Gene Expression Analysis of the Stress Response to Lithium, Nickel, and Zinc in Paracentrotus lividus Embryos
Abstract
1. Introduction
2. Materials and Methods
2.1. Embryo Culture
2.2. Metals Treatment of P. lividus Embryos
2.3. Indirect Immunofluorescence (IF)
2.4. nCounter NanoString Gene Expression Assay
2.5. Real-Time Quantitative PCR (qPCR)
2.6. KEGG Enrichment Analysis
2.7. Statistical Analysis
3. Results
3.1. Developmental Effects of Li, Ni and Zn on Sea Urchin Embryos
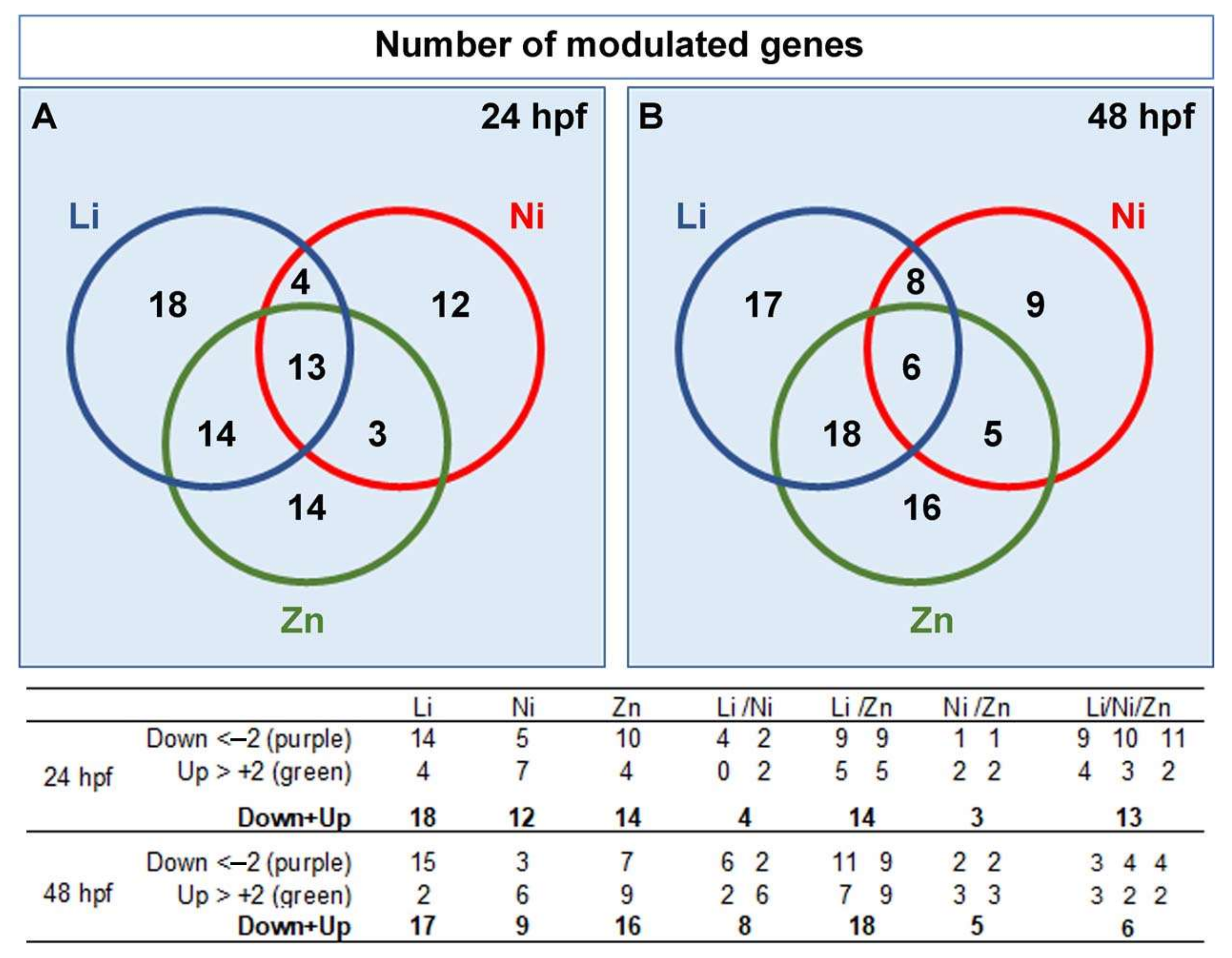
3.2. Profiling Transcriptional Outputs in Li, Ni, Zn Treated Embryos
3.3. Metal-Dependent Sensitive Genes as Potential Biomarkers
3.4. Metal-Dependent Sensitive Genes and Functional Enrichment
4. Discussion
4.1. Looking for Potential Biomarkers
4.2. Analysis of Pathways Affected by Metals
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caito, S.; Aschner, M. Neurotoxicity of Metals. Handb. Clin. Neurol. 2015, 131, 169–189. [Google Scholar] [PubMed]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Blewett, T.A.; Leonard, E.M. Mechanisms of nickel toxicity to fish and invertebrates in marine and estuarine waters. Environ. Pollut. 2017, 223, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Ansari, T.M.; Marr, I.L.; Tariq, N. Heavy Metals in Marine Pollution Perspective—A Mini Review. J. Appl. Sci. 2003, 4, 1–20. [Google Scholar] [CrossRef]
- Thompson, E.D.; Hogstrand, C.; Glover, C.N. From sea squirts to squirrelfish: Facultative trace element hyperaccumulation in animals. Metallomics 2018, 10, 777–793. [Google Scholar] [CrossRef] [PubMed]
- Yllmaz, A.B.; Yanar, A.; Alkan, E.N. Review of heavy metal accumulation on aquatic environment in Northern East Mediterrenean Sea part I: Some essential metals. Rev. Environ. Health 2017, 32, 119–163. [Google Scholar] [CrossRef]
- Deforest, D.K.; Schlekat, C.E. Species Sensitivity Distribution Evaluation for Chronic Nickel Toxicity to Marine Organisms. Integr. Environ. Assess. Manag. 2013, 9, 580–589. [Google Scholar] [CrossRef]
- Police, S.; Maity, S.; Chaudhary, D.K.; Dusane, C.K.; Sahu, S.K.; Kumar, A.V. Estimation of trace and toxic metals in marine biota and associated health risk assessment in Thane Creek, Mumbai, India. Environ. Chem. Ecotoxicol. 2021, 3, 234–240. [Google Scholar] [CrossRef]
- Kavanagh, L.; Keohane, J.; Cleary, J.; Cabellos, G.G.; Lloyd, A. Lithium in the natural waters of the south east of Ireland. Int. J. Environ. Res. Public Health 2017, 14, 561. [Google Scholar] [CrossRef]
