Technological and Safety Characterization of Coagulase-Negative Staphylococci Isolated from Sardinian Fermented Sausage Made by Ovine Meat
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Cultural Conditions
2.2. Strains Identification and Genetic Characterization
2.3. Staphylococci Technological Characterization
2.3.1. Nitrate Reductase Activity
2.3.2. Proteolytic Activity
2.3.3. Lipolytic Activity
2.3.4. Antibiotic Resistance
2.3.5. Biogenic Amines Production
3. Results and Discussion
3.1. Staphylococci Identification
3.2. Technological Characterization of Staphylococcus Strains
3.3. Strains’ Safety Assessment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mangia, N.P.; Cottu, M.; Mura, M.E.; Murgia, M.A.; Blaiotta, G. Technological parameters, anti-listeria activity, biogenic amines formation and degradation ability of L. plantarum strains isolated from sheep-fermented sausage. Microorganisms 2021, 9, 1895. [Google Scholar] [CrossRef]
- Mangia, N.P.; Murgia, M.A.; Garau, G.; Merella, R.; Deiana, P. Sardinian fermented sheep sausage: Microbial biodiversity resource for quality improvement. Options Méditerranéennes Ser. A 2008, 78, 273–277. Available online: http://om.ciheam.org/article.php?IDPDF=800276 (accessed on 22 December 2022).
- Bovolenta, S.; Boscolo, D.; Dovier, S.; Morgante, M.; Pallotti, A.; Piasentier, E. Effect of pork lard content on the chemical, microbiological and sensory properties of a typical fermented meat product (Pitina) obtained from Alpagota sheep. Meat Sci. 2008, 80, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.; Silva, S.; Guedes, C.; Rodrigues, S. Sheep and goat meat processed products quality: A review. Foods 2020, 9, 960. [Google Scholar] [CrossRef]
- Stajić, S.; Perunović, M.; Stanišić, N.; Žujović, M.; Živković, D. Sucuk (Turkish-style dry-fermented sausage) quality as an influence of recipe formulation and inoculation of starter cultures. J. Food Proc. Pres. 2013, 37, 870–880. [Google Scholar] [CrossRef]
- Helgesen, H.; Solheim, R.; Næs, T. Consumer purchase probability of dry fermented lamb sausages. Food Qual. Pref. 1998, 9, 295–301. [Google Scholar] [CrossRef]
