-Szanto, A.; Al-Saleem, T.; Cash, T.P.; Simon, M.C.; Henske, E.P. The role of the Birt-Hogg-Dubé protein in mTOR activation and renal tumorigenesis. Oncogene 2009, 28, 1594–1604. [Google Scholar] [CrossRef]
- Hudon, V.; Sabourin, S.; Dydensborg, A.B.; Kottuis, V.; Ghazi, A.; Paquet, M.; Crosby, K.; Pomerleau, V.; Ueatani, N.; Pause, A. Renal tumour suppressor function of the Birt-Hogg-Dubé syndrome gene product folliculin. J. Med. Genet. 2010, 47, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Si, S.; Li, Y.; Schoen, S.; Xiao, G.Q.; Li, X.; Teh, B.T.; Wu, G.; Chen, J. Plcn-deficient renal cells are tumorigenic and sensitive to mTOR suppression. Oncotarget 2015, 6, 32761–32773. [Google Scholar] [CrossRef]
- De Martin Garrido, N.; Aylett, C. Nutrient signaling and lysosome positioning crosstalk through a multifunctional protein folliculin. Front. Cell Dev. Biol. 2020, 8, 108. [Google Scholar] [CrossRef]
- Shen, K.; Huang, R.K.; Brignole, E.J.; Kondon, K.J.; Valenstein, M.L.; Chantranupong, L. Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature 2018, 556, 64–69. [Google Scholar] [CrossRef]
- Meng, J.; Ferguson, S.M. GATOR1-dependent recruitment of FLCN-FNIP to lysosomes coordinates rag GTPase heterodimer nucleotide status in response to amino acids. J. Cell. Biol. 2017, 217, 2765–2776. [Google Scholar] [CrossRef]
- Possik, E.; Jalali, S.; Nouet, Y.; Yan, M.; Gingras, M.C.; Schmeisser, K. Folliculin regulates AMPK-dependent autophagy and metabolic stress survival. PLoS Genet. 2014, 10, e1004273. [Google Scholar] [CrossRef]
- Yan, M.; Gingras, M.C.; Dunlop, E.A.; Nouet, Y.; Dupuy, F.; Jalali, J.A. The tumor suppressor folliculin regulates AMPK metabolic transformation. J. Clin. Investig. 2014, 124, 2640–2650. [Google Scholar] [CrossRef]
- El-Houjeiri, L.; Possik, E.; Vijayaraghavan, T.; Paquette, M.; Martina, J.A.; Kazan, J.M. The transcription factors TFEB and TFE3 link the FLCN-AMPK signaling axis to innate immune response and pathogen resistance. Cell Rep. 2019, 26, 3613–3628. [Google Scholar] [CrossRef] [PubMed]
- Siggs, O.M.; Stockenhuber, A.; Deobagkar-Lele, M.; Bull, K.R.; Crockford, T.L.; Kingston, B.L.; Crawford, G.; Anzilotti, C.; Steeples, V.; Ghaffari, S.; et al. Mutation of FNIP1 is associated with B-cell deficiency, cardiomyopathy, and elevated AMPK activity. Proc. Natl. Acad. Sci. USA 2016, 113, E3706–E3715. [Google Scholar] [CrossRef]
- Hasumi, H.; Baba, M.; Hasumi, Y.; Huang, Y.; Oh, H.; Hughes, R.M.; Klein, M.E.; Takikita, S.; Nagashima, K.; Schmidt, L.S.; et al. Regulation of mitochondrial oxidative metabolism by tumor suppressor FLCN. J. Natl. Cancer Inst. 2012, 104, 1750–1764. [Google Scholar] [CrossRef] [PubMed]
- Klomp, J.A.; Petillo, D.; Niemi, N.M.; Dykema, K.J.; Chen, J.; Yang, X.J.; Saaf, A.; Zickert, P.; Aly, M.; Bewrgerheim, U.; et al. Birt-Hogg-Dubé renal tumors are genetically distinct from other renal neoplasias and are associated with up-regulation of mitochondrial gene expression. BMC Med. Genom. 2010, 3, 59. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Sudet-Walsh, E.; Manteghi, S.; Dufour, C.R.; Walker, B.; Baba, M.; St.Pierre, J.; Giguère, V.; Pause, A. Chronic AMPK activation via loss of FLCN induces functional beige adipose through PGC-1α/ERRα. Genes Dev. 2016, 30, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- Bertolotto, C.; Lesuer, F.; Giuliano, S.; Strub, T.; De Lichy, M.; Bille, K.; Dessen, P.; D’Haver, B.; Mohamdi, H.; Remenieras, A.; et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011, 480, 94–98. [Google Scholar] [CrossRef]
- Stoehr, C.G.; Walter, B.; Denzinger, S.; Ghiorzo, P.; Sturm, R.A.; Hinze, R.; Moch, H.; Junker, K.; Hartmann, A.; Stoehr, R. The microphtalmia-associated transcription factor p.E318K mutation does not play a major role in sporadic renal cell tumors from Caucasian patients. Pathobiology 2016, 85, 165–169. [Google Scholar] [CrossRef]
- Davis, C.F.; Ricketts, C.J.; Wang, M.; Yang, L.; Cherniack, A.D.; Shen, H.; Buhay, C.; Kang, H.; Kim, S.C.; Fahey, C.C.; et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014, 26, 319–330. [Google Scholar] [CrossRef]
- Martignoni, G.; Pea, M.; Chilosi, M.; Brunelli, M.; Scarpa, A.; Colato, C.; Tardanico, R.; Zamboni, G.; Bonetti, F. Parvalbumin is constantly expressed in chromophobe renal carcinoma. Mod. Pathol. 2001, 14, 760–767. [Google Scholar] [CrossRef]
- Speicher, M.R.; Schoell, B.; Du Manoir, S.; Schrock, E.; Ried, T.; Cremer, T.; Storkel, S.; Kovacs, A.; Kovacs, G. Specific loss of chromosomes 1, 2, 6, 10, 13, 17 and 21 in chromophobe renal cell carcinomas revealed by comparative genomic hybridization. Am. J. Pathol. 1994, 145, 356–364. [Google Scholar]
