Development of a Rapid and Cost-Effective Multiplex PCR Assay for the Simultaneous Identification of Three Commercially Important Sea Squirt Species (Halocynthia spp.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Acquisition and Genomic DNA Extraction
2.2. Mitochondrial Cytochrome Coxidase Subunit I (COX1) Amplification
2.3. Species-Specific Primer Design
2.4. Multiplex PCR of Species-Specific Primer Sets for Three Species of Sea Squirts
2.5. Confirmation of Multiplex PCR Amplification According to DNA Concentration
3. Results and Discussion
3.1. Mitochondrial Cytochrome Coxidase Subunit I (COX1) Amplification
3.2. Species-Specific Primer Design
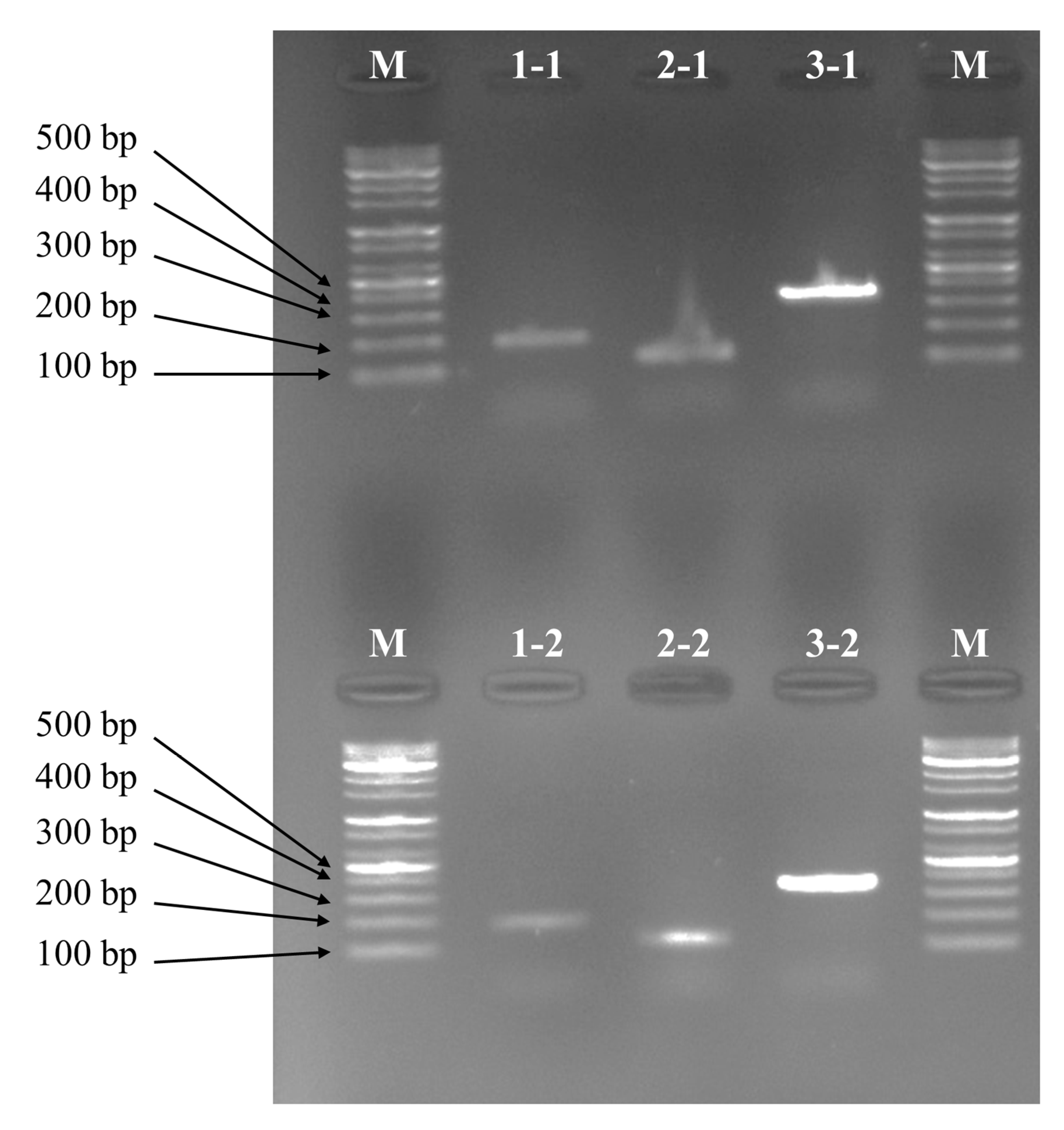
3.3. Multiplex PCR Set Amplification PCR Band
3.4. Multiplex PCR Set Amplification Sensitivity by DNA Concentration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lambert, G. Ecology and Natural History of the Protochordates. Can. J. Zool. 2005, 83, 34–50. [Google Scholar] [CrossRef]
- Hayashi, T.; Yamaguchi, K.; Konosu, S. Sensory Analysis of Taste Active Components in the Extract of Boiled Snow Crab Meat. J. Food Sci. 1981, 46, 479–483. [Google Scholar] [CrossRef]
- Nishikawa, T. The Ascidians of the Japan Sea. II. Publ. Seto Mar. Biol. Lab. 1991, 35, 25–170. [Google Scholar] [CrossRef]
- Jacquet, J.L.; Pauly, D. Trade Secrets: Renaming and Mislabeling of Seafood. Mar. Policy 2008, 32, 309–318. [Google Scholar] [CrossRef]
- Lee, G.Y.; Suh, S.M.; Lee, Y.M.; Kim, H.Y. Multiplex PCR Assay for Simultaneous Identification of Five Types of Tuna (Katsuwonus pelamis, Thunnus alalonga, T. albacares, T. obesus and T. thynnus). Foods 2022, 11, 280. [Google Scholar] [CrossRef]
- Lee, Y.M.; Lee, S.; Kim, H.Y. A Multiplex PCR Assay Combined with Capillary Electrophoresis for the Simultaneous Identification of Atlantic Cod, Pacific Cod, Blue Whiting, Haddock, and Alaska Pollock. Foods 2021, 10, 2631. [Google Scholar] [CrossRef]
- Lin, W.F.; Hwang, D.F. A Multiplex PCR Assay for Species Identification of Raw and Cooked Bonito. Food Control 2008, 19, 879–885. [Google Scholar] [CrossRef]
