Identifying the Relationship between PM2.5 and Hyperlipidemia Using Mendelian Randomization, RNA-seq Data and Model Mice Subjected to Air Pollution
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Mendelian Randomization Analysis
2.3. Enrichment Analysis
2.4. Estimation of Immune Cells Infiltration in Mice Livers
2.5. Preparation of PM2.5
2.6. Animal Model
2.7. The Assays of Plasma Lipids and Lipoproteins
2.8. qPCR
2.9. Statistical Analysis
3. Results
3.1. Mendelian Randomization Indicates Genetic Association with PM2.5 and Hyperlipidemia
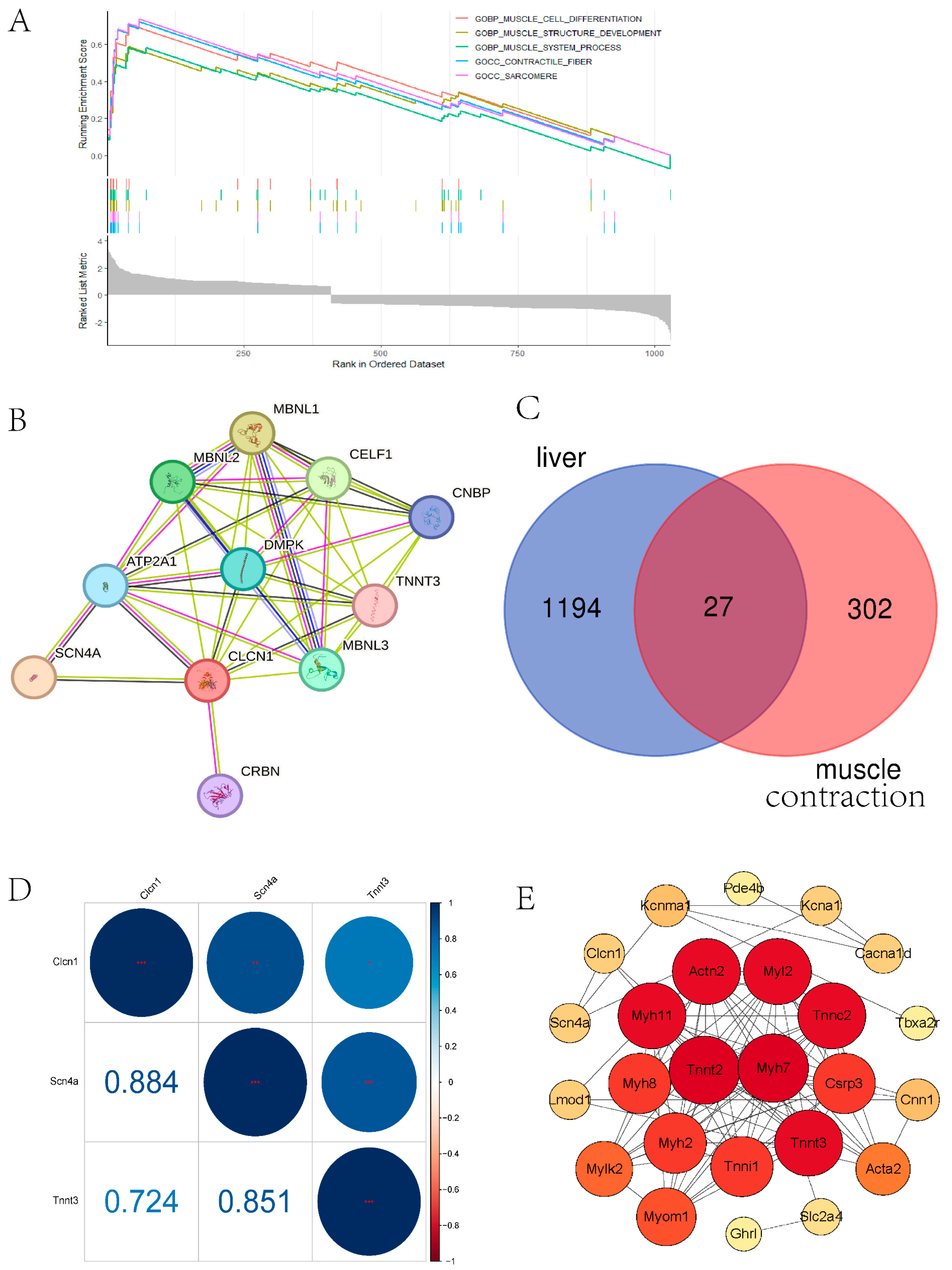
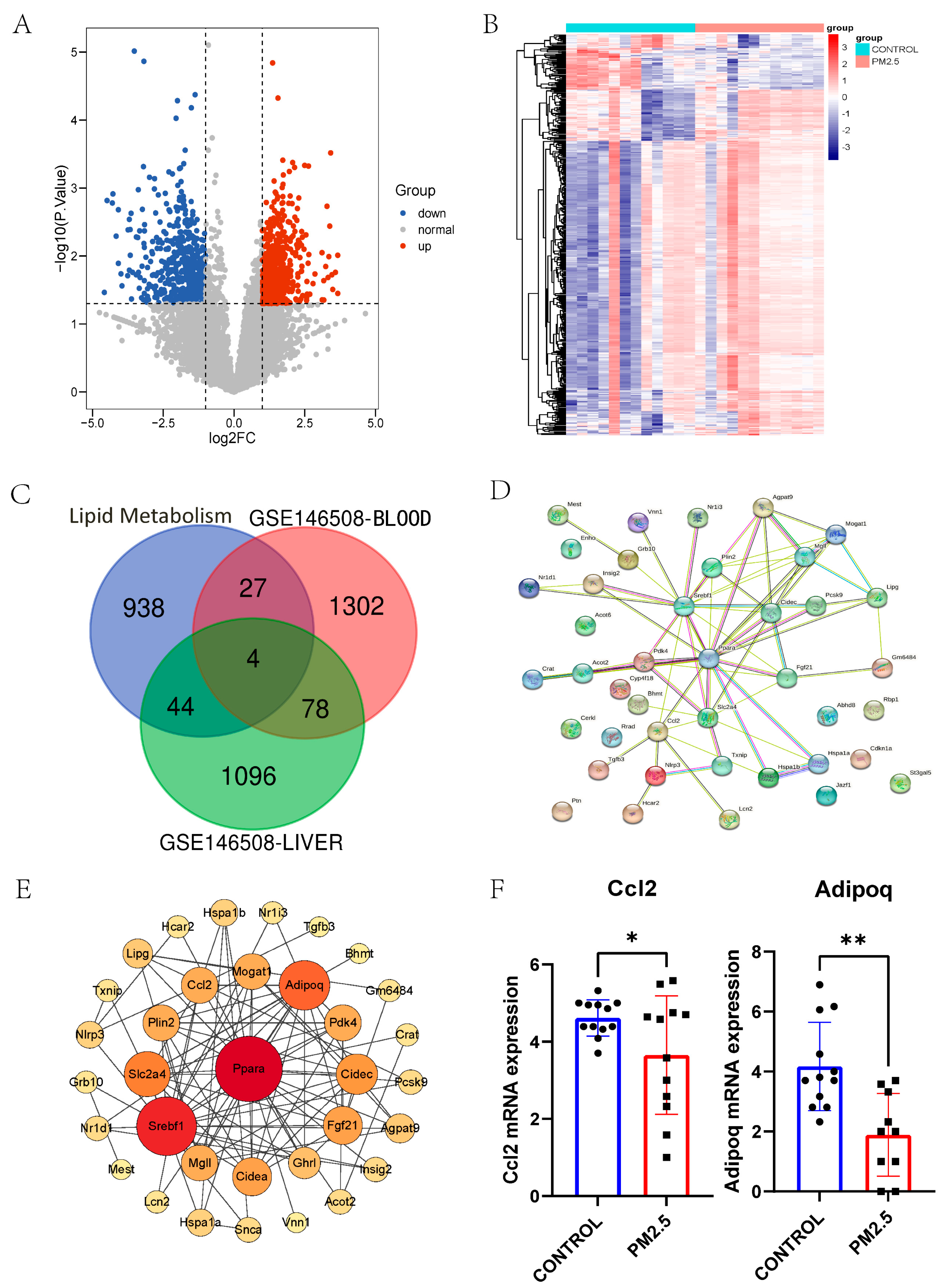
3.2. RNA-seq Data of Liver and Plasma in Air Pollution Model Mice Revealed Hub Genes in Muscle Contraction and Lipid Metabolism
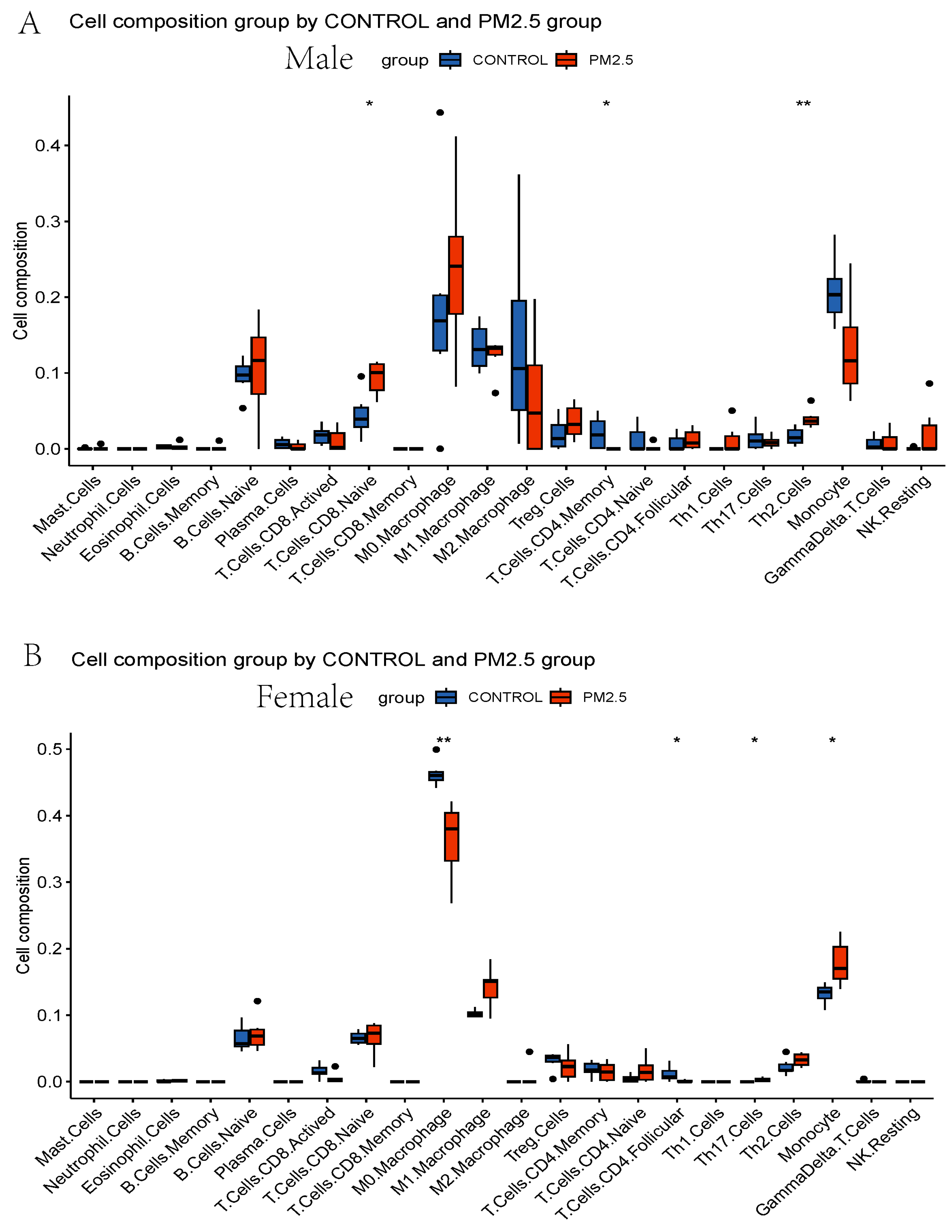
3.3. Immune Infiltration Analysis
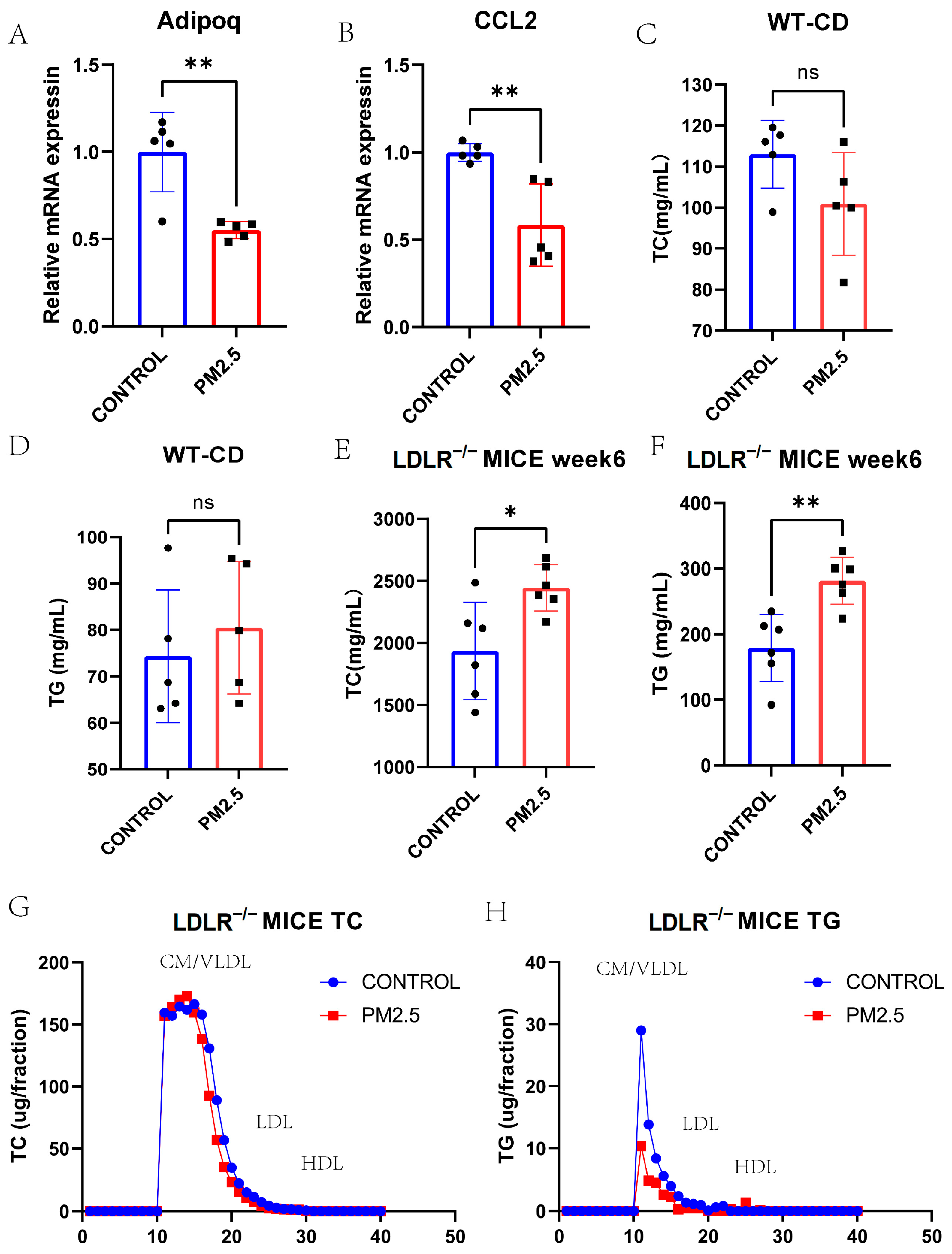
3.4. Lipid and Lipoprotein Profiles in Model Mice Exposed to Intranasal Instillation-Induced Air Pollution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Srimuruganandam, B.; Shiva Nagendra, S.M. Source characterization of PM10 and PM2.5 mass using a chemical mass balance model at urban roadside. Sci. Total Environ. 2012, 433, 8–19. [Google Scholar] [CrossRef]
- Cheung, K.; Daher, N.; Kam, W.; Shafer, M.M.; Ning, Z.; Schauer, J.J.; Sioutas, C. Spatial and temporal variation of chemical composition and mass closure of ambient coarse particulate matter (PM10–2.5) in the Los Angeles area. Atmos. Environ. 2011, 45, 2651–2662. [Google Scholar] [CrossRef]
- Löndahl, J.; Pagels, J.; Swietlicki, E.; Zhou, J.; Ketzel, M.; Massling, A.; Bohgard, M. A set-up for field studies of respiratory tract deposition of fine and ultrafine particles in humans. J. Aerosol Sci. 2006, 37, 1152–1163. [Google Scholar] [CrossRef]
- Volckens, J.; Quinn, C.; Leith, D.; Mehaffy, J.; Henry, C.S.; Miller-Lionberg, D. Development and evaluation of an ultrasonic personal aerosol sampler. Indoor Air 2017, 27, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, S.; Hawken, S.; Ôunpuu, S.; Dans, T.; Avezum, A.; Lanas, F.; McQueen, M.; Budaj, A.; Pais, P.; Varigos, J.; et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): Case-control study. Lancet 2004, 364, 937–952. [Google Scholar] [CrossRef] [PubMed]