- Brunelli, M.; Eble, J.N.; Zhang, S.; Martignoni, G.; Delahunt, B.; Cheng, L. Eosinophilic and classic chromophobe renal cell carcinomas have similar frequent losses of multiple chromosomes from among chromosomes 1, 2, 6, 10, and 17, and this pattern of genetic abnormality is not present in renal oncocytoma. Mod. Pathol. 2005, 18, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, R.; Schraml, P.; Angori, S.; Batavia, A.A.; Rupp, N.J.; Ohe, C.; Otsuki, Y.; Kawasaki, T.; Kobayashi, H.; Kobayashi, K.; et al. Classic chromophobe renal cell carcinoma incur a larger number of chromosomal losses than seen in the eosinophilic subtype. Cancers 2019, 11, 1492. [Google Scholar] [CrossRef] [PubMed]
- Tam, M.H.; Wong, C.F.; Tan, H.L.; Yang, X.J.; Ditlev, J.; Matsuda, D.; Khoo, S.K.; Sugimura, J.; Fujioka, T.; Furge, K.A.; et al. Genomic expression and single-nucleotide polymorphism discriminates chromophobe renal cell carcinoma and oncocytoma. BMC Cancer 2010, 10, 196. [Google Scholar]
- Brunelli, M.; Gobbo, S.; Cossu-Rocca, P.; Cheng, L.; Hse, O.; Delahunt, B.; Pea, M.; Bonetti, F.; Mina, M.M.; Ficarra, V.; et al. Chromosomal gains in the sarcomatoid transformation of chromophobe renal cell carcinoma. Mod. Pathol. 2007, 20, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Cornejo, K.M.; Cheng, L.; Hutchinson, L.; Wang, M.; Zhang, S.; Tomaszewicz, K.; Cosar, E.F.; Woda, B.A.; Jiang, Z. Next-generation sequencing to detect deletion of RB1 and ERBB4 genes in chromophobe renal cell carcinoma. A potential role in distinguishing chromophobe renal cell carcinoma from renal oncocytoma. Am. J. Pathol. 2018, 188, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Stawiski, E.W.; Pavia-Jimenez, A.; Modrusan, Z.; Kapur, P.; Jaiswal, B.S.; Zhang, N.; Tofessi-Tcheuyap, V.; Nguyen, T.T.; Pabujia, K.B.; et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat. Genet. 2015, 47, 13–21. [Google Scholar] [CrossRef]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Berookhim, R.; et al. The cancer genome atlas comprehensive molecular characterization of renal carcinoma. Cell Rep. 2018, 23, 313–326. [Google Scholar] [CrossRef]
- Casuscelli, J.; Weinhold, N.; Gundem, G.; Wung, L.; Zaboor, E.C.; Drill, E.; Wang, P.I.; Nanjangud, G.J.; Redzematovic, A.; Nargund, A.M.; et al. Genomic landscape and evolution of metastatic chromophobe and renal cell carcinoma. JCI Insight 2017, 2, e92688. [Google Scholar] [CrossRef]
- Pan, C.C.; Chen, P.C.H.; Chiang, H. Overexpression of KIT (CD117) in chromophobe renal cell carcinoma and renal oncocytoma. Am. J. Clin. Pathol. 2004, 121, 878–883. [Google Scholar] [CrossRef]
- Przybycin, C.G.; Cronin, A.M.; Darvishian, F.; Gopalan, A.; Ll-Ahmadie, H.A.; Fine, S.W.; Chen, Y.B.; Bernstein, M.; Russo, P.; Reuter, V.E.; et al. Chromophobe renal cell carcinoma: A clinicopathologic study of 203 tumors in 200 patients with primary resection at a single institution. Am. J. Surg. Pathol. 2011, 35, 962–970. [Google Scholar] [CrossRef]
- Volpe, A.; Novara, G.; Antonelli, A.; Bertini, R.; Billia, M.; Carmignani, G.; Cunico, S.C.; Longo, N.; Martignoni, G.; Minervini, A.; et al. Chromophobe renal cell carcinoma (RCC): Oncological outcomes and prognostic factors in a large multicentre series. BJU Int. 2012, 110, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.M.; Ruiz Morales, J.M.; Donskov, F.; Fraccon, A.; Basso, U.; Rini, B.I.; Lee, J.L.; Bjarnason, J.A.; Sim, H.W.; Beuselinck, B.; et al. Outcomes of metastatic chromophobe renal cell carcinoma (chrRCC) in the targeted therapy era: Results from the international metastatic renal cell cancer database consortium (IMDC). Kidney Cancer 2017, 1, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Ged, Y.; Chen, Y.B.; Knezevic, A.; Casuscelli, J.; Redzematovic, A.; DiNatale, R.G.; Carlo, M.I.; Lee, C.H.; Feldman, D.R.; Patil, S.; et al. Metastatic chromophobe renal cell carcinoma: Presence or absence of sarcomatoid differentiation determines clinical course and treatment outcomes. Clin. Genitourin. Cancer 2019, 17, e678–e688. [Google Scholar] [CrossRef] [PubMed]
- Casuscelli, J.; Becerra, M.F.; Seier, K.; Menley, B.J.; Benfants, N.; Redzematovic, A.; Stief, C.G.; Hsieh, J.J.; Tichoo, S.K.; Reuter, V.E.; et al. Chromophobe renal cell carcinoma: Results from a largeinstitution series. Clin. Genitourin. Cancer 2019, 17, 373–379. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17009. [Google Scholar] [CrossRef]
- Delahunt, B.; Eble, J.N. Papillary renal cell carcinoma: A clinicopathologic and immunohistochemical study of 105 tumors. Mod. Pathol. 1997, 10, 537–544. [Google Scholar]
- Lee, B.H. Commentary on: “Comprehensive molecular characterization of papillary renal-cell carcinoma.” Cancer genome atlas research network.: N Eng J Med 2016 Jan 374(2): 135–142. Urol. Oncol. 2017, 35, 578–579. [Google Scholar] [CrossRef]
- Jiang, F.; Richter, J.; Schraml, P.; Budendorf, L.; Gasser, T.; Sauter, G.; Mihatsch, M.J.; Moch, H. Chromosomal imbalances in papillary renal cell carcinoma: Genetic differences between histological subtypes. Am. J. Pathol. 1998, 153, 1467–1473. [Google Scholar] [CrossRef]