- Leite, A.; Rodrigues, S.; Pereira, E.; Paulos, K.; Oliveira, A.F.; Lorenzo, J.M.; Teixeira, A. Physicochemical properties, fatty acid profile and sensory characteristics of sheep and goat meat sausages manufactured with different pork fat levels. Meat Sci. 2015, 105, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Paulos, K.; Rodrigues, S.; Oliveira, A.F.; Leite, A.; Pereira, E.; Teixeira, A. Sensory characterization and consumer preference mapping of fresh sausages manufactured with goat and sheep meat. J. Food Sci. 2015, 80, S1568–S1573. [Google Scholar] [CrossRef]
- Mangia, N.P.; Trani, A.; Di Luccia, A.; Faccia, M.; Gambacorta, G.; Fancello, F.; Deiana, P. Effect of the use of autochthonous Lactobacillus curvatus, Lactobacillus plantarum and Staphylococcus xylosus strains on microbiological and biochemical properties of the Sardinian fermented sausage. Eur. Food Res. Technol. 2013, 236, 557–566. [Google Scholar] [CrossRef]
- Lopez, C.M.; Callegari, M.L.; Patrone, V.; Rebecchi, A. Assessment of antibiotic resistance in staphylococci involved in fermented meat product processing. Curr. Opin. Food Sci. 2020, 31, 17–23. [Google Scholar] [CrossRef]
- Sun, J.; Cao, C.C.; Feng, M.Q.; Xu, X.L.; Zhou, G.H. Technological and safety characterization of coagulase-negative staphylococci with high protease activity isolated from Traditional Chinese fermented sausages. LWT Food Sci. Technol. 2019, 114, 108371. [Google Scholar] [CrossRef]
- Feng, L.; Qiao, Y.; Zou, Y.; Huang, M.; Kang, Z.; Zhou, G. Effect of Flavourzyme on proteolysis, antioxidant capacity and sensory attributes of Chinese sausage. Meat Sci. 2014, 98, 34–40. [Google Scholar] [CrossRef]
- Berardo, A.; Devreese, B.; De Maere, H.; Stavropoulou, D.A.; Van Royen, G.; Leroy, F.; De Smet, S. Actin proteolysis during ripening of dry fermented sausages at different pH values. Food Chem. 2017, 221, 1322–1332. [Google Scholar] [CrossRef]
- Rebecchi, A.; Miragoli, F.; Lopez, C.; Bassi, D.; Fontana, C. Exploring coagulase-negative Staphylococci diversity from Artisanal Llama sausages: Assessment of technological and safety traits. Microorganisms 2020, 8, 629. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, Q.; Chen, X.; Zhou, J.; Wu, J. Isolation and characterization of coagulase-negative staphylococci with high proteolytic activity from dry fermented sausages as a potential starter culture. Food Res. Int. 2022, 162, 111957. [Google Scholar] [CrossRef] [PubMed]
- Miralles, M.C.; Flores, J.; Pèrez-Martìnez, G. Biochemical test for the selection of Staphylococcus strains as potential meat starter cultures. Food Microbiol. 1996, 13, 227–236. [Google Scholar] [CrossRef]