- Shen, J.; Li, X.; Shi, X.; Wang, W.; Zhou, H.; Wu, J.; Wang, X.; Li, J. The toxicity of lithium to human cardiomyocytes. Environ. Sci. Eur. 2020, 32, 59. [Google Scholar] [CrossRef]
- Pandey, G.N.; Davis, J.M. Biology of the Lithium Ion. In Lithium Effects on Granulopoiesis and Immune Function; Rossof, A.H., Robinson, W.A., Eds.; Springer: Boston, MA, USA, 1980; pp. 15–59. ISBN 978-1-4757-0259-0. [Google Scholar]
- Manji, H.K.; Lenox, R.H. Signaling: Cellular insights into the pathophysiology of bipolar disorder. Biol. Psychiatry 2000, 48, 518–530. [Google Scholar] [CrossRef]
- Thibon, F.; Weppe, L.; Vigier, N.; Churlaud, C.; Lacoue-Labarthe, T.; Metian, M.; Cherel, Y.; Bustamante, P. Large-scale survey of lithium concentrations in marine organisms. Sci. Total Environ. 2021, 751, 141453. [Google Scholar] [CrossRef] [PubMed]
- Horstadius, S. Vegetalization of the sea-urchin egg by dinitrophenol and animalization by trypsin and ficin. J. Embryol. Exp. Morph. 1953, 1, 327–348. [Google Scholar] [CrossRef]
- Davidson, E.H.; Cameron, R.A.; Ransick, A. Specification of cell fate in the sea urchin embryo: Summary and some proposed mechanisms. Development 1998, 125, 3269–3290. [Google Scholar] [CrossRef]
- Martik, M.L.; Mcclay, D.R. Deployment of a retinal determination gene network drives directed cell migration in the sea urchin embryo. eLife 2015, 4, e08827. [Google Scholar] [CrossRef]
- Davidson, E.H.; Rast, J.P.; Oliveri, P.; Ransick, A.; Calestani, C.; Yuh, C.H.; Minokawa, T.; Amore, G.; Hinman, V.; Arenas-Mena, C.; et al. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev. Biol. 2002, 246, 162–190. [Google Scholar] [CrossRef]
- McClay, D.R.; Martik, M.L.; Lyons, D.C. Developmental gene regulatory networks in sea urchins and what we can learn from them. F1000Research 2016, 5, 203. [Google Scholar]
- Poustka, A.J.; Kühn, A.; Groth, D.; Weise, V.; Yaguchi, S.; Burke, R.D.; Herwig, R.; Lehrach, H.; Panopoulou, G. A global view of gene expression in lithium and zinc treated sea urchin embryos: New components of gene regulatory networks. Genome Biol. 2007, 8, R85. [Google Scholar] [CrossRef]
- Ghiglione, C.; Lhomond, G.; Lepage, T.; Gache, C. Cell-autonomous expression and position-dependent repression by Li+ of two zygotic genes during sea urchin early development. EMBO J. 1993, 12, 87–96. [Google Scholar] [CrossRef]