- Karr, S. Epidemiology and management of hyperlipidemia. Am. J. Manag. Care 2017, 23 (Suppl. 9), S139–S148. [Google Scholar]
- Bell, G.; Mora, S.; Greenland, P.; Tsai, M.; Gill, E.; Kaufman, J.D. Association of Air Pollution Exposures with High-Density Lipoprotein Cholesterol and Particle Number: The Multi-Ethnic Study of Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 976–982. [Google Scholar] [CrossRef]
- Bind, M.A.; Peters, A.; Koutrakis, P.; Coull, B.; Vokonas, P.; Schwartz, J. Quantile Regression Analysis of the Distributional Effects of Air Pollution on Blood Pressure, Heart Rate Variability, Blood Lipids, and Biomarkers of Inflammation in Elderly American Men: The Normative Aging Study. Environ. Health Perspect. 2016, 124, 1189–1198. [Google Scholar] [CrossRef]
- Cai, Y.; Hansell, A.L.; Blangiardo, M.; Burton, P.R.; de Hoogh, K.; Doiron, D.; Fortier, I.; Gulliver, J.; Hveem, K.; Mbatchou, S.; et al. Long-term exposure to road traffic noise, ambient air pollution, and cardiovascular risk factors in the HUNT and lifelines cohorts. Eur. Heart J. 2017, 38, 2290–2296. [Google Scholar] [CrossRef]
- Chuang, K.J.; Yan, Y.H.; Chiu, S.Y.; Cheng, T.J. Long-term air pollution exposure and risk factors for cardiovascular diseases among the elderly in Taiwan. Occup. Environ. Med. 2011, 68, 64–68. [Google Scholar] [CrossRef]
- Jiang, S.; Bo, L.; Gong, C.; Du, X.; Kan, H.; Xie, Y.; Song, W.; Zhao, J. Traffic-related air pollution is associated with cardio-metabolic biomarkers in general residents. Int. Arch. Occup. Environ. Health 2016, 89, 911–921. [Google Scholar] [CrossRef]
- Poursafa, P.; Mansourian, M.; Motlagh, M.E.; Ardalan, G.; Kelishadi, R. Is air quality index associated with cardiometabolic risk factors in adolescents? The CASPIAN-III Study. Environ. Res. 2014, 134, 105–109. [Google Scholar] [CrossRef]
- Shanley, R.P.; Hayes, R.B.; Cromar, K.R.; Ito, K.; Gordon, T.; Ahn, J. Particulate Air Pollution and Clinical Cardiovascular Disease Risk Factors. Epidemiology 2016, 27, 291–298. [Google Scholar] [CrossRef]
- Sørensen, M.; Hjortebjerg, D.; Eriksen, K.T.; Ketzel, M.; Tjønneland, A.; Overvad, K.; Raaschou-Nielsen, O. Exposure to long-term air pollution and road traffic noise in relation to cholesterol: A cross-sectional study. Environ. Int. 2015, 85, 238–243. [Google Scholar] [CrossRef]