- Yoon, J.M.; Kim, J.Y. Genetic Differences and Variation of Ascidians, Halocynthia roretzi von Drasche and H. hilgendorfi Oka Identified by PCR Analysis. Dev. Reprod. 2011, 15, 359–364. [Google Scholar]
- Mafra, I.; Ferreira, I.M.; Oliveira, M.B.P. Food Authentication by PCR-Based Methods. Eur. Food Res. Technol. 2008, 227, 649–665. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Hebert, P.D.; Ratnasingham, S.; deWaard, J.R. Barcoding Animal Life: Cytochrome c Oxidase Subunit 1 Divergences among Closely Related Species. Proc. R. Soc. B Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [PubMed]
- Lockley, A.K.; Bardsley, R.G. DNA-Based Methods for Food Authentication. Trends Food Sci. Technol. 2000, 11, 67–77. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. DNA Barcodes: Genes, Genomics, and Bioinformatics. Proc. Natl. Acad. Sci. USA 2008, 105, 2761–2762. [Google Scholar] [CrossRef]
- Gostel, M.R.; Kress, W.J. The Expanding Role of DNA Barcodes: Indispensable Tools for Ecology, Evolution, and Conservation. Diversity 2022, 14, 213. [Google Scholar] [CrossRef]
- Dawan, J.; Ahn, J. Application of DNA Barcoding for Ensuring Food Safety and Quality. Food Sci. Biotechnol. 2022, 31, 1355–1364. [Google Scholar] [CrossRef]
- Rasmussen, R.S.; Morrissey, M.T. DNA-Based Methods for the Identification of Commercial Fish and Seafood Species. Compr. Rev. Food Sci. Food Saf. 2008, 7, 280–295. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Bonants, P.; Groenewald, E.; Rasplus, J.Y.; Maes, M.; De Vos, P.; Frey, J.; Boonham, N.; Nicolaisen, M.; Bertacini, A.; Robert, V.; et al. QBOL: A New EU Project Focusing on DNA Barcoding of Quarantine Organisms. EPPO Bull. 2010, 40, 30–33. [Google Scholar] [CrossRef]
- Bottero, M.T.; Dalmasso, A. Animal Species Identification in Food Products: Evolution of Biomolecular Methods. Vet. J. 2011, 190, 34–38. [Google Scholar] [CrossRef]
- Pfenninger, M.; Cordellier, M.; Streit, B. Comparing the Efficacy of Morphologic and DNA-Based Taxonomy in the Freshwater Gastropod Genus Radix (Basommatophora, Pulmonata). BMC Evol. Biol. 2006, 6, 100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhai, X.; He, L.; Wang, Z.; Cao, H.; Wang, P.; Ma, W. Morphological Description and DNA Barcoding Research of Nine Syringa Species. Front. Genet. 2025, 16, 1544062. [Google Scholar] [CrossRef] [PubMed]