- Hardin, J.; Coffman, J.A.; Black, S.D.; MCClay, D.R. Commitment along the Dorsoventral Axis of the Sea-Urchin Embryo Is Altered in Response To NiCl2. Development 1992, 116, 671–685. [Google Scholar] [CrossRef]
- Goldstone, J.V.; Hamdoun, A.; Cole, B.J.; Howard-Ashby, M.; Nebert, D.W.; Scally, M.; Dean, M.; Epel, D.; Hahn, M.E.; Stegeman, J.J. The chemical defensome: Environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev. Biol. 2006, 300, 366–384. [Google Scholar] [CrossRef] [PubMed]
- López-Landavery, E.A.; Amador-Cano, G.; Alejandri, N.; Ramirez-Álvarez, N.; Montelongo, I.; Díaz, F.; Galindo-Sánchez, C.E. Transcriptomic response and hydrocarbon accumulation in the eastern oyster (Crassostrea virginica) exposed to crude oil. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 225, 108571. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Suh, S.S.; Park, M.; Park, S.Y.; Lee, S.; Lee, T.K. Differential gene expression patterns during embryonic development of sea urchin exposed to triclosan. Environ. Toxicol. 2017, 32, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Bonaventura, R.; Zito, F.; Russo, R.; Costa, C. A preliminary gene expression analysis on Paracentrotus lividus embryos exposed to UVB, Cadmium and their combination. Aquat. Toxicol. 2021, 232, 105770. [Google Scholar] [CrossRef]
- Russo, R.; Bonaventura, R.; Chiaramonte, M.; Costa, C.; Matranga, V.; Zito, F. Response to metals treatment of Fra1, a member of the AP-1 transcription factor family, in P. lividus sea urchin embryos. Mar. Environ. Res. 2018, 139, 99–112. [Google Scholar] [CrossRef]
- Zito, F.; Burke, R.D.; Matranga, V. Pl-nectin, a discoidin family member, is a ligand for beta C integrins in the sea urchin embryo. Matrix Biol. 2010, 29, 341–345. [Google Scholar] [CrossRef]
- Costa, C.; Karakostis, K.; Zito, F.; Matranga, V. Phylogenetic analysis and expression patterns of p16 and p19 in Paracentrotus lividus embryos. Dev. Genes Evol. 2012, 222, 245–251. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Russo, R.; Zito, F.; Costa, C.; Bonaventura, R.; Matranga, V. Transcriptional increase and misexpression of 14-3-3 epsilon in sea urchin embryos exposed to UV-B. Cell Stress Chaperones 2010, 15, 993–1001. [Google Scholar] [CrossRef]
- Russo, R.; Bonaventura, R.; Matranga, V. Time- and dose-dependent gene expression in sea urchin embryos exposed to UVB. Mar. Environ. Res. 2014, 93, 85–92. [Google Scholar] [CrossRef]