- Wallwork, R.S.; Colicino, E.; Zhong, J.; Kloog, I.; Coull, B.A.; Vokonas, P.; Schwartz, J.D.; Baccarelli, A.A. Ambient Fine Particulate Matter, Outdoor Temperature, and Risk of Metabolic Syndrome. Am. J. Epidemiol. 2017, 185, 30–39. [Google Scholar] [CrossRef]
- Yeatts, K.; Svendsen, E.; Creason, J.; Alexis, N.; Herbst, M.; Scott, J.; Kupper, L.; Williams, R.; Neas, L.; Cascio, W.; et al. Coarse particulate matter (PM2.5–10) affects heart rate variability, blood lipids, and circulating eosinophils in adults with asthma. Environ. Health Perspect. 2007, 115, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Yitshak Sade, M.; Kloog, I.; Liberty, I.F.; Schwartz, J.; Novack, V. The Association Between Air Pollution Exposure and Glucose and Lipids Levels. J. Clin. Endocrinol. Metab. 2016, 101, 2460–2467. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.Y.; Kim, J.H.; Lee, G.; Choi, S.; Kim, S.R.; Hong, Y.C.; Park, S.M. Exposure to ambient fine particulate matter is associated with changes in fasting glucose and lipid profiles: A nationwide cohort study. BMC Public Health 2020, 20, 430. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Li, S.; Wang, C.; Liu, Y.; Li, N.; Liu, F.; Huang, S.; Liu, S.; Lu, Y.; Mao, Z.; et al. Is long-term PM1 exposure associated with blood lipids and dyslipidemias in a Chinese rural population? Environ. Int. 2020, 138, 105637. [Google Scholar] [CrossRef]
- Roswall, N.; Poulsen, A.H.; Hvidtfeldt, U.A.; Hendriksen, P.F.; Boll, K.; Halkjær, J.; Ketzel, M.; Brandt, J.; Frohn, L.M.; Christensen, J.H.; et al. Exposure to ambient air pollution and lipid levels and blood pressure in an adult, Danish cohort. Environ. Res. 2023, 220, 115179. [Google Scholar] [CrossRef]
- Rajkumar, S.; Young, B.N.; Clark, M.L.; Benka-Coker, M.L.; Bachand, A.M.; Brook, R.D.; Nelson, T.L.; Volckens, J.; Reynolds, S.J.; L’Orange, C.; et al. Household air pollution from biomass-burning cookstoves and metabolic syndrome, blood lipid concentrations, and waist circumference in Honduran women: A cross-sectional study. Environ. Res. 2019, 170, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zheng, S.; Nie, Y.; Weng, J.; Cheng, N.; Hu, X.; Ren, X.; Pei, H.; Bai, Y. Association