- Akhtar, M.; Al-Bozom, I.A.; Al Hussain, T. Papillary ernal cell carcinoma (PRCC): An update. Adv. Anat. Pathol. 2019, 26, 124–132. [Google Scholar] [CrossRef]
- Ooi, A. Advances in hereditary leiomyomatosis and renal cell carcinoma (HLRCC) research. Semin. Cancer Biol. 2020, 61, 158–166. [Google Scholar] [CrossRef]
- Pal, S.K.; Ali, S.M.; Yakirevich, E.; Geynisman, D.M.; Karam, J.A.; Elvin, J.A.; Frampton, G.M.; Huang, X.; Lin, D.I.; Rosenzweig, M.; et al. Characterization of clinical cases of advanced papillary renal cell carcinoma via comprehensive genomic profiling. Eur. Oncol. 2018, 73, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Such, B.M.; Gerstein, M.B. Whole-genome analysis of papillary kidney cancer finds significant noncoding alterations. PLoS Genet. 2017, 13, e1006685. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Poeta, M.L.; Costantini, M.; Zhang, T.; Shi, J.; Sentinelli, S.; Zhao, W.; Pompeo, V.; Cardelli, M.; Alexandrov, B.S.; et al. The genomic and epigenomic evolutionary history of papillary renal cell carcinomas. BioRxiv 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Vera-Badillo, F.E.; Templeton, A.J.; Duran, I.; Ocana, A.; De Gouveia, P.; Aneja, P.; Knox, J.J.; Tannock, I.F.; Escudier, B.; Amir, E. Systemic therapy for non-clear cell renal cell carcinomas: A systematic review and meta-analysis. Eur. Urol. 2015, 67, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.C.; Donskov, F.; Fraccon, A.P.; Pasini, F.; Bjarnason, G.A.; Beuselinck, B.; Knox, J.J.; Rha, S.Y.; Agarwal, N.; Bowman, I.A.; et al. Characterizing the outcomes of metastatic papillary renal cell carcinoma. Cancer Med. 2017, 6, 902–909. [Google Scholar] [CrossRef]
- Rhoades Smith, K.E.; Bilen, M.A. A review of papillary renal cell carcinoma and MET inhibitors. Kidney Cancer J. 2019, 3, 151–161. [Google Scholar] [CrossRef]
- Campbell, M.T.; Bilen, M.A.; Shah, A.Y.; Leruke, E.; Jouasch, E.; Venka, A.M.; Aetinmakas, E.; Duran, C.; Msaouel, P.; Tannir, N.M. Cabozantinib for the treatment of patients with metastatic non-clear renal cell carcinoma: A retrospective analysis. Eur. J. Cancer 2018, 104, 188–194. [Google Scholar] [CrossRef]
- Chanza, N.M.; Xie, W.; Bilen, M.A.; Dzimitrowicz, H.; Burkert, J.; Geynisman, D.M.; Balakrishnan, A.; Bowman, I.A.; Jain, R.; Stadler, W.; et al. Cabozantinib in advanced non-clear-cell renal cell carcinoma: A multicentre, retrospective, cohort study. Lancet Oncol. 2019, 20, 581–590. [Google Scholar] [CrossRef]
- Choueiri, T.K.; Heng, D.; Lee, J.L.; Cancel, M.; Verheijen, R.B.; Mellemgaard, A.; Ottesen, L.H.; Frigault, M.M.; L’Hernault, A.; Szijgyarto, Z.; et al. Efficacy of savolitinib vs sunitinib in patients with MET-driven papillary renal cell carcinoma: The SAVOIR phase 3 randomized clinical trial. JAMA Oncol. 2020, in press. [Google Scholar] [CrossRef]
- Powles, T.; Larkin, J.; Patel, P.; Perez-Valderrama, B.; Rodriguez-Vida, A.; Glen, H.; Thistlethwaite, F.; Ralph, C.; Srinivasan, G.; Mendez-Vidal, M.J.; et al. A phase II study investigating the safety and efficacy of savolitinib and durvalumab in metastatic papillary renal cancer (CALYPSO). J. Clin. Oncol. 2019, 37, 545. [Google Scholar] [CrossRef]
- Gnarra, J.R.; Tory, K.; Weng, Y.; Schmidt, L.; Wei, M.H.; Li, H.; Latif, F.; Liu, S.; Chen, F.; Duh, F.M.; et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 1994, 7, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Gallou, C.; Joly, D.; Méjean, A.; Staroz, F.; Martin, N.; Tarlet, G.; Orfanelli, M.T.; Bouvier, R.; Droz, D.; Chrétien, C.; et al. Mutations of the VHL gene in sporadic renal cell carcinoma: Definition of a risk factor for VHL patients to develop an RCC. Hum. Mutat. 1999, 13, 464–475. [Google Scholar] [CrossRef]
- Schrmal, P.; Struckman, K.; Hatz, F.; Sonnet, S.; Kully, C.; Gasser, T.; Sauter, G.; Mihatsch, M.J.; Moch, H. VHL mutations and their correlation with tumour cell proliferation, microvessel density, and patient prognosis in clear cell renal cell carcinoma. J. Pathol. 2002, 196, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.G.; Latif, F.; Weng, Y.; Lerman, M.I.; Zbar, B.; Liu, S.; Samod, D.; Duan, D.S.; Gnarra, J.R.; Linehan, W.M.; et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl. Acad. Sci. USA 1994, 91, 9700–9704. [Google Scholar] [CrossRef]
- Varela, I.; Tarpey, P.; Raine, K.; Huang, D.; Ong, C.K.; Stephens, P.; Davies, H.; Jones, D.; Lin, M.L.; Teague, J.; et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011, 469, 539–542. [Google Scholar] [CrossRef]
- Dalgliesh, G.L.; Furge, K.; Greenman, C.; Chen, L.; Bignell, G.; Butler, A.; Davies, H.; Edlins, S.; Hardy, C.; Latimer, C.; et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2000, 463, 360–363. [Google Scholar] [CrossRef]