- Russo, R.; Pinsino, A.; Costa, C.; Bonaventura, R.; Matranga, V.; Zito, F. The newly characterized Pl-jun is specifically expressed in skeletogenic cells of the Paracentrotus lividus sea urchin embryo. FEBS J. 2014, 281, 3828–3843. [Google Scholar] [CrossRef] [PubMed]
- Bonaventura, R.; Zito, F.; Chiaramonte, M.; Costa, C.; Russo, R. Nickel toxicity in P. lividus embryos: Dose dependent effects and gene expression analysis. Mar. Environ. Res. 2018, 139, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Wakamatsu, Y.; Uchikawa, M. The many faces of Sox2 function in neural crest development. Dev. Growth Differ. 2021, 63, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Hinman, V.F.; Burke, R.D. Embryonic neurogenesis in echinoderms. WIREs Dev. Biol. 2018, 7, e316. [Google Scholar] [CrossRef]
- Prokopec, S.D.; Watson, J.D.; Waggott, D.M.; Smith, A.B.; Wu, A.H.; Okey, A.B.; Pohjanvirta, R.; Boutros, P.C. Systematic evaluation of medium-throughput mRNA abundance platforms. RNA 2013, 19, 51–62. [Google Scholar] [CrossRef]
- Nemer, M. An altered series of ectodermal gene expressions accompanying the reversible suspension of differentiation in the zinc-animalized sea urchin embryo. Dev. Biol. 1986, 114, 214–224. [Google Scholar] [CrossRef]
- Ryu, T.K.; Lee, G.; Rhee, Y.; Park, H.S.; Chang, M.; Lee, S.; Lee, J.; Lee, T.K. Identification of nickel response genes in abnormal early developments of sea urchin by differential display polymerase chain reaction. Ecotoxicol. Environ. Saf. 2012, 84, 18–24. [Google Scholar] [CrossRef]
- Molina, M.D.; Lepage, T. Maternal factors regulating symmetry breaking and dorsal–ventral axis formation in the sea urchin embryo. In Current Topics in Developmental Biology; Marlow, F.L., Ed.; Academic Press Inc. Elsevier Science: San Diego CA, USA, 2020; Volume 140, pp. 283–316. ISBN 9780128152201. [Google Scholar]
- Anishchenko, E.; Arnone, M.I.; D’Aniello, S. SoxB2 in sea urchin development: Implications in neurogenesis, ciliogenesis and skeletal patterning. EvoDevo 2018, 9, 5. [Google Scholar] [CrossRef]
- Shimada, T.; Fournier, A.E.; Yamagata, K. Neuroprotective function of 14-3-3 proteins in neurodegeneration. BioMed Res. Int. 2013, 2013, 564534. [Google Scholar] [CrossRef]
- Solek, C.M.; Oliveri, P.; Loza-Coll, M.; Schrankel, C.S.; Ho, E.C.H.; Wang, G.; Rast, J.P. An ancient role for Gata-1/2/3 and Scl transcription factor homologs in the development of immunocytes. Dev. Biol. 2013, 382, 280–292. [Google Scholar] [CrossRef]