between Short-Term Exposure to Air Pollution and Dyslipidemias among Type 2 Diabetic Patients in Northwest China: A Population-Based Study. Int. J. Environ. Res. Public Health 2018, 15, 631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, H.; He, W.; Chen, G.; Lu, P.; Xu, R.; Yu, P.; Ye, T.; Li, S.; Xie, Y.; et al. The association between ambient air pollution and blood lipids: A longitudinal study in Shijiazhuang, China. Sci. Total Environ. 2021, 752, 141648. [Google Scholar] [CrossRef] [PubMed]
- Gui, Z.H.; Yang, B.Y.; Zou, Z.Y.; Ma, J.; Jing, J.; Wang, H.J.; Dong, G.H.; Ma, Y.H.; Guo, Y.M.; Chen, Y.J. Exposure to ambient air pollution and blood lipids in children and adolescents: A national population based study in China. Environ. Pollut. 2020, 266 Pt 3, 115422. [Google Scholar] [CrossRef]
- Yang, B.Y.; Bloom, M.S.; Markevych, I.; Qian, Z.M.; Vaughn, M.G.; Cummings-Vaughn, L.A.; Li, S.; Chen, G.; Bowatte, G.; Perret, J.L.; et al. Exposure to ambient air pollution and blood lipids in adults: The 33 Communities Chinese Health Study. Environ. Int. 2018, 119, 485–492. [Google Scholar] [CrossRef]
- Kim, K.N.; Ha, B.; Seog, W.; Hwang, I.U. Long-term exposure to air pollution and the blood lipid levels of healthy young men. Environ. Int. 2022, 161, 107119. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Chen, G.; Liu, F.; Li, N.; Wang, C.; Liu, Y.; Liu, S.; Lu, Y.; Xiang, H.; Guo, Y.; et al. Long-term effects of ambient air pollutants to blood lipids and dyslipidemias in a Chinese rural population. Environ. Pollut. 2020, 256, 113403. [Google Scholar] [CrossRef]
- Wang, L.; Chen, G.; Pan, Y.; Xia, J.; Chen, L.; Zhang, X.; Silang, Y.; Chen, J.; Xu, H.; Zeng, C.; et al. Association of long-term exposure to ambient air pollutants with blood lipids in Chinese adults: The China Multi-Ethnic Cohort study. Environ. Res. 2021, 197, 111174. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.; Lu, J.; Li, Z.; Martinez, L.; Tao, B.; Wang, C.; Zhu, L.; Lu, W.; Zhu, B.; et al. Effects of short-term PM2.5 exposure on blood lipids among 197,957 people in eastern China. Sci. Rep. 2023, 13, 4505. [Google Scholar] [CrossRef]