- Pena-Lopis, S.; Vega-Rubin-de-Celis, S.; Liao, A.; Leng, N.; Pavia-Jimenez, A.; Wang, S.; Yamasaki, T.; Zhrebker, L.; Sivanand, S.; Spence, P.; et al. BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 2012, 44, 751–759. [Google Scholar] [CrossRef]
- Guo, G.; Gui, Y.; Gao, S.; Tang, A.; Hu, X.; Huang, Y.; Jia, W.; Li, Z.; He, M.; Sun, L.; et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat. Genet. 2011, 44, 17–19. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshizato, T.; Shiraishi, Y.; Maekawa, S.; Okuno, Y.; Kamura, T.; Shimamura, T.; Sato.Otsubo, A.; Nagae, G.; Suzuki, H.; et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013, 45, 860–867. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–48. [Google Scholar] [CrossRef]
- Li, L.; Shen, C.; Nakamura, E.; Ando, K.; Signoretti, S.; Beroukhim, R.; Cowley, G.S.; Lizotte, P.; Liberzon, E.; Bair, S.; et al. SQSTM1 is a pathogenic target of 5q copy number gains in kidney carcer. Cancer Cell 2013, 24, 738–750. [Google Scholar] [CrossRef] [PubMed]
- Tippu, Z.; Au, L.; Turajilic, S. Evolution of renal cell carcinoma. Eur. Urol. Focus 2020, 29, S2405–S4569. [Google Scholar] [CrossRef] [PubMed]
- Psutka, S.P.; Cheville, J.C.; Costello, B.A.; Stewart-Merrill, S.B.; Lohse, C.M.; Leibovich, B.C.; Boorijian, S.A.; Thompson, R.H. Concordance of pathologic features between metastatic sites and the primary tumor in surgically resected metastatic renal cell carcinoma. Urology 2016, 96, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.H.; Parasramka, M. Differential gene expression of matched primary renal clear cell carcinoma and metastases reveals upregulation of extracellular matrix genes. Annals of Oncology 2017, 28, 604–610. [Google Scholar] [CrossRef]
- De Velasco, G.; Wankowicz, S.A.; Madison, R.; Ali, S.M.; Norton, C.; Duquette, A.; Ross, J.S.; Bossé, D.; Lalani, A.K.; Miller, V.A.; et al. Targeted genomic landscape of metastases compared to primary tumours in clear cell metastatic renal cell carcinoma. Br. J. Cancer 2018, 118, 1238–1242. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Matthews, N.; Stewart, A.; Tarpey, P.; Varela, I.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Gerlinger, M.; Horswell, S.; Larkin, J.; Rowan, A.J.; Salm, M.P.; Varela, I.; Fisher, R.; McGranahan, N.; Matthews, N. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014, 46, 225–233. [Google Scholar] [CrossRef]
- Turajilic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Horswell, S.; Chambers, T.; O’Brien, T.; Lopez, J.I.; Watkins, T.; Nicol, D.; et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 2018, 173, 595–610. [Google Scholar] [CrossRef]
- Turajilic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Chambers, T.; Lopez, J.I.; Nicol, D.; O’Brien, T.; Larkin, J.; Horswell, S.; et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 2018, 173, 581–594. [Google Scholar]
- Huang, Y.; Wang, J.; Jia, P.; Li, X.; Pei, G.; Wang, C.; Fang, X.; Zhao, Z.; Cai, Z.; Yi, X.; et al. Clonal architectures predict clinical outcome in clear cell renal cell carcinoma. Nat. Commun. 2019, 10, 1245. [Google Scholar] [CrossRef]
- Clark, D.J.; Dhaansekaran, S.M.; Petralia, F.; Pan, J.; Song, X.; Hu, Y.; Da Veiga Leprevost, F.; Reva, B.; Lih, T.S.; Chang, H.Y. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 2019, 179, 964–983. [Google Scholar] [CrossRef] [PubMed]
- Hakimi, A.A.; Reznik, E.; Lee, C.H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An integrated metabolic atlas of clear cell renal carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Kotecha, R.R.; Motzer, R.J.; Voss, M.H. Towards individualized therapy for metastatic renal cell carcinoma. Nat. Rev. Clin. Oncol. 2019, 16, 621–633. [Google Scholar] [CrossRef]
- Carill-Ajuria, L.; Santos, M.; Roldàn-Romero, J.M.; Rodriguez-Antona, C.; De Velasco, G. Prognostic and predictive value of PBRM1 in clear cell renal cell carcinoma. Cancers 2020, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Voss, M.H.; Reising, A.; Cheng, Y.; Patel, P.; Marker, M.; Kuo, F.; Chan, T.A.; Choueiri, T.K.; Hsieh, J.J.; Hakimi, A.A.; et al. Genomically annotated risk model for advanced renal-cell carcinoma: A retrospective cohort study. Lancet Oncol. 2018, 19, 1688–1698. [Google Scholar] [CrossRef]
- McDermott, D.F.; Huseni, M.A.; Atkins, M.B.; Motzer, R.J.; Rini, B.I.; Escudier, B.; Fong, L.; Joseph, R.W.; Pai, S.K.; Reeves, J.A.; et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 2018, 24, 749–757. [Google Scholar] [CrossRef]
- Miao, D.; Margolis, C.A.; Gao, W.; Voss, M.H.; Li, W.; Martini, D.J.; Norton, C.; Bossé, D.; Wancowicz, S.M.; Cullen, D.; et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018, 359, 801–806. [Google Scholar] [CrossRef]