- Shokralla, S.; Porter, T.M.; Gibson, J.F.; Dobosz, R.; Janzen, D.H.; Hallwachs, W.; Hajibabaei, M. Massively Parallel Multiplex DNA Sequencing for Specimen Identification Using an Illumina MiSeq Platform. Sci. Rep. 2015, 5, 9687. [Google Scholar] [CrossRef]
- Stein, E.D.; Martinez, M.C.; Stiles, S.; Miller, P.E.; Zakharov, E.V. Is DNA Barcoding Actually Cheaper and Faster than Traditional Morphological Methods? Results from a Survey of Freshwater Bioassessment Efforts in the United States. PLoS ONE 2014, 9, e95525. [Google Scholar] [CrossRef]
- Zouganelis, G.D.; Tairis, N. Low Throughput Direct Cycle Sequencing of Polymerase Chain Reaction (PCR) Products. In DNA Manipulation and Analysis; Springer: New York, NY, USA, 2023; pp. 195–211. [Google Scholar]
- Rossen, J.W.; Friedrich, A.W.; Moran-Gilad, J. Practical Issues in Implementing Whole Genome Sequencing in Routine Diagnostic Microbiology. Clin. Microbiol. Infect. 2018, 24, 355–360. [Google Scholar] [CrossRef]
- Rathi, U.; Prabhavathi, V.; Meena, D.S.; Bamel, K. Understanding the Drifts in DNA Barcoding: A Systematic Review. Manuscript in preparation. Plant Sci. Today 2025, in press. [Google Scholar] [CrossRef]
- Bai, W.; Xu, W.; Huang, K.; Yuan, Y.; Cao, S.; Luo, Y. A Novel Common Primer Multiplex PCR (CPMPCR) Method for the Simultaneous Detection of Meat Species. Food Control 2009, 20, 366–370. [Google Scholar] [CrossRef]
- Hanapi, U.K.; Desa, M.N.M.; Ismail, A.; Mustafa, S. A Higher Sensitivity and Efficiency of Common Primer Multiplex PCR Assay in Identification of Meat Origin Using NADH Dehydrogenase Subunit 4 Gene. J. Food Sci. Technol. 2015, 52, 4166–4175. [Google Scholar] [CrossRef]
- Kitpipit, T.; Sittichan, K.; Thanakiatkrai, P. Direct Multiplex PCR Assay for Meat Species Identification in Food Products. Food Chem. 2014, 163, 77–82. [Google Scholar] [CrossRef]