- Fadda, S.; Sanz, Y.; Vignolo, G.M.; Aristoy, C.; Oliver, G.; Toldra, F. Characterization of muscle sarcoplasmic and myofibrillar protein hydrolysis caused by Lactobacillus plantarum. Appl. Environ. Microbiol. 1999, 65, 3540–3546. [Google Scholar] [CrossRef] [PubMed]
- Drosinos, E.H.; Paramithiotis, S.; Kolovos, G.; Tsikouras, I. Phenotypic and technological diversity of lactic acid bacteria and staphylococci isolated from traditionally fermented sausages in Southern Greece. Food Microbiol. 2007, 24, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Mauriello, G.; Casaburi, A.; Blaiotta, G.; Villani, F. Isolation and technological properties of coagulase-negative staphylococci from fermented sausages of Southern Italy. Meat Sci. 2004, 67, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing (32nd ed.), CLSI supplement M100. CLSI. 2022. Available online: https://clsi.org/about/press-releases/clsi-publishes-m100-performance-standards-for-antimicrobial-susceptibility-testing-32nd-edition/ (accessed on 22 December 2022).
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters, Version 12.0. 2022. Available online: http://www.policlinico.pa.it/portal/pdf/news/CIO/Breakpoint%20EUCAST%20per%20l_interpretazione%20delle%20MIC%20per%20farmaci%20antibatterici%20v_12.0%202022.pdf (accessed on 22 December 2022).
- Yüceer, Ö.; Özden Tuncer, B. Determination of antibiotic resistance and biogenic amine production of Lactic Acid Bacteria isolated from Fermented Turkish sausage (Sucuk). J. Food Saf. 2015, 35, 276–285. [Google Scholar] [CrossRef]
- Landeta, G.; De las Rivas, B.; Carrascosa, A.V.; Muñoz, R. Screening of biogenic amine production by coagulase-negative staphylococci isolated during industrial Spanish dry-cured ham processes. Meat Sci. 2007, 77, 556–561. [Google Scholar] [CrossRef][Green Version]
- Blaiotta, G.; Pennacchia, C.; Villani, F.; Ricciardi, A.; Tofalo, R.; Parente, E. Diversity and dynamics of communities of coagulase–negative staphylococci in traditional fermented sausages. J. Appl. Microbiol. 2004, 97, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Coppola, S.; Mauriello, G.; Aponte, M.; Moschetti, G.; Villani, F. Microbial succession during ripening of Naples-type salami, a southern Italian fermented sausage. Meat Sci. 2000, 56, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Iacumin, L.; Osualdini, M.; Bovolenta, S.; Boscolo, D.; Chiesa, L.; Panseri, S.; Comi, G. Microbial, chemico-physical and volatile aromatic compounds characterization of Pitina PGI, a peculiar sausage-like product of North East Italy. Meat Sci. 2020, 163, 108081. [Google Scholar] [CrossRef] [PubMed]