- Suh, S.S.; Hwang, J.; Park, M.; Park, S.Y.; Ryu, T.K.; Lee, S.; Lee, T.K. Hypoxia-modulated gene expression profiling in sea urchin (Strongylocentrotus nudus) immune cells. Ecotoxicol. Environ. Saf. 2014, 109, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Zito, F.; Tesoro, V.; McClay, D.R.; Nakano, E.; Matranga, V. Ectoderm cell-ECM interaction is essential for sea urchin embryo skeletogenesis. Dev. Biol. 1998, 196, 184–192. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barsi, J.C.; Tu, Q.; Calestani, C.; Davidson, E.H. Genome-wide assessment of differential effector gene use in embryogenesis. Development 2015, 142, 3892–3901. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.F.Z.; Reuille, R.L.; Ming, L.J.; Livingston, B.T. Overexpression and mechanistic characterization of blastula protease 10, a metalloprotease involved in sea urchin embryogenesis and development. J. Biol. Chem. 2006, 281, 10737–10744. [Google Scholar] [CrossRef]
- Kozmik, Z. Pax genes in eye development and evolution. Curr. Opin. Genet. Dev. 2005, 15, 430–438. [Google Scholar] [CrossRef]
- Lapraz, F.; Röttinger, E.; Duboc, V.; Range, R.; Duloquin, L.; Walton, K.; Wu, S.Y.; Bradham, C.; Loza, M.A.; Hibino, T.; et al. RTK and TGF-β signaling pathways genes in the sea urchin genome. Dev. Biol. 2006, 300, 132–152. [Google Scholar] [CrossRef]
- Saudemont, A.; Haillot, E.; Mekpoh, F.; Bessodes, N.; Quirin, M.; Lapraz, F.; Duboc, V.; Röttinger, E.; Range, R.; Oisel, A.; et al. Ancestral regulatory circuits governing ectoderm patterning downstream of nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet. 2010, 6, e1001259. [Google Scholar] [CrossRef]
- Zito, F.; Costa, C.; Sciarrino, S.; Poma, V.; Russo, R.; Angerer, L.M.; Matranga, V. Expression of univin, a TGF-β growth factor, requires ectoderm–ECM interaction and promotes skeletal growth in the sea urchin embryo. Dev. Biol. 2003, 264, 217–227. [Google Scholar] [CrossRef]
- Martino, C.; Costa, C.; Roccheri, M.C.; Koop, D.; Scudiero, R.; Byrne, M. Gadolinium perturbs expression of skeletogenic genes, calcium uptake and larval development in phylogenetically distant sea urchin species. Aquat. Toxicol. 2018, 194, 57–66. [Google Scholar] [CrossRef]
- Mcintyre, D.C.; Seay, N.W.; Croce, J.C.; McClay, D.R. Short-range Wnt5 signaling initiates specification of sea urchin posterior ectoderm. Development 2013, 140, 4881–4889. [Google Scholar] [CrossRef]