- Song, L.; Lei, L.; Jiang, S.; Pan, K.; Zeng, X.; Zhang, J.; Wang, C.; Zhu, L.; Lu, W.; Zhu, B.; et al. NLRP3 inflammasome is involved in ambient PM2.5-related metabolic disorders in diabetic model mice but not in wild-type mice. Inhal. Toxicol. 2021, 33, 260–267. [Google Scholar] [CrossRef]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Gavett, S.H.; Haykal-Coates, N.; Highfill, J.W.; Ledbetter, A.D.; Chen, L.C.; Cohen, M.D.; Harkema, J.R.; Wagner, J.G.; Costa, D.L. World Trade Center fine particulate matter causes respiratory tract hyperresponsiveness in mice. Environ. Health Perspect. 2003, 111, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Arifin, W.N.; Zahiruddin, W.M. Sample Size Calculation in Animal Studies Using Resource Equation Approach. Malays. J. Med. Sci. 2017, 24, 101–105. [Google Scholar] [PubMed]
- Zeng, Y.; Jin, H.; Wang, J.; Guo, C.; Chen, W.; Tan, Y.; Wang, L.; Zhou, Z. An optimized method for intratracheal instillation in mice. J. Pharmacol. Toxicol. Methods 2022, 118, 107230. [Google Scholar] [CrossRef]
- Morimoto, Y.; Izumi, H.; Yoshiura, Y.; Fujishima, K.; Yatera, K.; Yamamoto, K. Usefulness of Intratracheal Instillation Studies for Estimating Nanoparticle-Induced Pulmonary Toxicity. Int. J. Mol. Sci. 2016, 17, 165. [Google Scholar] [CrossRef]
- Li, J.; Hu, Y.; Liu, L.; Wang, Q.; Zeng, J.; Chen, C. PM2.5 exposure perturbs lung microbiome and its metabolic profile in mice. Sci. Total Environ. 2020, 721, 137432. [Google Scholar] [CrossRef]
- Happo, M.S.; Hirvonen, M.R.; Hälinen, A.I.; Jalava, P.I.; Pennanen, A.S.; Sillanpää, M.; Hillamo, R.; Salonen, R.O. Seasonal variation in chemical composition of size-segregated urban air particles and the inflammatory activity in the mouse lung. Inhal. Toxicol. 2010, 22, 17–32. [Google Scholar] [CrossRef]
- Park, E.-J.; Roh, J.; Kim, Y.; Park, K.; Kim, D.-S.; Yu, S.-D. PM 2.5 collected in a residential area induced Th1-type inflammatory responses with oxidative stress in mice. Environ. Res. 2011, 111, 348–355. [Google Scholar] [CrossRef]
- Bide, R.W.; Armour, S.J.; Yee, E.; Defence Research Establishment (Suffield, Ralston, Alberta). Estimation of Human Toxicity from Animal Inhalation Toxicity Data. I. Minute Volume-Body Weight Relationships between Animals and Man; Accession No ADA336351; U.S. Department of Defense: Washington, DC, USA, 1997. [Google Scholar]