- Amadoro, C.; Rossi, F.; Poltronieri, P.; Marino, L.; Colavita, G. Diversity and safety aspects of Coagulase-Negative Staphylococci in Ventricina del Vastese Italian dry fermented sausage. Appl. Sci. 2022, 12, 13042. [Google Scholar] [CrossRef]
- Iacumin, L.; Manzano, M.; Comi, G. Catalase-positive cocci in fermented sausage: Variability due to different pork breeds, breeding systems and sausage production technology. Food Microbiol. 2012, 29, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Rebecchi, A.; Pisacane, V.; Miragoli, F.; Polka, J.; Falasconi, I.; Morelli, L.; Puglisi, E. High-throughput assessment of bacterial ecology in hog, cow and ovine casings used in sausages production. Int. J. Food Microbiol. 2015, 212, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Mainar, M.; Leroy, F. Process-driven bacterial community dynamics are key to cured meat colour formation by coagulase-negative staphylococci via nitrate reductase or nitric oxide synthase activities. Int. J. Food Microbiol. 2015, 212, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Cachaldora, A.; Fonseca, S.; Da Franco, I.; Carballo, J. Technological and safety characteristics of Staphylococcaceae isolated from Spanish traditional dry-cured sausages. Food Microbiol. 2013, 33, 61–68. [Google Scholar] [CrossRef]
- Kanmani, P.; Kumaresan, K.; Aravind, J. Utilization of coconut oil mill waste as a substrate for optimized lipase production, oil biodegradation and enzyme purification studies in Staphylococcus pasteuri. Electron. J. Biotechnol. 2015, 18, 20–28. [Google Scholar] [CrossRef]
- Villani, F.; Casaburi, A.; Pennacchia, C.; Filosa, L.; Russo, F.; Ercolini, D. Microbial ecology of the Soppressata of Vallo di Diano, a traditional dry fermented sausage from Southern Italy, and in vitro and in situ selection of autochthonous starter cultures. Appl. Environ. Microbiol. 2007, 73, 5453–5463. [Google Scholar] [CrossRef]