- Croce, J.; Lhomond, G.; Lozano, J.C.; Gache, C. ske-T, a T-box gene expressed in the skeletogenic mesenchyme lineage of the sea urchin embryo. Mech. Dev. 2001, 107, 159–162. [Google Scholar] [CrossRef]
- Croce, J.; Lhomond, G.; Gache, C. Coquillette, a sea urchin T-box gene of the Tbx2 subfamily, is expressed asymmetrically along the oral-aboral axis of the embryo and is involved in skeletogenesis. Mech. Dev. 2003, 120, 561–572. [Google Scholar] [CrossRef]
- Beere, H.M. Death versus survival: Functional interaction between the apoptotic and stress-inducible heat shock protein pathways. J. Clin. Investig. 2005, 115, 2633–2639. [Google Scholar] [CrossRef] [PubMed]
- Bonaventura, R.; Zito, F.; Costa, C.; Giarrusso, S.; Celi, F.; Matranga, V. Stress response gene activation protects sea urchin embryos exposed to X-rays. Cell Stress Chaperones 2011, 16, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Hirate, Y.; Hiyama, T.; Yamasu, K.; Suyemitsu, T.; Ishihara, K. Purification of EGIP-D-Binding Protein from the Embryos of the Sea Urchin Anthocidaris crassispina. Zool. Sci. 1997, 14, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Duloquin, L.; Lhomond, G.; Gache, C. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development 2007, 134, 2293–2302. [Google Scholar] [CrossRef]
- Karakostis, K.; Costa, C.; Zito, F.; Brümmer, F.; Matranga, V. Characterization of an Alpha Type Carbonic Anhydrase from Paracentrotus lividus Sea Urchin Embryos. Mar. Biotechnol. 2016, 18, 384–395. [Google Scholar] [CrossRef]
- Piacentino, M.L.; Zuch, D.T.; Fishman, J.; Rose, S.; Speranza, E.E.; Li, C.; Yu, J.; Chung, O.; Ramachandran, J.; Ferrell, P.; et al. RNA-Seq identifies SPGs as a ventral skeletal patterning cue in sea urchins. Development 2016, 142, 4217–4229. [Google Scholar] [CrossRef]
- Ho, E.C.H.; Buckley, K.M.; Schrankel, C.S.; Schuh, N.W.; Hibino, T.; Solek, C.M.; Bae, K.; Wang, G.; Rast, J.P. Perturbation of gut bacteria induces a coordinated cellular immune response in the purple sea urchin larva. Immunol. Cell Biol. 2016, 94, 861–874. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, R.; Wei, X.; Lv, M.; Jiang, Z. Metalloimmunology: The metal ion-controlled immunity. Adv. Immunol. 2020, 145, 187–241. [Google Scholar] [CrossRef]