- Sultana, S.; Hossain, M.M.; Naquiah, N.N.A.; Ali, M.E. Novel Multiplex PCR-RFLP Assay Discriminates Bovine, Porcine and Fish Gelatin Substitution in Asian Pharmaceuticals Capsule Shells. Food Addit. Contam. Part A 2018, 35, 1662–1673. [Google Scholar] [CrossRef] [PubMed]
- Boyrusbianto, F.T.; Utama, D.T.; Khikmawati, I.; Fadhila, A.; Volkandari, S.D.; Abdurrahman, Z.H.; Cahyadi, M. A Simplex and Multiplex PCR Assay for Simultaneous Detection of Beef, Pork, and Chicken Meat in Sausages Based on Mitochondrial DNA Cytochrome Oxidase Subunit I. Food Res. 2023, 7, 188–193. [Google Scholar] [CrossRef] [PubMed]
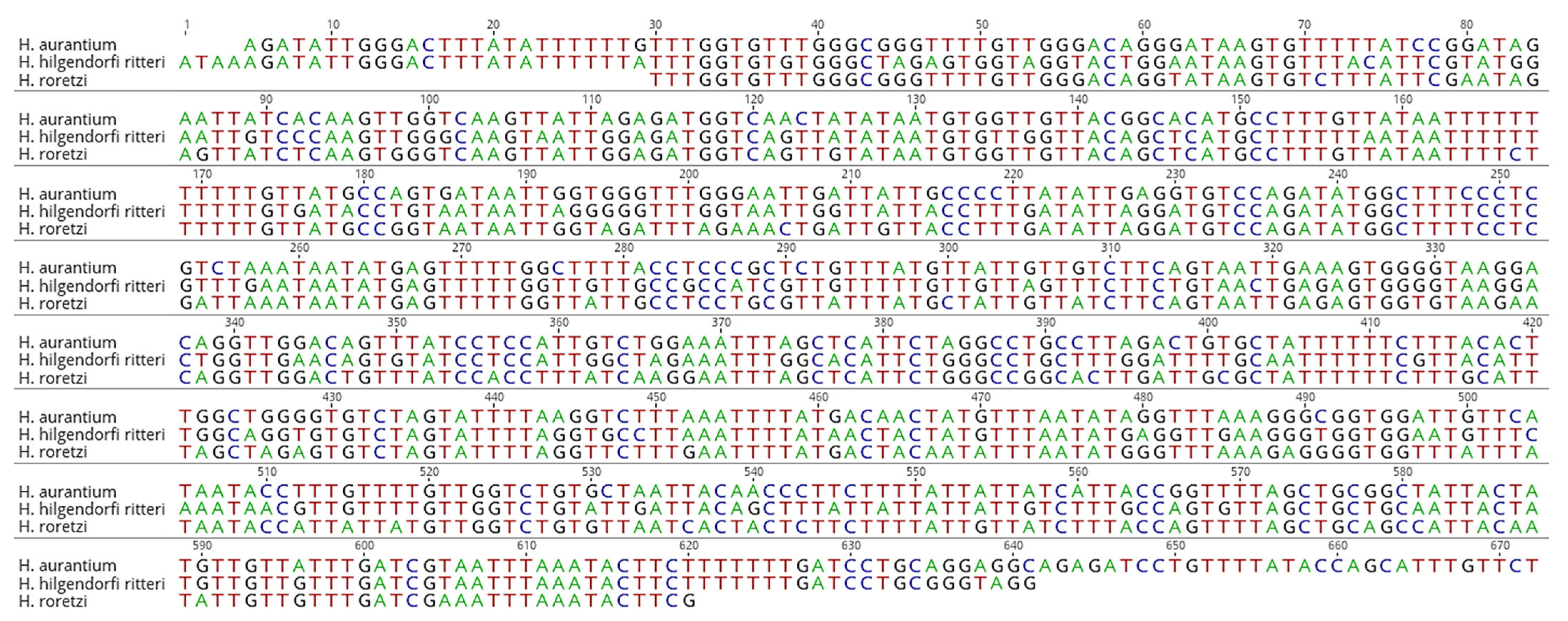


| Primer Name | Species | Sequence(5′-3′) | Primer Direction | Product Size (bp) |
|---|---|---|---|---|
| Hhr_CO1_F | H. hilgendorfi ritteri | TTGGTGTGTGGGCTAGAGTG | Forward | 360 |
| Ha_CO1_F | H. aurantium | GATTATTGCCCCTTATATTGAGG | Forward | 184 |
| Hr_CO1_F | H. roretzi | TGGTTATTGCCTCCTGCGT | Forward | 118 |
| Pyuridae_CO1_R | - | CMGGCCYAGAATGWGCYAA | Reverse | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-R.; Kim, H.-J.; Bang, I.-C. Development of a Rapid and Cost-Effective Multiplex PCR Assay for the Simultaneous Identification of Three Commercially Important Sea Squirt Species (Halocynthia spp.). Foods 2025, 14, 3003. https://doi.org/10.3390/foods14173003
Kim K-R, Kim H-J, Bang I-C. Development of a Rapid and Cost-Effective Multiplex PCR Assay for the Simultaneous Identification of Three Commercially Important Sea Squirt Species (Halocynthia spp.). Foods. 2025; 14(17):3003. https://doi.org/10.3390/foods14173003
Chicago/Turabian StyleKim, Kang-Rae, Hye-Jin Kim, and In-Chul Bang. 2025. "Development of a Rapid and Cost-Effective Multiplex PCR Assay for the Simultaneous Identification of Three Commercially Important Sea Squirt Species (Halocynthia spp.)" Foods 14, no. 17: 3003. https://doi.org/10.3390/foods14173003
APA StyleKim, K.-R., Kim, H.-J., & Bang, I.-C. (2025). Development of a Rapid and Cost-Effective Multiplex PCR Assay for the Simultaneous Identification of Three Commercially Important Sea Squirt Species (Halocynthia spp.). Foods, 14(17), 3003. https://doi.org/10.3390/foods14173003