- Zeng, X.; He, L.; Guo, X.; Deng, L.; Yang, W.; Zhu, Q.; Duan, Z. Predominant processing adaptability of Staphylococcus xylosus strains isolated from Chinese traditional low-salt fermented whole fish. Int. J. Food Microbiol. 2017, 242, 141–151. [Google Scholar] [CrossRef]
- Mauriello, G.; Casaburi, A.; Villani, F. Proteolytic activity of Staphylococcus xylosus strains on pork myofibrillar and sarcoplasmic proteins and use of selected strains in the production of ‘Naples type’salami. J. Appl. Microbiol. 2002, 92, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Van der Veken, D.; Benhachemi, R.; Charmpi, C.; Ockerman, L.; Poortmans, M.; Van Reckem, E.; Michiels, C.; Leroy, F. Exploring the ambiguous status of coagulase-negative staphylococci in the biosafety of fermented meats: The case of antibacterial activity versus biogenic amine formation. Microorganisms 2020, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Even, S.; Leroy, S.; Charlier, C.; Zakour, N.B.; Chacornac, J.P.; Lebert, I.; Jamet, E.; Desmonts, M.H.; Coton, E.; Pochet, S.; et al. Low occurrence of safety hazards in coagulase-negative staphylococci isolated from fermented foodstuffs. Int. J. Food Microbiol. 2010, 139, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Marty, E.; Bodenmann, C.; Buchs, J.; Hadorn, R.; Eugster-Meier, E.; Lacroix, C.; Meile, L. Prevalence of antibiotic resistance in coagulase-negative staphylococci from spontaneously fermented meat products and safety assessment for new starters. Int. J. Food Microbiol. 2012, 159, 74–83. [Google Scholar] [CrossRef]
- Fontana, C.; Patrone, V.; Lopez, C.M.; Morelli, L.; Rebecchi, A. Incidence of tetracycline and erythromycin resistance in meat-associated bacteria: Impact of different livestock management strategies. Microorganisms 2021, 9, 2111. [Google Scholar] [CrossRef]
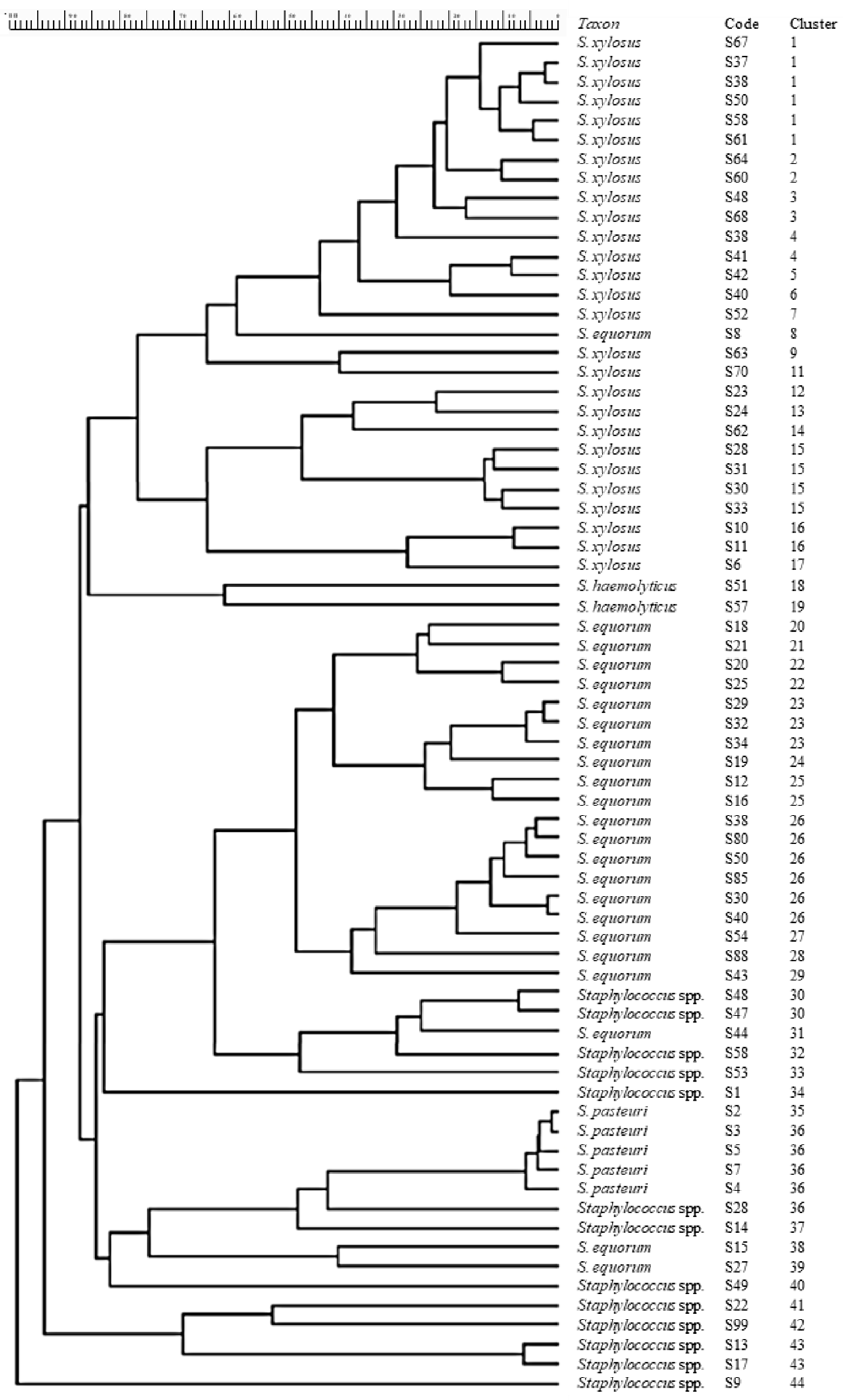
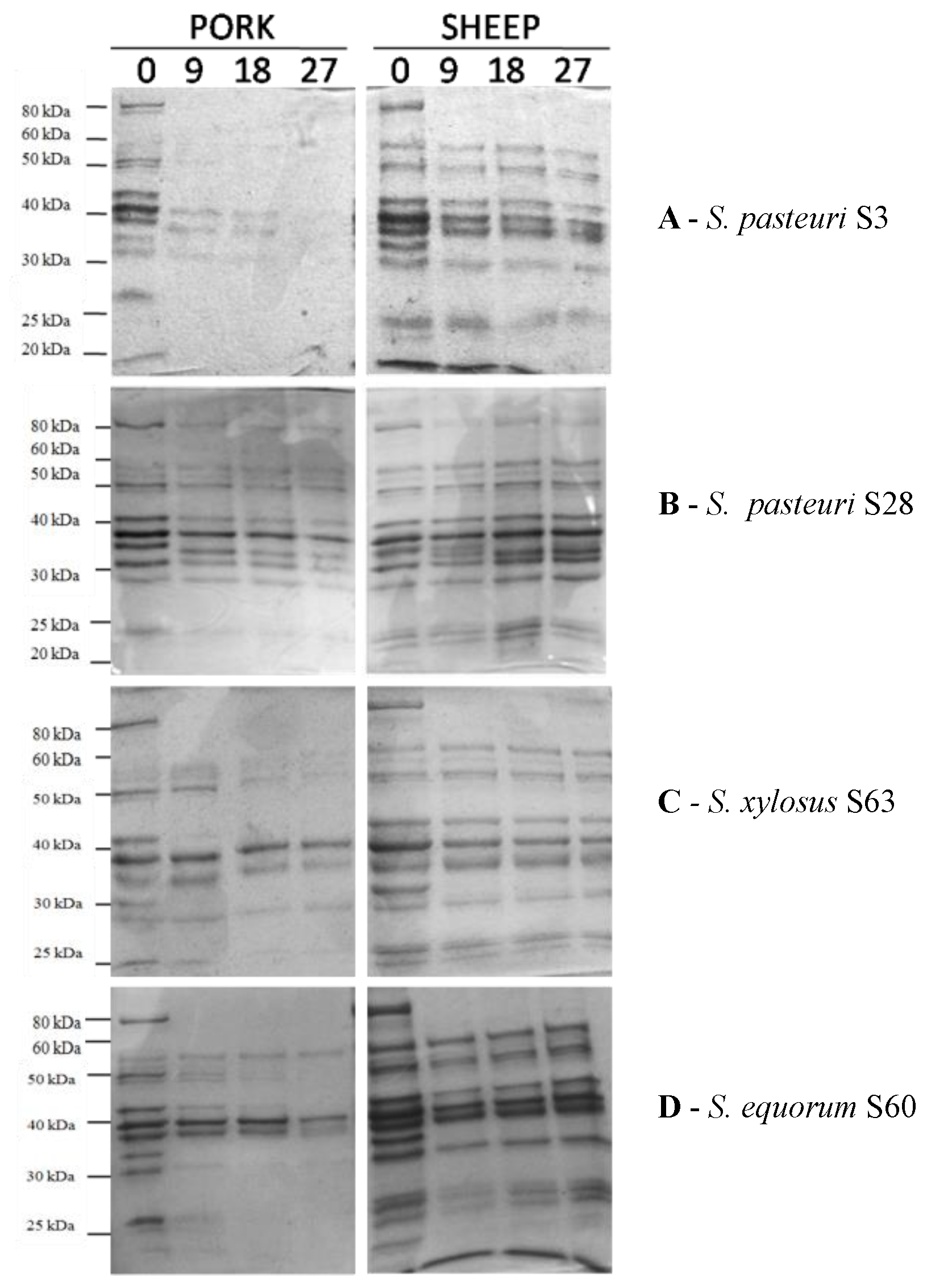
| Taxon | Strain Code | Lipolysis | Proteolysis on Sarcoplasmic Proteins | ||
|---|---|---|---|---|---|
| Tributyrin a | % Oleic Acid b | Pork c | Sheep | ||
| S. xylosus | S41 | 1.4 ± 0.10 | 3.91 ± 0.85 | - | - |
| S48 | 1.25 ± 0.05 | 3.83 ± 0.00 | - | - | |
| S63 | 1.95 ± 0.04 | 1.91 ± 0.06 | 1.2 | 1.05 | |
| S. equorum | S8 | 1.29 ± 0.07 | 3.22 ± 0.01 | - | - |
| S29 | 1.30 ± 0.01 | 2.90 ± 0.24 | - | - | |
| S59 | - | - | 1.1 | 1.10 | |
| S60 | 1.63 ± 0.18 | 1.69 ± 0.04 | 1.2 | 1.00 | |
| S65 | - | - | 1.1 | 1.10 | |
| S. pasteuri | S2 | 1.55 ± 0.05 | 3.93 ± 0.70 | - | - |