- Arshinoff, B.I.; Cary, G.A.; Karimi, K.; Foley, S.; Agalakov, S.; Delgado, F.; Lotay, V.S.; Ku, C.J.; Pells, T.J.; Beatman, T.R.; et al. Echinobase: Leveraging an extant model organism database to build a knowledgebase supporting research on the genomics and biology of echinoderms. Nucleic Acids Res. 2022, 50, D970–D979. [Google Scholar] [CrossRef] [PubMed]
- Andrikou, C.; Iovene, E.; Rizzo, F.; Oliveri, P.; Arnone, M.I. Myogenesis in the Sea Urchin Embryo: The Molecular Fingerprint of the Myoblast Precursors. EvoDevo 2013, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Oliveri, P.; Walton, K.D.; Davidson, E.H.; McClay, D.R. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development 2006, 133, 4173–4181. [Google Scholar] [CrossRef] [PubMed]
- Robert, N.; Lhomond, G.; Schubert, M.; Croce, J.C. A comprehensive survey of wnt and frizzled expression in the sea urchin Paracentrotus lividus. Genesis 2014, 52, 235–250. [Google Scholar] [CrossRef]
- Cui, M.; Siriwon, N.; Li, E.; Davidson, E.H.; Peter, I.S. Specific functions of the Wnt signaling system in gene regulatory networks throughout the early sea urchin embryo. Proc. Natl. Acad. Sci. USA 2014, 111, E5029–E5038. [Google Scholar] [CrossRef]
- Range, R.C.; Angerer, R.C.; Angerer, L.M. Integration of Canonical and Noncanonical Wnt Signaling Pathways Patterns the Neuroectoderm along the Anterior-Posterior Axis of Sea Urchin Embryos. PLoS Biol. 2013, 11, e1001467. [Google Scholar] [CrossRef]
- Pascual-Vargas, P.; Salinas, P.C. A Role for Frizzled and Their Post-Translational Modifications in the Mammalian Central Nervous System. Front. Cell Dev. Biol. 2021, 9, 692888. [Google Scholar] [CrossRef]
- Poustka, A.J.; Kü, A.; Radosavljevic, V.; Wellenreuther, R.; Lehrach, H.; Panopoulou, G. On the origin of the chordate central nervous system: Expression of onecut in the sea urchin embryo. Evol. Dev. 2004, 6, 227–236. [Google Scholar] [CrossRef]
- Tu, Q.; Brown, C.T.; Davidson, E.H.; Oliveri, P. Sea urchin Forkhead gene family: Phylogeny and embryonic expression. Dev. Biol. 2006, 300, 49–62. [Google Scholar] [CrossRef]
- Molina, M.D.; de Crozé, N.; Haillot, E.; Lepage, T. Nodal: Master and commander of the dorsal-ventral and left-right axes in the sea urchin embryo. Curr. Opin. Genet. Dev. 2013, 23, 445–453. [Google Scholar] [CrossRef]
- Bonaventura, R.; Russo, R.; Zito, F.; Matranga, V. Combined effects of cadmium and UVB radiation on sea urchin embryos: Skeleton impairment parallels p38 MAPK activation and stress genes overexpression. Chem. Res. Toxicol. 2015, 28, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