- Li, X.; Geng, J.; Chen, Y.; Chen, F.; Liu, C.; Xu, Q.; Zhao, J.; Hu, J.; Xie, J.; Xu, B. Exposure to particulate matter induces cardiomyocytes apoptosis after myocardial infarction through NFκB activation. Biochem. Biophys. Res. Commun. 2017, 488, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.C.; Steinmeyer, K.; Lorenz, C.; Ricker, K.; Wolf, F.; Otto, M.; Zoll, B.; Lehmann-Horn, F.; Grzeschik, K.H.; Jentsch, T.J. The skeletal muscle chloride channel in dominant and recessive human myotonia. Science 1992, 257, 797–800. [Google Scholar] [CrossRef]
- Gao, Y.; Peng, L.; Zhao, C. MYH7 in cardiomyopathy and skeletal muscle myopathy. Mol. Cell Biochem. 2023. [Google Scholar] [CrossRef] [PubMed]
- Bosè, F.; Renna, L.V.; Fossati, B.; Arpa, G.; Labate, V.; Milani, V.; Botta, A.; Micaglio, E.; Meola, G.; Cardani, R. TNNT2 Missplicing in Skeletal Muscle as a Cardiac Biomarker in Myotonic Dystrophy Type 1 but Not in Myotonic Dystrophy Type 2. Front. Neurol. 2019, 10, 992. [Google Scholar] [CrossRef]
- Singh, S.; Anshita, D.; Ravichandiran, V. MCP-1: Function, regulation, and involvement in disease. Int. Immunopharmacol. 2021, 101 Pt B, 107598. [Google Scholar] [CrossRef]
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte chemoattractant protein-1 (MCP-1): An overview. J. Interferon Cytokine Res. 2009, 29, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Presa, M.; Bustos, C.; Ortego, M.; Tuñon, J.; Renedo, G.; Ruiz-Ortega, M.; Egido, J. Angiotensin-converting enzyme inhibition prevents arterial nuclear factor-kappa B activation, monocyte chemoattractant protein-1 expression, and macrophage infiltration in a rabbit model of early accelerated atherosclerosis. Circulation 1997, 95, 1532–1541. [Google Scholar] [CrossRef]
- Achari, A.E.; Jain, S.K. Adiponectin, a Therapeutic Target for Obesity, Diabetes, and Endothelial Dysfunction. Int. J. Mol. Sci. 2017, 18, 1321. [Google Scholar] [CrossRef] [PubMed]