- Braun, D.A.; Ishii, Y.; Van Allen, E.M.; Wu, C.J.; Shukla, S.A.; Choueiri, T.K. Clin ical validation of PBRM1 alterations as a marker of immune check inhibitor response in renal cell carcinoma. JAMA Oncol. 2019, 5, 1631–1633. [Google Scholar] [CrossRef]
- Braun, D.A.; Hou, Y.; Bakouni, Z.; Ficial, M.; Sant’Angelo, M.; Forman, J.; Ross-MacDonald, P.; Berger, A.C.; Jegede, O.A.; Elagina, L.; et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat. Med. 2020, in press. [Google Scholar] [CrossRef]
- Davis, C.J.; Mostofi, F.K.; Sesterhenn, I.A. Renal medullary carcinoma. The seventh sickle cell nephropathy. Am. J. Surg. Pathol. 1995, 19, 1–11. [Google Scholar] [CrossRef]
- Calderaro, J.; Moorch, J.; Pierron, G.; Pedentour, F.; Grison, C.; Maillé, P.; Sayeux, P.; De la Taille, A.; Coutourier, J.; Viellefond, A.; et al. SMARCB1/INI1 inactivation in renal medullary carcinoma. Histopathology 2012, 61, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, J.; Masliah-Planchon, J.; Richer, W.; Maillot, L.M.; Mansuy, L.; Bastien, C.; De la Taille, A.; Boussion, H.; Carpy, C.; Jourdain, A.; et al. Balanced translocations disrupting SMARCB1 are hallmark recurrent genetic alterations in renal medullary carcinomas. Eur. Urol. 2016, 69, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Carlo, M.I.; Chaim, J.; Patil, S.; Kemel, Y.; Schram, A.M.; Woo, K.; Coskey, D.; Nanjangud, G.J.; Voss, M.H.; Feldman, D.R.; et al. Genomic characterization of renal medullary carcinoma and treatment outcomes. Clin. Genitourin. Cancer 2017, 15, e987–e994. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Carlo, M.I.; Khan, H.; Nanjangud, G.J.; Rana, S.; Cimera, R.; Zhang, Y.; Ari Hakimi, A.; Verma, A.K.; Al-Ahmadie, H.A.; et al. Distinctive mechanisms underlie the loss of SMARCB1 protein expression in renal medullary carcinoma: Morphologic and molecular analysis of 20 cases. Mod. Pathol. 2019, 32, 1329–1343. [Google Scholar] [CrossRef] [PubMed]
- Hong, A.L.; Tseng, Y.Y.; Wala, J.A.; Kim, W.J.; Kynnap, B.D.; Doshi, M.B.; Kugener, G.; Sandoval, G.J.; Howard, T.P.; Li, J.; et al. Renal medullary carcinomas depend upon SMARCB1 loss and are sensitive to proteasome inhibition. Elife 2019, 8, e44161. [Google Scholar] [CrossRef]
- Mac Lennan, G.T.; Farrow, G.M.; Bostwick, D.G. Low-grade collecting duct carcinoma of the kidney: Report of 13 cases of low-grade mucinous tubulocystic renal carcinoma of possible collecting duct origin. Urology 1997, 50, 679–684. [Google Scholar] [CrossRef]
- Amin, M.B.; MacLennan, G.T.; Gupta, R.; Grignon, D.; Paraf, F.; Vielliefond, A.; Paner, G.P.; Stovsky, M.; Young, A.N.; Srigley, J.R.; et al. Tubulocystic carcinoma of the kidney: Clinicopathologic analysis of 31 cases of a distinctive rare subtype of renal cell carcinoma. Am. J. Surg. Pathol. 2009, 33, 384–392. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Armesto, M.; Fernandez-Mercado, M.; Arestìn, M.; Manterola, L.; Giocoechea, I.; Larrea, E.; Caffarel, M.M.; Araujo, A.M.; Sole, C.; et al. Noncoding RNA expression and targeted next-generation sequencing distinguish tubulocystic renal cell carcinoma (TC-RCC) from other renal neoplasms. J. Mol. Diagn. 2018, 20, 34–45. [Google Scholar] [CrossRef]
- Sarungbam, J.; Mehra, R.; Tomlins, S.A.; Smith, S.C.; Jayakumaran, G.; Al-Ahmadie, H.; Gopalan, A.; Sirintrapun, S.J.; Fine, S.W.; Zhang, Y.; et al. Tubulocystic renal cell carcinoma: A distinct clinicopathologic entity with a characteristic genomic profile. Mod. Pathol. 2019, 32, 701–709. [Google Scholar] [CrossRef]
- Gadd, S.; Huff, V.; Huang, C.C.; Ruteshouser, E.C.; Dome, J.S.; Grundy, P.E.; Breslow, N.; Jenniong, L.; Green, D.M.; Beckwith, J.B.; et al. Clinically relevant subsets identified by gene expression patterns support a revised ontogenic model of Wilms tumor: A children’s oncology group study. Neoplasia 2012, 14, 742–756. [Google Scholar] [CrossRef]
- Torrezan, G.T.; Ferreira, E.N.; Nakahata, A.M.; Barros, B.; Castro, M.; Correa, B.R.; Krepischi, A.C.; Olivieri, E.; Cunha, I.W.; Tabori, U.; et al. Recurrent mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat. Commun. 2014, 5, 4039. [Google Scholar] [CrossRef] [PubMed]
- Weigert, J.; IOshaque, N.; Vardapour, R.; Georg, C.; Gu, Z.; Bieg, M.; Ziegler, B.; Bausenwein, S.; Nourkami, N.; Ludwig, N.; et al. Mutations in SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell 2015, 27, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Rakheja, D.; Chen, K.S.; Liu, Y.; Shukla, A.A.; Schmid, V.; Chang, T.C.; Khokhar, S.; Wickiser, J.E.; Karandikar, N.J.; Malter, J.S.; et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mecahnisms in Wilms tumours. Nat. Commun. 2014, 2, 4802. [Google Scholar] [CrossRef] [PubMed]
- Walz, A.L.; Ooms, A.; Gadd, S.; Gerhard, D.S.; Smith, M.A.; Auvil, J.; Meerzaman, D.; Chen, Q.R.; Hsu, C.H.; Yan, C.; et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 2015, 27, 286–297. [Google Scholar] [CrossRef]