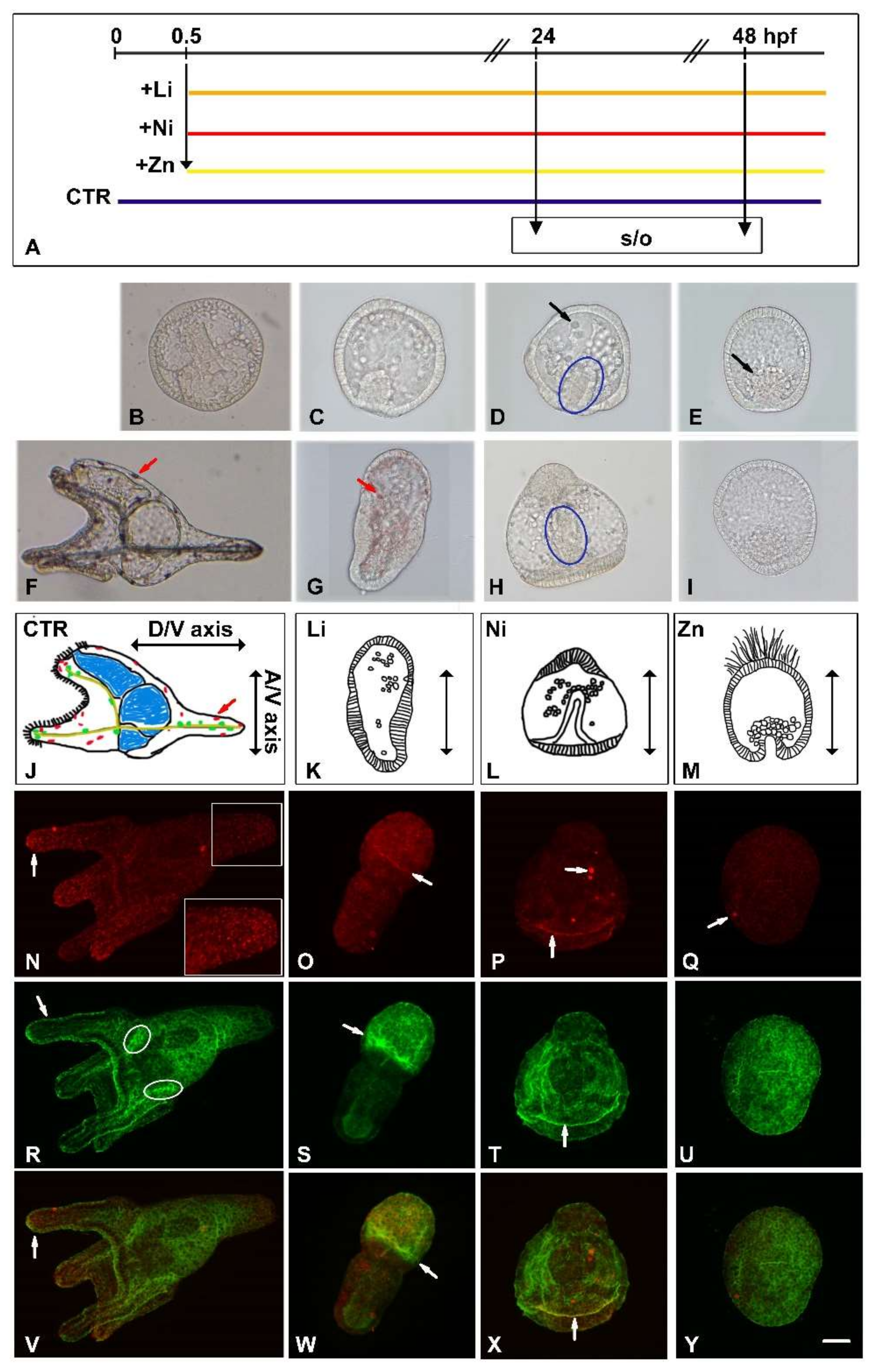
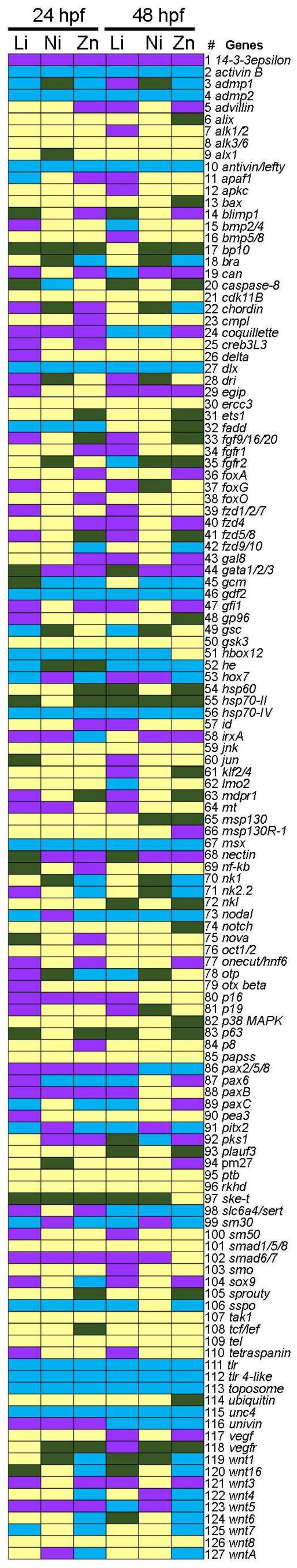
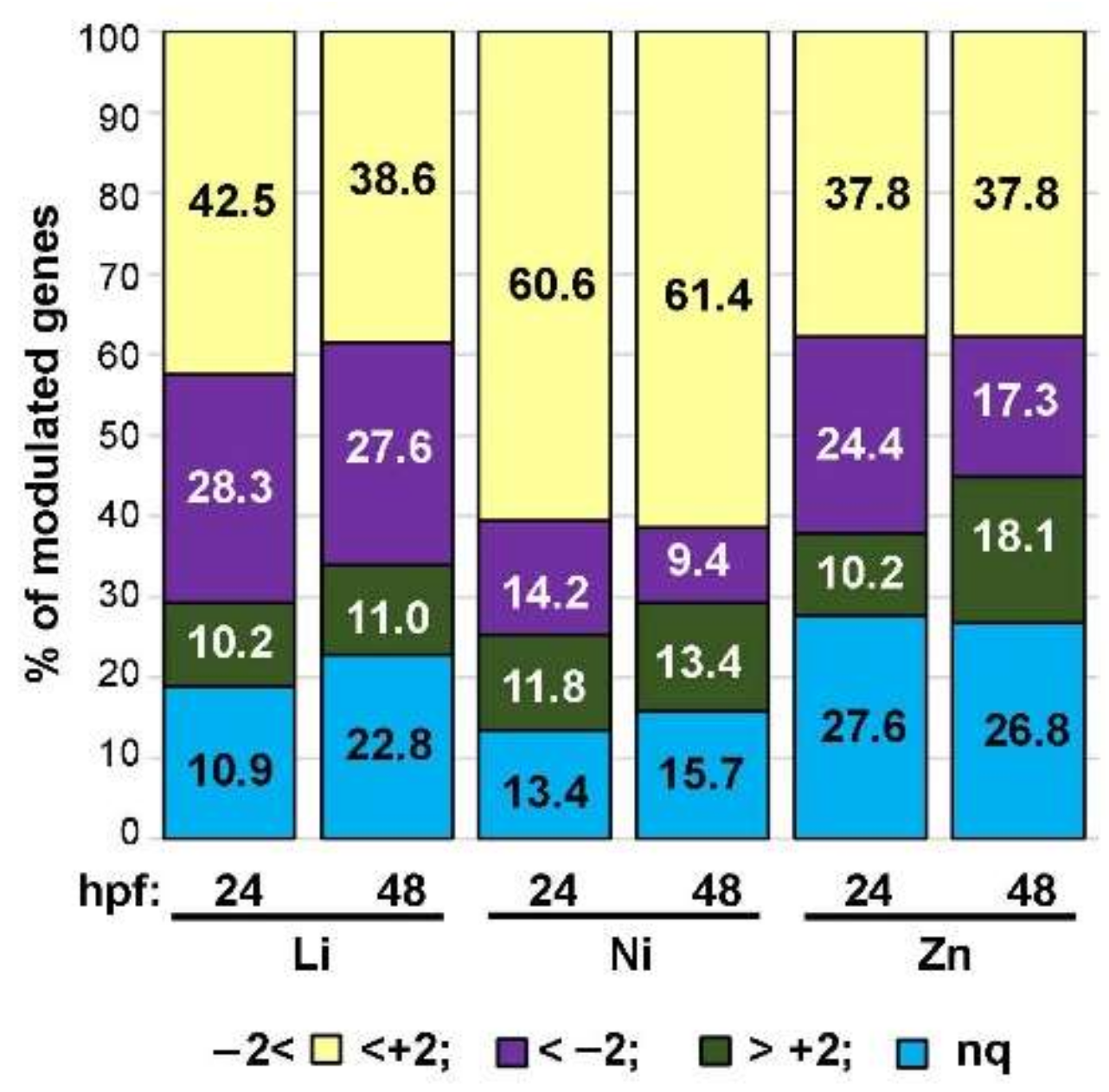


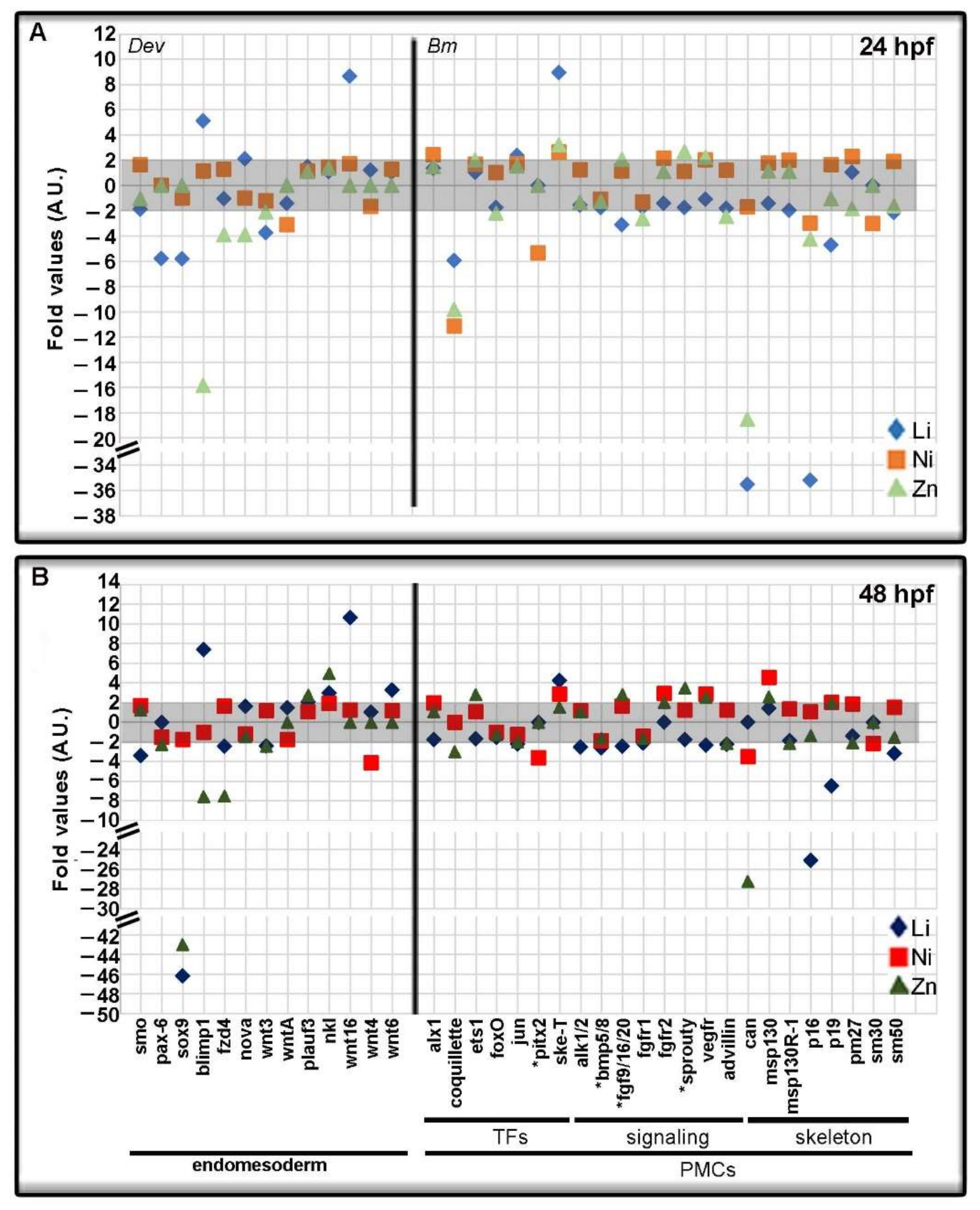

| Endpoint | Gene | Accession Number | Category | Function |
|---|---|---|---|---|
| 24 hpf | 14-3-3epsilon | AJ493680.2 | Defense | protein adaptor |
| gata1/2/3 | GQ377404.1 | Immune | TF | |
| chordin | FJ976182.1 | Nervous | extracellular modulator | |
| pax2/5/8 | AF016884.1 | Nervous | TF | |
| bp10 | X56224.1 | Development | Enzyme | |
| nectin | AJ578435.2 | Development | extracellular matrix protein | |
| paxB | AF016885.1 | Development | TF | |
| smad6/7 | HM449802.1 | Development | TF | |
| univin | DQ536195.1 | Development | TGF-β ligand | |
| wnt5 | HM449806.1 | Development | secreted glycoprotein | |
| p16 | FR693763.1 | Biomineralization | spicule matrix protein | |
| coquillette | AJ508929.1 | Biomineralization | TF | |
| ske-t | AJ309216.1 | Biomineralization | TF | |
| 48 hpf | 14-3-3epsilon | AJ493680.2 | Defense | protein adaptor |
| hsp70-II | X16544.1 | Defense | hsp family | |
| gata1/2/3 | GQ377404.1 | Immune | TF | |
| egip precursor | HM449819.1 | Development | Polypeptide | |
| nectin | AJ578435.2 | Development | extracellular matrix protein | |
| vegfr | AM419057.1 | Biomineralization | VEGF receptor |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonaventura, R.; Costa, C.; Deidda, I.; Zito, F.; Russo, R. Gene Expression Analysis of the Stress Response to Lithium, Nickel, and Zinc in Paracentrotus lividus Embryos. Toxics 2022, 10, 325. https://doi.org/10.3390/toxics10060325
Bonaventura R, Costa C, Deidda I, Zito F, Russo R. Gene Expression Analysis of the Stress Response to Lithium, Nickel, and Zinc in Paracentrotus lividus Embryos. Toxics. 2022; 10(6):325. https://doi.org/10.3390/toxics10060325
Chicago/Turabian StyleBonaventura, Rosa, Caterina Costa, Irene Deidda, Francesca Zito, and Roberta Russo. 2022. "Gene Expression Analysis of the Stress Response to Lithium, Nickel, and Zinc in Paracentrotus lividus Embryos" Toxics 10, no. 6: 325. https://doi.org/10.3390/toxics10060325
APA StyleBonaventura, R., Costa, C., Deidda, I., Zito, F., & Russo, R. (2022). Gene Expression Analysis of the Stress Response to Lithium, Nickel, and Zinc in Paracentrotus lividus Embryos. Toxics, 10(6), 325. https://doi.org/10.3390/toxics10060325