| S3 | 1.40 ± 0.03 | 5.02 ± 0.18 | 1.0 | 1.00 | |
| S4 | 1.45 ± 0.02 | 4.51 ± 0.32 | 1.0 | 1.00 | |
| S5 | 1.37 ± 0.50 | 4.31 ± 0.16 | 1.0 | 1.00 | |
| S7 | 1.58 ± 0.03 | 3.12 ± 0.10 | 0.9 | 0.85 | |
| S28 | - | - | 1.1 | 1.10 | |
| Taxon | Strains | Antibiotics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gen 1 | Kan | Tet | Clo | Amp | Van | Ery | Amx | Pen | Cly | ||
| S. xylosus | S41 | S 2 | R | S | S | S | ND | S | R | R | S |
| S48 | S | R | S | S | S | ND | S | R | R | S | |
| S63 | S | R | S | S | S | ND | S | R | R | S | |
| S69 | S | R | S | S | S | ND | S | R | R | S | |
| S. pasteuri | S2 | S | R | R | S | R | ND | S | R | R | S |
| S3 | S | R | R | S | R | ND | S | R | R | S | |
| S4 | S | R | R | S | R | ND | S | R | R | S | |
| S5 | S | R | R | S | R | ND | S | R | R | S | |
| S7 | S | R | I | S | R | ND | S | R | R | S | |
| S. equorum | S8 | S | R | S | S | S | ND | S | R | S | S |
| S29 | S | R | S | S | S | ND | S | R | S | S | |
| S59 | S | R | S | S | S | ND | S | R | S | S | |
| S60 | S | R | S | S | S | ND | S | R | S | S | |
| S65 | S | R | S | S | S | ND | S | R | S | S | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mangia, N.P.; Cottu, M.; Aponte, M.; Murgia, M.A.; Mura, M.E.; Blaiotta, G. Technological and Safety Characterization of Coagulase-Negative Staphylococci Isolated from Sardinian Fermented Sausage Made by Ovine Meat. Foods 2024, 13, 633. https://doi.org/10.3390/foods13040633
Mangia NP, Cottu M, Aponte M, Murgia MA, Mura ME, Blaiotta G. Technological and Safety Characterization of Coagulase-Negative Staphylococci Isolated from Sardinian Fermented Sausage Made by Ovine Meat. Foods. 2024; 13(4):633. https://doi.org/10.3390/foods13040633
Chicago/Turabian StyleMangia, Nicoletta P., Michele Cottu, Maria Aponte, Marco A. Murgia, Maria E. Mura, and Giuseppe Blaiotta. 2024. "Technological and Safety Characterization of Coagulase-Negative Staphylococci Isolated from Sardinian Fermented Sausage Made by Ovine Meat" Foods 13, no. 4: 633. https://doi.org/10.3390/foods13040633
APA StyleMangia, N. P., Cottu, M., Aponte, M., Murgia, M. A., Mura, M. E., & Blaiotta, G. (2024). Technological and Safety Characterization of Coagulase-Negative Staphylococci Isolated from Sardinian Fermented Sausage Made by Ovine Meat. Foods, 13(4), 633. https://doi.org/10.3390/foods13040633