- Ooms, A.A.; Gadd, S.; Gerhard, D.S.; Smith, M.A.; Auvil, J.; Meerzman, D.; Chen, Q.R.; Hsu, C.H.; Yan, C.; Nguyen, C.; et al. Significance of TP53 mutation in Wilms tumors with diffuse anaplasia: A report from the children’s oncology group. Clin. Cancer Res. 2016, 22, 5582–5591. [Google Scholar] [CrossRef]
- Perlman, E.J.; Gadd, S.; Arold, S.T.; Radhakrishnan, A.; Gerhard, D.S.; Jenning, L.; Huff, V.; Auvil, J.; Davidsen, T.M.; Dome, J.S.; et al. MLLT1 YEATS domain mutations in clinically distinctive favourable histology Wilms tumours. Nat. Commun. 2015, 6, 10013. [Google Scholar] [CrossRef]
- Gadd, S.; Huff, V.; Walz, A.L.; Ooms, A.; Armstrong, A.E.; Gerhard, D.S.; Smith, M.A.; Auvil, J.; Meerzaman, D.; Chen, Q.R.; et al. Children Oncology Group and TARGET initiative exploring the genetic landscaope of Wilms Tumors. Nat. Genet. 2017, 49, 1487–1494. [Google Scholar] [CrossRef]
- Wan, L.; Chong, S.; Xuan, F.; Liang, A.; Cui, X.; Gates, L.; Carroll, T.S.; Li, Y.; Feng, L.; Chen, G.; et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 2020, 577, 121–126. [Google Scholar] [CrossRef]
- Coorens, T.; Treger, T.D.; Al-Saadi, R.; Moore, L.; Tran, M.; Mitchell, T.J.; Tugnait, S.; Thevanesan, C.; Young, M.D.; Oliver, T.; et al. Embryonal precursors of Wilms tumor. Science 2019, 366, 1247–1251. [Google Scholar] [CrossRef]
- Jones, T.D.; Eble, J.N.; Wang, M.; Maclennan, G.T.; Jain, S.; Cheng, I. Clonal divergence and genetic heterogeneity in clear cell renal cell carcinomas with sarcomatoid transformation. Cancer 2005, 104, 1195–1203. [Google Scholar] [CrossRef]
- Cheville, J.C.; Lohse, C.M.; Zincke, H.; Weaver, A.L.; Leibovich, B.C.; Franck, I.; Blute, M.L. Sarcomatoid renal cell carcinoma: An examination of underlying histologic subtype and an analysis of associationswith patient outcome. Am. J. Surg. Pathol. 2004, 28, 435–441. [Google Scholar] [CrossRef] [PubMed]
- De Peralta-Venturina, M.; Moch, H.; Amin, M.; Tamboli, P.; Hallemariam, S.; Mihatsch, M.; Javidan, J.; Stricker, H.; Ro, J.Y.; Amin, M.B. Sarcomatoid differentiation in renal cell carcinoma: A study of 101 cases. Am. J. Surg. Pathol. 2001, 25, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Sircar, K.; Yoop, S.Y.; Majewski, T.; Wani, K.; Patel, L.R.; Voicu, H.; Torres-Garcia, W.; Verhaak, R.G.W.; Tannir, N.; Karam, J.A.; et al. Biphasic components of sarcomatoid clear cell renal cell carcinomas are molecularly similar to each other, but distinct fron, non-sarcomatoid renal carcinomas. J. Pathol. Clin. Res. 2015, 1, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Bi, M.; Zhao, S.; Said, J.W.; Merino, M.J.; Adeniran, A.J.; Xie, Z.; Nawaf, C.B.; Choi, J.; Belldegrun, A.S.; Pantuck, A.J.; et al. Genomic characterization of sarcomatoid transformation in clear cell renal cell carcinoma. Proc. Natl. Acad. Sci. USA 2016, 113, 2170–2175. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kim, T.B.; Peng, B.; Karam, J.A.; Creighton, C.J.; Joon, A.Y.; Kawakami, F.; Trevisan, P.; Jonash, E.; Chow, C.W.; et al. Sarcomatoid renal cell carcinoma has a distinct molecular pathogenesis, driver mutation profile and transcriptional landscape. Clin. Cancer Res. 2017, 23, 6686–6696. [Google Scholar] [CrossRef]
- Pal, S.K.; He, M.; Tong, T.; Wu, H.; Liu, X.; Lau, C.; Wang, J.H.; Warden, C.; Wu, X.; Signoretti, S.; et al. RNA-seq reveals aurora kinase-driven mTOR pathway activation in patients with sarcomatoid metastatic renal cell carcinoma. Mol. Cancer Res. 2015, 13, 130–137. [Google Scholar] [CrossRef]
- Malouf, G.G.; Ali, S.M.; Wang, K.; Balasubramanian, S.; Ross, J.S.; Miller, V.A.; Stephens, P.J.; Khayat, D.; Pal, S.K.; Su, X.; et al. Genomic characterization of renal cell carcinoma with sarcomatoid dedifferentiation pinpoints recurrent genomic alterations. Eur. Urol. 2016, 70, 348–357. [Google Scholar] [CrossRef]
- Malouf, G.G.; Filippot, R.; Dong, Y.; Dinatale, R.G.; Chen, Y.B.; Su, X.; Comperat, E.; Roupret, M.; Mano, R.; Blum, K.A.; et al. Molecular characterization of sarcomatoid clear cell renal cell carcinoma unveils new candidate oncogenic drivers. Sci. Rep. 2020, 10, 701. [Google Scholar] [CrossRef]
- Ito, T.; Pei, J.; Dulaimi, E.; Menges, C.; Abbosh, P.H.; Smaldone, M.C.; Chen, D.Y.; Greenberg, R.E.; Kutikov, A.; Viterbo, R.; et al. Genomic copy number alterations in renal cell carcinoma with sarcomatoid features. J. Urol. 2016, 195, 852–858. [Google Scholar] [CrossRef]
- White, S.M.; Avantaggiatio, M.L.; Nemazanyy, I.; Di Poto, C.; Yang, Y.; Pende, M.; Gibney, G.; Ressom, H.; Field, J.; Atkins, M.B.; et al. YAP/TAZ inhibition induces metabolic and signaling rewiring resulting in targetable vulnerabilities in NF2-deficient tumor cells. Dev. Cell 2019, 49, 425–443. [Google Scholar] [CrossRef]
