RNA-Seq Reveals Pathways Responsible for Meat Quality Characteristic Differences between Two Yunnan Indigenous Chicken Breeds and Commercial Broilers
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experimentation Ethical Statement
2.2. Chicken, Diet and Housin
2.3. Measurement of Growth Performance
2.4. Slaughter Procedure and Sample Collecting
2.5. Measurement of Muscle Physical Parameters
2.6. Measurement of Muscle Chemical Parameters
2.7. Measurement of AA
2.8. Measurement of FA
2.9. Cryosectioning and Hematoxylin and Eosin Staining
2.10. RNA-Seq Library Preparation and Data Analysis
2.11. qPCR Verification
2.12. Data and Statistical Analysis
3. Results
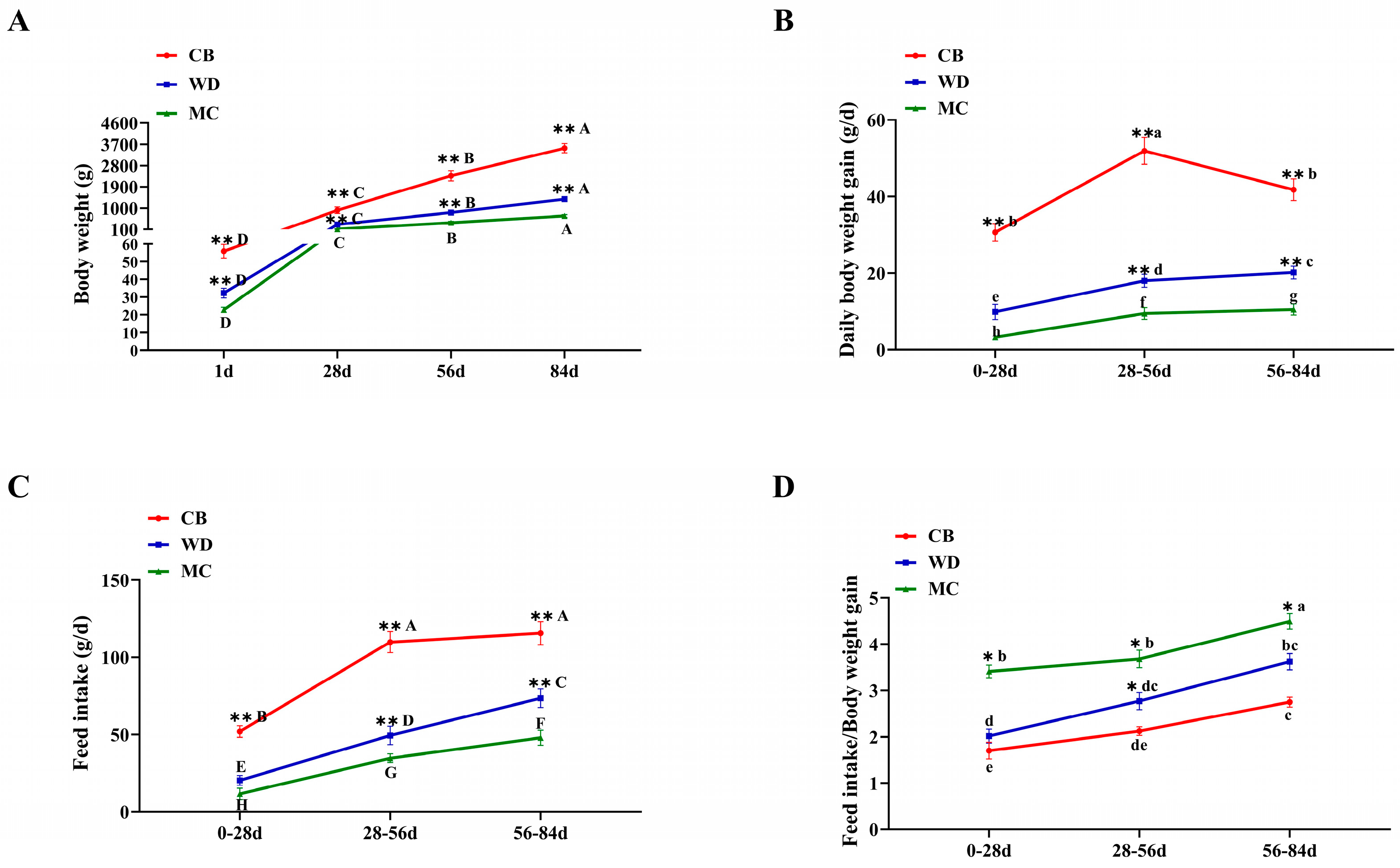