- Sattar, N.; Wannamethee, G.; Sarwar, N.; Tchernova, J.; Cherry, L.; Wallace, A.M.; Danesh, J.; Whincup, P.H. Adiponectin and coronary heart disease: A prospective study and meta-analysis. Circulation 2006, 114, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Ouchi, N.; Kihara, S.; Walsh, K.; Kumada, M.; Abe, Y.; Funahashi, T.; Matsuzawa, Y. Selective suppression of endothelial cell apoptosis by the high molecular weight form of adiponectin. Circ. Res. 2004, 94, e27–e31. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, M.; Shimomura, I.; Sata, M.; Arita, Y.; Nishida, M.; Maeda, N.; Kumada, M.; Okamoto, Y.; Nagaretani, H.; Nishizawa, H.; et al. Role of adiponectin in preventing vascular stenosis. The missing link of adipo-vascular axis. J. Biol. Chem. 2002, 277, 37487–37491. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Wang, Y.; Keshaw, H.; Xu, L.Y.; Lam, K.S.; Cooper, G.J. The fat-derived hormone adiponectin alleviates alcoholic and nonalcoholic fatty liver diseases in mice. J. Clin. Investig. 2003, 112, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Kamei, Y.; Ezaki, O. Mest/Peg1 imprinted gene enlarges adipocytes and is a marker of adipocyte size. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E117–E124. [Google Scholar] [CrossRef] [PubMed]
- Seidah, N.G.; Awan, Z.; Chrétien, M.; Mbikay, M. PCSK9: A key modulator of cardiovascular health. Circ. Res. 2014, 114, 1022–1036. [Google Scholar] [CrossRef] [PubMed]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Eur. Heart J. 2020, 41, 111–188. [Google Scholar] [CrossRef]
- Luderer, U.; Lim, J.; Ortiz, L.; Nguyen, J.D.; Shin, J.H.; Allen, B.D.; Liao, L.S.; Malott, K.; Perraud, V.; Wingen, L.M.; et al. Exposure to environmentally relevant concentrations of ambient fine particulate matter (PM2.5) depletes the ovarian follicle reserve and causes sex-dependent cardiovascular changes in apolipoprotein E null mice. Part. Fibre Toxicol. 2022, 19, 5. [Google Scholar] [CrossRef]
- Kubes, P.; Jenne, C. Immune Responses in the Liver. Annu. Rev. Immunol. 2018, 36, 247–277. [Google Scholar] [CrossRef]
- Huang, L.-R.; Wohlleber, D.; Reisinger, F.; Jenne, C.N.; Cheng, R.-L.; Abdullah, Z.; Schildberg, F.A.; Odenthal, M.; Dienes, H.-P.; van Rooijen, N.; et al. Intrahepatic myeloid-cell aggregates enable local proliferation of CD8+ T cells and successful immunotherapy against chronic viral liver infection. Nat. Immunol. 2013, 14, 574–583. [Google Scholar] [CrossRef]
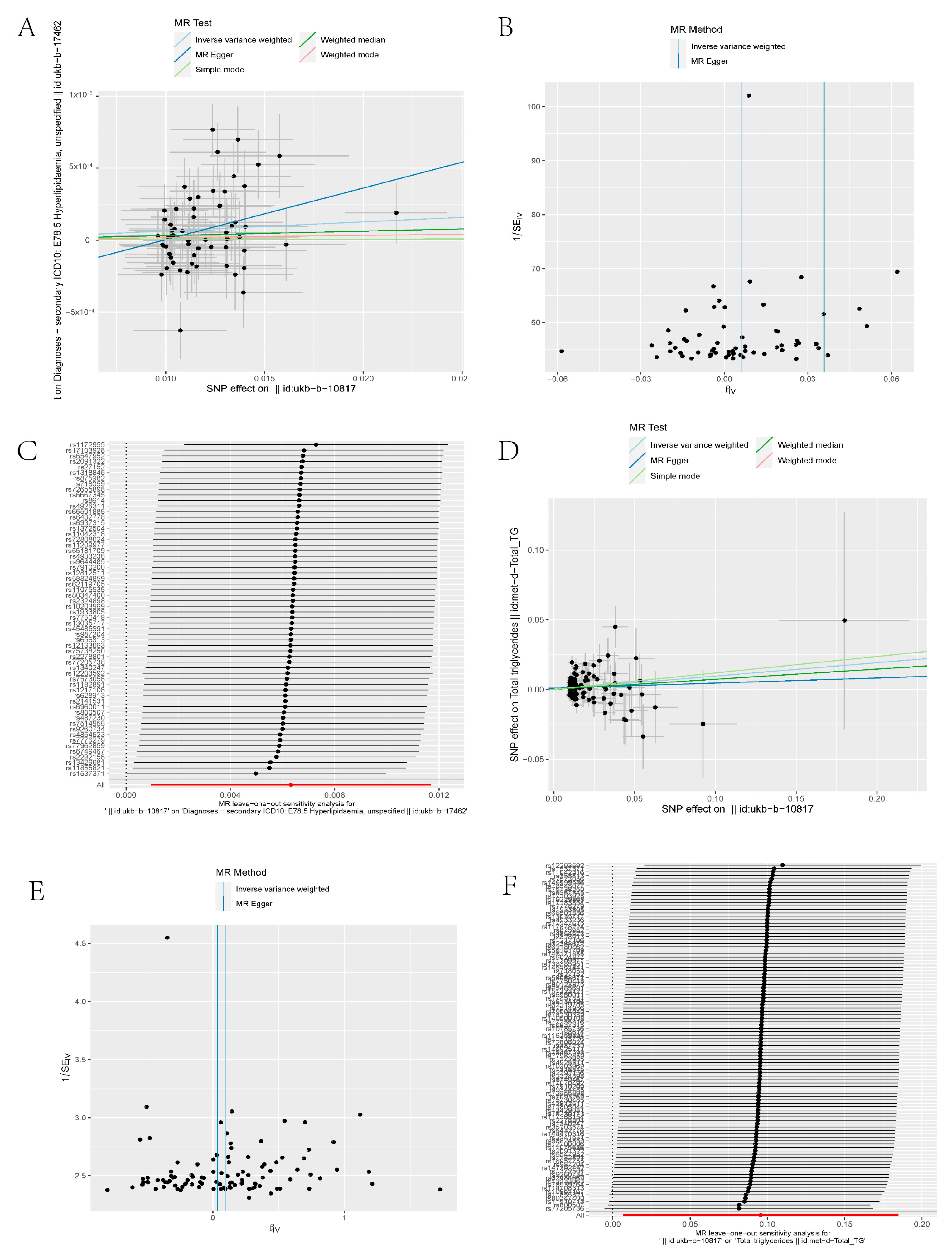