- Kawakami, F.; Sircar, K.; Rodriguez-Canales, J.; Fellman, B.L.; Urbauer, D.L.; Tamboli, P.; Tannir, N.M.; Jonasch, E.; Wistuba, L.L.; Wooid, C.G.; et al. Programmed cell death ligand 1 and tumor-infiltrating lymphocyte status in patients with renal cell carcinoma and sarcomatoid dedifferentiation. Cancer 2017, 123, 4823–4831. [Google Scholar] [CrossRef] [PubMed]
- Joseph, R.W.; Millis, S.Z.; Carballido, E.M.; Bryant, D.; Gatalica, Z.; Reddy, S.; Bryce, A.H.; Vogelzang, N.J.; Stanton, M.L.; Castle, E.P.; et al. PD-1 and PD-L1 expression in renal cell carcinoma with sarcomatoid differentiation. Cancer Immunol. Res. 2015, 3, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Flippot, R.; McGregor, B.A.; Flaifel, A.; Gray, K.P.; Signoretti, S.; Steinharter, J.A.; Van Allen, E.M.; Walsh, M.K.; Gundy, K.; Wei, X.X.; et al. Atezolizumab plus bevacizumab in non-clear cell renal cell carcinoma (NccRCC) and clear cell renal cell carcinoma with sarcomatoid differentiation (ccRCCsd): Updated results of activity and predictive biomarkers from a phase II study. J. Clin. Oncol. 2019, 3, 4583. [Google Scholar] [CrossRef]
- Rini, B.I.; Motzer, R.J.; Powles, T.; McDermott, D.F.; Escudier, B.; Donskov, F.; Hawkins, R.E.; Bracarda, S.; Bedke, J.; De Giorgi, U.; et al. Atezolizumab (atezo) + bevacizumab (bev) versus sunitinib (sun) in pts with untreated metastatic renal cell carcinoma (mRCC) and sarcomatoid histology: IMmotion151 subgroup analysis. J. Clin. Oncol. 2019, 37, 4512. [Google Scholar] [CrossRef]
- Gupta, S.; Cheville, J.C.; Jungbluth, A.A.; Zhang, Y.; Zhang, L.; Chen, Y.B.; Tickoo, S.K.; Fine, S.W.; Gopalan, A.; Al-Ahmadie, H.A.; et al. JAK2/PD-L1/PD-L2 (9p24.1) amplifications in renal cell carcinomas with sarcomatoid transformation: Implications for clinical management. Mod. Pathol. 2019, 32, 1344–1358. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.I.; Powles, T.; Atkins, M.B.; Escudier, B.; McDermott, D.F.; Suarez, C.; Bracarda, S.; Stadler, W.M.; Donskov, F.; Lee, J.L.; et al. Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (IMmotion151): A multicenter, open-label, phase 3, randomized controlled trial. Lancet 2019, 393, 2404–2415. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, Y.; Senbabaoglu, Y.; Ciriello, G.; Yang, L.; Reznik, E.; Shuch, B.; Micevic, G.; De Velasco, N.; Shinbrot, E.; et al. Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep. 2016, 14, 2476–2489. [Google Scholar] [CrossRef]
- Poehlman, W.L.; Hsieh, J.J.; Feltus, F.A. Linking binary gene relationships to drivers of renal cell carcinoma reveals convergent function in alternate tumor progression paths. Sci. Rep. 2019, 9, 2899. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Le, V.; Cao, D.; Cheng, E.H.; Creighton, C.J. Genomic classifications of renal cell carcinoma: A critical step towards the future application of personalized kidney cancer care with pan-omics precision. J. Pathol. 2018, 244, 525–537. [Google Scholar] [CrossRef]
- Huang, J.J.; Hsieh, J.J. The therapeutic landscape of renal cell carcinoma: From the dark age to the golden age. Semin. Nephrol. 2020, 40, 28–41. [Google Scholar] [CrossRef]
- Rini, B.I.; Plimack, E.R.; Stus, V.; Gafanov, R.; Hawkins, R.; Nosov, D.; Pouliot, F.; Alekseev, B.; Soulieres, D.; Melichar, B.; et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2019, 380, 1116–1127. [Google Scholar] [CrossRef] [PubMed]
- Pilmack, E.R.; Rini, B.I.; Stus, V.; Gafanov, R.; Waddell, T.; Nosov, D.; Pouliot, F.; Soulieres, D.; Meichar, B.; Vynnychenko, I.; et al. Pembrolizumab plus axitinib as first-line therapy for advanced renal cell carcinoma (RCC): Updated amalysis of KEYNOTE-426. J. Clin. Oncol. 2020, 38, 5001. [Google Scholar] [CrossRef]
- Motzer, R.J.; Penkov, K.; Haanen, J.; Rini, B.I.; Albiges, L.; Campbell, M.T.; Venugopal, B.; Kollmannsberger, C.; Negrier, S.; Uemura, M.; et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2019, 380, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Motzer, R.J.; Rini, B.I.; Haanen, J.; Campbell, M.T.; Vengogal, B.; Kollmannsberger, C.; Grais-Mescom, G.; Uemura, M.; Lee, J.L.; et al. Updated efficacy results from the JAVELIN Renal 101 trial: First-line avelumab plus axitinib versus sunitinib in patients with advanced renal cell carcinoma. Ann. Oncol. 2020; in press. [Google Scholar]
- Choueiri, T.K.; Plimack, E.R.; Bauer, T.M.; Merchan, J.R.; Papadopoulos, K.; McDermott, D.F.; Michaelson, D.; Appleman, L.J.; Zojwalle, N.; Jonasch, E. Phase I/II study of the oral HIF-2α inhibitor MK-6482 in patients with advanced clear cell renal cell carcinoma (RCC). J. Clin. Oncol. 2020, 38, 611. [Google Scholar] [CrossRef]
- Jonasch, E.; Donskov, F.; Iliopoulos, O.; Rathmell, W.K.; Narayan, V.; Maughan, B.L.; Oudard, S.; Else, T.; Marouchie, J.K.; Walsh, S.J.; et al. Phase II study of the oral HIF-2α inhibitor MK-6482 for von Hippel-Lindau disease-associated renal cell carcinoma. J. Clin. Oncol. 2020, 38, 5003. [Google Scholar] [CrossRef]