3.1. Comparative Analysis of Growth Performance
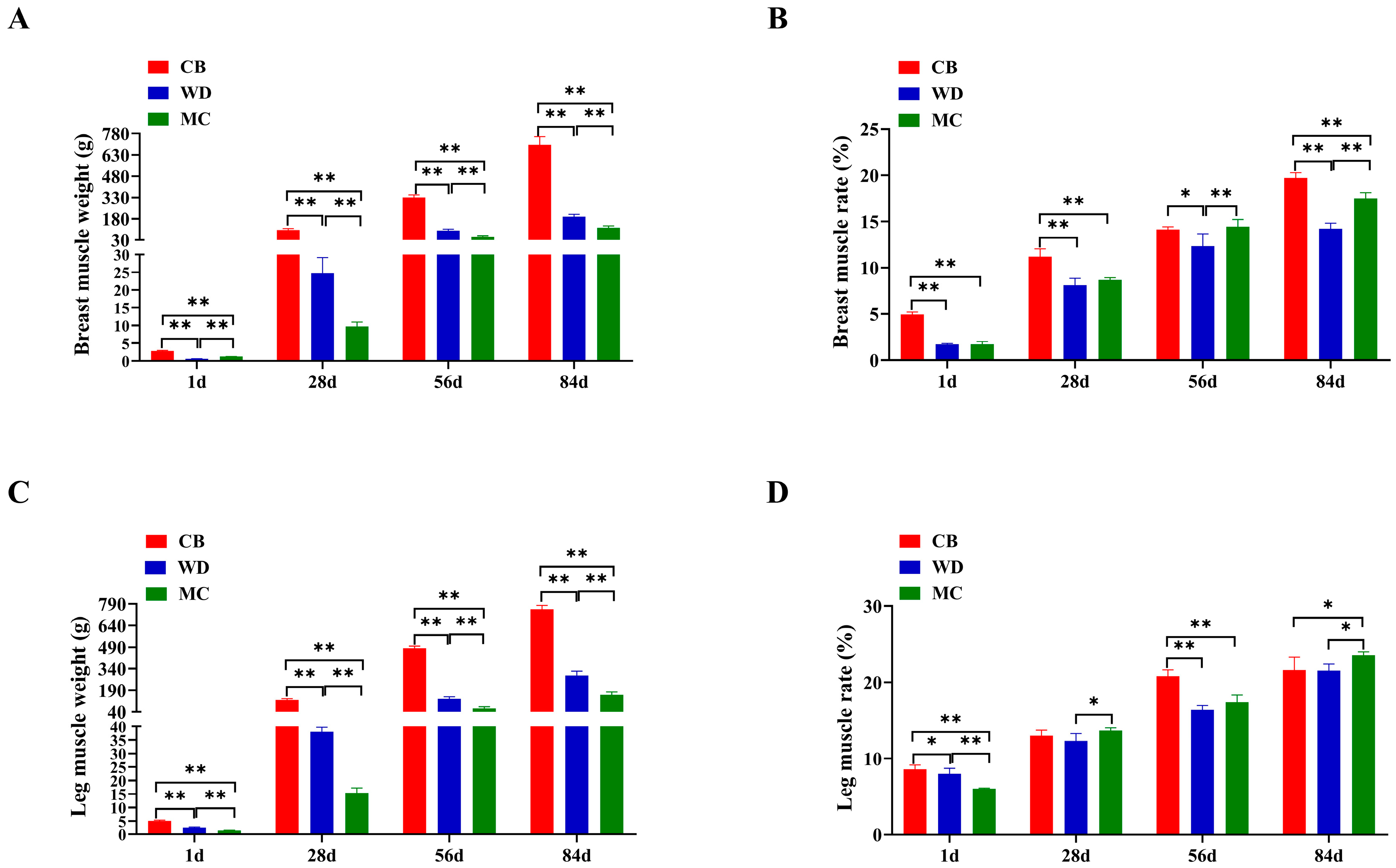
3.2. Comparative Analysis of Development in Skeletal Muscle
3.3. Comparative Analysis of Meat Quality Physical Characteristics
3.4. Comparative Analysis of Meat Quality Chemical Parameters
3.5. Comparative Analysis of AA Contents
3.6. Comparative Analysis of FA Contents
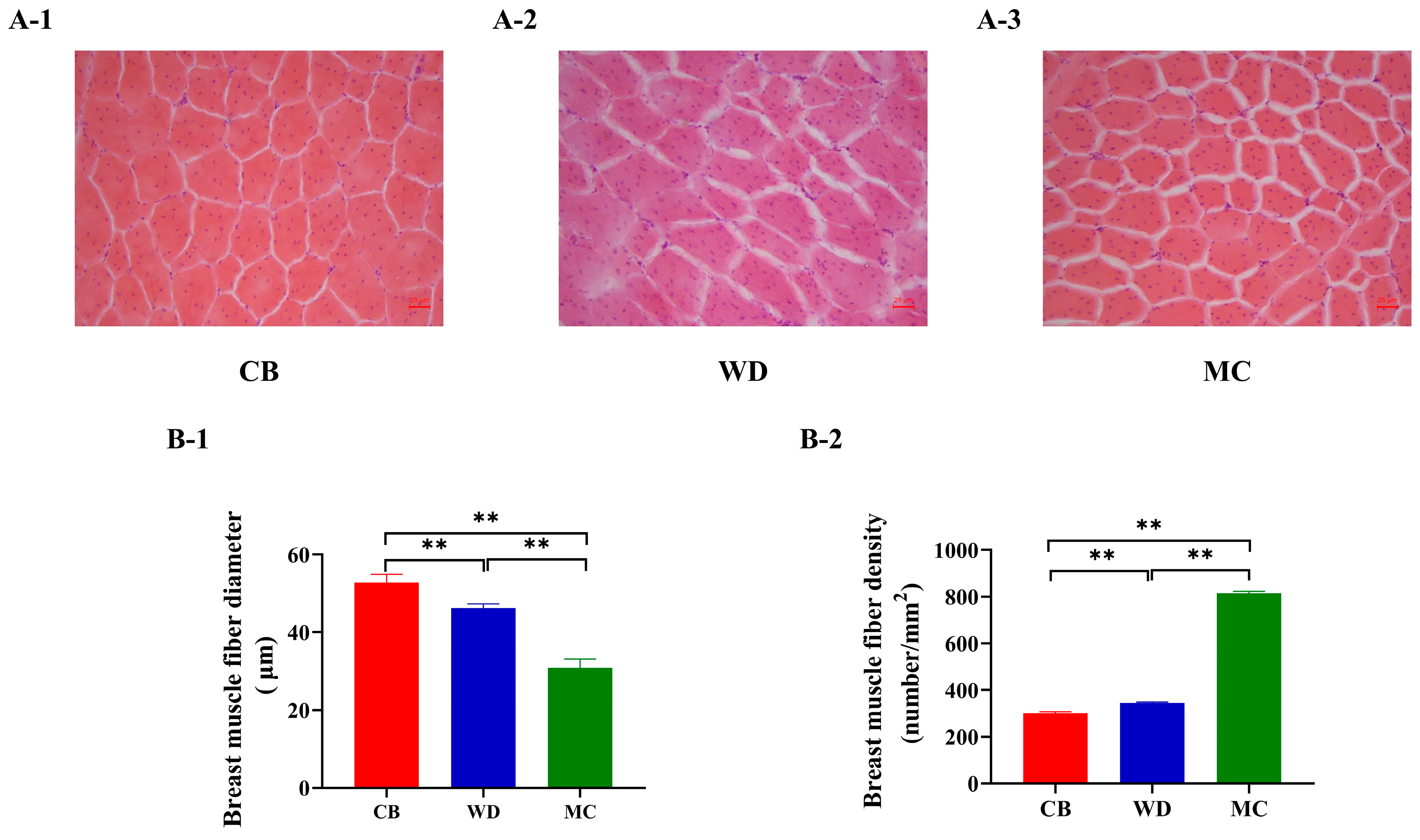
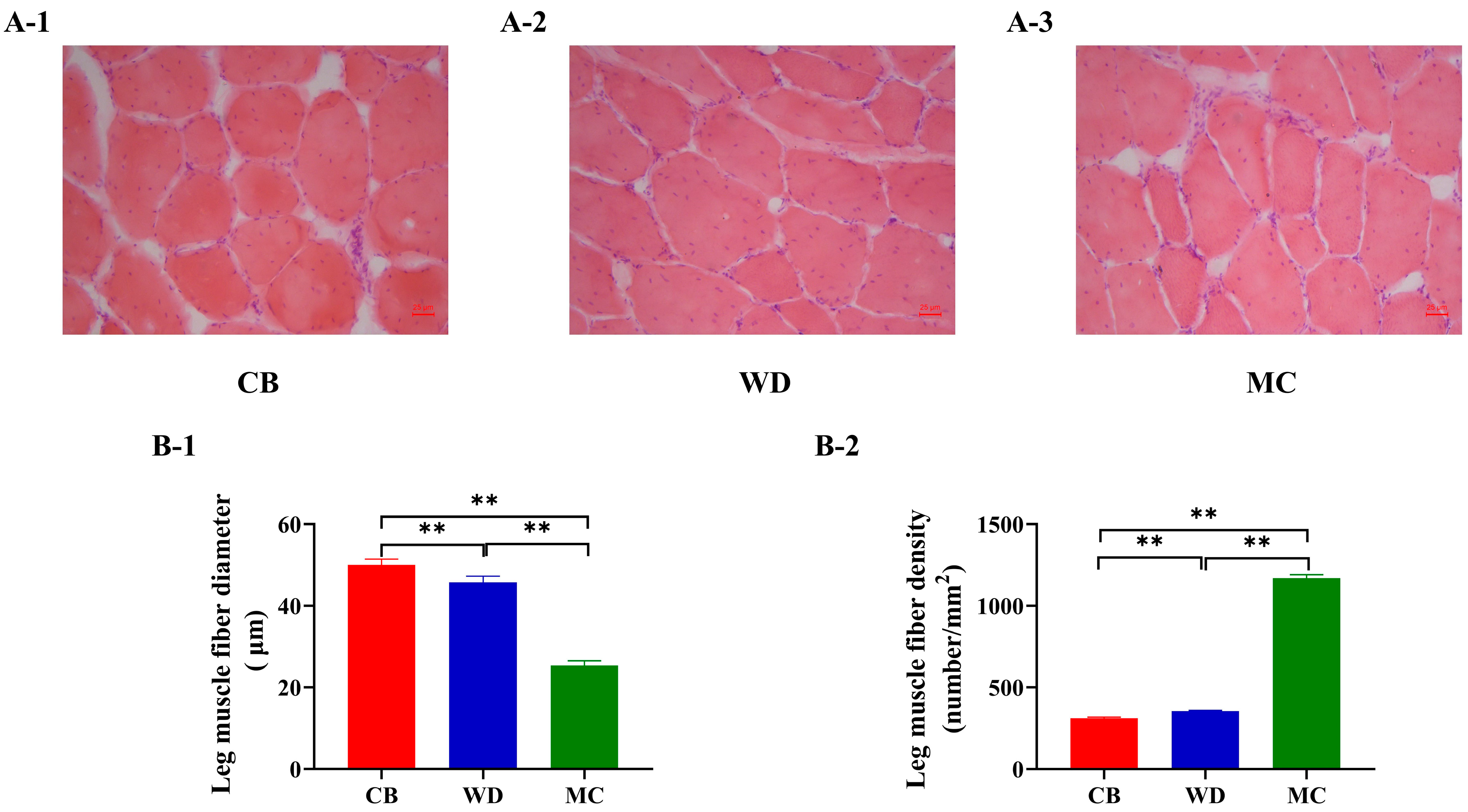
3.7. Comparative Analysis of Skeletal Muscle Fiber
3.8. Comparative Analysis of RNA-Seq in Breast Muscles
3.9. qPCR
4. Discussion
4.1. Muscle Meat Quality Characteristics
4.2. Candidate Genes Associated with Muscle Growth and Meat Quality