| Exposure | Outcome | Method | snp | OR | SE | p Value |
|---|---|---|---|---|---|---|
| ukb-b-10817 | ukb-b-17462 | MR Egger | 60 | 1.036497 | 0.016453 | 0.033427 |
| ukb-b-10817 | ukb-b-17462 | Weighted median | 60 | 1.003108 | 0.003327 | 0.351026 |
| ukb-b-10817 | ukb-b-17462 | Inverse variance weighted | 60 | 1.006341 | 0.002738 | 0.020954 |
| ukb-b-10817 | ukb-b-17462 | Simple mode | 60 | 1.000313 | 0.007391 | 0.966405 |
| ukb-b-10817 | ukb-b-17462 | Weighted mode | 60 | 1.001635 | 0.006905 | 0.813814 |
| Exposure | Outcome | Method | snp | OR | SE | p Value |
|---|---|---|---|---|---|---|
| ukb-b-10817 | met-d-Total_TG | MR Egger | 102 | 1.036998 | 0.116365 | 0.75553 |
| ukb-b-10817 | met-d-Total_TG | Weighted median | 102 | 1.075608 | 0.05714 | 0.202108 |
| ukb-b-10817 | met-d-Total_TG | Inverse variance weighted | 102 | 1.100426 | 0.045397 | 0.035029 |
| ukb-b-10817 | met-d-Total_TG | Simple mode | 102 | 1.125343 | 0.177939 | 0.508431 |
| ukb-b-10817 | met-d-Total_TG | Weighted mode | 102 | 1.125343 | 0.1713 | 0.492174 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Shen, G.; Lin, X.; Zhang, L.; Fan, F.; Zhang, Y.; Li, J. Identifying the Relationship between PM2.5 and Hyperlipidemia Using Mendelian Randomization, RNA-seq Data and Model Mice Subjected to Air Pollution. Toxics 2023, 11, 823. https://doi.org/10.3390/toxics11100823
Zhao Y, Shen G, Lin X, Zhang L, Fan F, Zhang Y, Li J. Identifying the Relationship between PM2.5 and Hyperlipidemia Using Mendelian Randomization, RNA-seq Data and Model Mice Subjected to Air Pollution. Toxics. 2023; 11(10):823. https://doi.org/10.3390/toxics11100823
Chicago/Turabian StyleZhao, Yixue, Geng Shen, Xipeng Lin, Long Zhang, Fangfang Fan, Yan Zhang, and Jianping Li. 2023. "Identifying the Relationship between PM2.5 and Hyperlipidemia Using Mendelian Randomization, RNA-seq Data and Model Mice Subjected to Air Pollution" Toxics 11, no. 10: 823. https://doi.org/10.3390/toxics11100823
APA StyleZhao, Y., Shen, G., Lin, X., Zhang, L., Fan, F., Zhang, Y., & Li, J. (2023). Identifying the Relationship between PM2.5 and Hyperlipidemia Using Mendelian Randomization, RNA-seq Data and Model Mice Subjected to Air Pollution. Toxics, 11(10), 823. https://doi.org/10.3390/toxics11100823