| Syndrome | Gene (chromosome) | Protein | Clinical Manifestations | Histology |
|---|---|---|---|---|
| Von Hippel-Lindau Syndrome | VHL (3p25) | pVHL | CCRCC, Pheochromocytoma, pancreatic endocrine tumors, CNS, and retinal hemangioblastomas | CCRCC Clear cell papillary |
| Hereditary Papillary RCC (HPRCC) | MET (7q31) | MET | Type 1 papillary RCC | Papillary type 1 |
| Cowden Syndrome | PTEN (10q23.31) | Phosphatase and tensin homolog | Dermatological lesions. breast cancer, thyroid cancer, endometrial cancer | Papillary Chromophobe CCRCC |
| BAP1 Hereditary Syndrome | BAP1 (3p21) | BRCA1-associated protein-1 | Uveal and cutaneous, melanoma, malignant mesothelioma, and/or lung adenocarcinoma | Undefined |
| Hereditary paraganglioma-pheochromocytoma syndromes | SDHA (5p15.33) SDHB (1p36.1-p35) SDHC (1q23.3) SDHD (11q23,.1) | Succinate dehydrogenase | Bilateral and extra-adrenal pheochromocytoma, paraganglioma, RCC, and other malignancies | SDH-deficient RCC (solid nests or tubular architecture with variable cysts; vacuolated cells with eosinophilic cytoplasm) |
| Hereditary leiomyomatosis and renal cell carcinoma (HLRCC) | FH (1q42,.1) | Fumarate hydratase | RCC, leiomyomas of skin and uterus (leiomyosarcoma), malignant pheochromocytoma/paraganglioma | HLRCC-associated RCC papillary type 2 |
| Birt-Hogg-Dubé (BHD) Syndrome | FLCN (17p11.2) | Folliculin | RCC (hybrid oncocytic and other types), fibrofolliculomas, pulmonary cysts | Chromophobe Oncocytoma Hybrid CCRCC |
| MITF-associated susceptibility to melanoma and RCC syndrome | MITF (3p14.1) | Microphtalmia- associated transcription factor | Melanoma, pancreatic cancer, and/or pheochromocytoma | Undefined |
| RCC Subtype | Somatic Mutations or Alterations | Copy Number Variations or Translocations | Prognostic Implications of Genomic Alterations |
|---|---|---|---|
| CCRCC | Mutations in VHL, PBMR1, SETD2, BAP1, KDM5C, TERT promoter, MTOR | Loss of chromosomes 3p, 14q, 9p, 6q, 8p,15q Gain of chromosome 5q | VHL: no association PBMR1: greater survival/no benefit BAP1, SETD2, CDKN2A, TP53: reduced survival PDH genes, Ribose sugar metabolism genes: reduced survival |
| PRCC, type I | Mutations in MET, NRF2, CUL3 | Gains of chromosomes 3, 7, 16, 17 | CDKN2A, PBMR1, TP53: reduced survival DKK1/SFRP1: unmethylation: reduced survival |
| PRCC, type II | Mutations in CDKN2A, CDKN2B, TERT, NF2, FH, MET, SETD2 | Gains of chromosomes 7, 16, 17, 5q Loss of chromosomes: 3p, 14q, 22q Translocation of TFE3 | CDKN2A, TP53: reduced survival DKK1/SFRP1: unmethylation: reduced survival |
| CHRCC | Mutations in TP53, PTEN | Loss of chromosomes 1, 2, 6, 10, 13, 17, 21 | PTEN, CDKN2A: reduced survival DKK1/SFRP1: unmethylation: reduced survival Metabolically divergent tumors: highly reduced survival |
| RMC | Mutations in SMARCB1 | Amplification of ABL | Unknown |
| TCRCC | Mutations in ABL1, PDGFRA | Gains of chromosomes: 7,17 | Unknown |
| Wilms Nephroblastoma | Mutations in TP53, AMER1, CTNNB1, WT1, DROSHA, DGGR8, DICER1, SIX1/SIX2, SMARCA-4, MLTT1 | Loss of chromosomes 1p, 16q, 1q, 17p, 4q, 14q, 11q, 11p15. | TP53, SIX1/SIX2, DROSHA/DGGR8: reduced survival Loss of chromosomes 1p, 1q, 11p15, and 16q: reduced survival |
| Evolution Pattern | Early Events | Primary Tumor | Genomic Characterization | Metastatic Potential |
|---|---|---|---|---|
| Linear | Chr 3p loss VHL inactivation Initial clonal expansion |  | Low GII Low ITH | Non-Metastatic |
| Branched | Chr 3p loss VHL inactivation Initial clonal expansion | 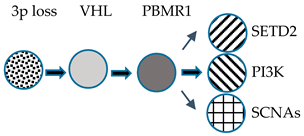 | High GII High GII | Slow Progression |
| Punctuated | Chr 3p loss VHL inactivation Initial clonal expansion | 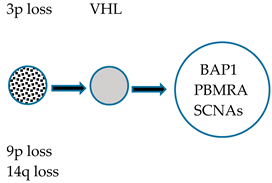 | High GII Low ITH | Rapid Progression |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Testa, U.; Pelosi, E.; Castelli, G. Genetic Alterations in Renal Cancers: Identification of The Mechanisms Underlying Cancer Initiation and Progression and of Therapeutic Targets. Medicines 2020, 7, 44. https://doi.org/10.3390/medicines7080044
Testa U, Pelosi E, Castelli G. Genetic Alterations in Renal Cancers: Identification of The Mechanisms Underlying Cancer Initiation and Progression and of Therapeutic Targets. Medicines. 2020; 7(8):44. https://doi.org/10.3390/medicines7080044
Chicago/Turabian StyleTesta, Ugo, Elvira Pelosi, and Germana Castelli. 2020. "Genetic Alterations in Renal Cancers: Identification of The Mechanisms Underlying Cancer Initiation and Progression and of Therapeutic Targets" Medicines 7, no. 8: 44. https://doi.org/10.3390/medicines7080044
APA StyleTesta, U., Pelosi, E., & Castelli, G. (2020). Genetic Alterations in Renal Cancers: Identification of The Mechanisms Underlying Cancer Initiation and Progression and of Therapeutic Targets. Medicines, 7(8), 44. https://doi.org/10.3390/medicines7080044