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, T.; Zhang, Y.; Guo, Y.; Zhang, X.; Yang, H.; Tian, X.; Zhu, M.; Guo, Z.; Zeng, S.; Luo, K.; et al. RNA-sequence reveals differentially expressed genes affecting the crested trait of Wumeng crested chicken. Poult. Sci. 2021, 100, 101357. [Google Scholar] [CrossRef] [PubMed]
- Piórkowska, K.; Żukowski, K.; Nowak, J.; Połtowicz, K.; Ropka-Molik, K.; Gurgul, A. Genome-wide RNA-Seq analysis of breast muscles of two broiler chicken groups differing in shear force. Anim. Genet. 2016, 47, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Dou, T.; Li, Z.; Wang, K.; Liu, L.; Rong, H.; Xu, Z.; Huang, Y.; Gu, D.; Chen, X.; Hu, W.; et al. Regulation of myostatin expression is associated with growth and muscle development in commercial broiler and DMC muscle. Mol. Biol. Rep. 2018, 45, 511–522. [Google Scholar] [PubMed]
- Dou, T.; Zhao, S.; Rong, H.; Gu, D.; Li, Q.; Huang, Y.; Xu, Z.; Chu, X.; Tao, L.; Liu, L.; et al. Biological mechanisms discriminating growth rate and adult body weight phenotypes in two Chinese indigenous chicken breeds. BMC Genom. 2017, 18, 469. [Google Scholar]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, Z.Q.; Luo, Y.Y.; Zhang, H.B.; Gao, N.; He, J.L.; Ji, C.L.; Zhang, D.X.; Li, J.Q.; Zhang, X.Q. Whole genomic prediction of growth and carcass traits in a Chinese quality chicken population. J. Anim. Sci. 2017, 95, 72–80. [Google Scholar] [PubMed]
- Park, W.; Rengaraj, D.; Kil, D.Y.; Kim, H.; Lee, H.K.; Song, K.D. RNA-seq analysis of the kidneys of broiler chickens fed diets containing different concentrations of calcium. Sci. Rep. 2017, 7, 11740. [Google Scholar]
- Willson, N.L.; Forder, R.E.A.; Tearle, R.; Williams, J.L.; Hughes, R.J.; Nattrass, G.S.; Hynd, P.I. Transcriptional analysis of liver from chickens with fast (meat bird), moderate (F1 layer x meat bird cross) and low (layer bird) growth potential. BMC Genom. 2018, 19, 309. [Google Scholar]
- Yi, B.; Chen, L.; Sa, R.; Zhong, R.; Xing, H.; Zhang, H. High concentrations of atmospheric ammonia induce alterations of gene expression in the breast muscle of broilers (Gallus gallus) based on RNA-Seq. BMC Genom. 2016, 17, 598. [Google Scholar]
- Liu, Y.; Liang, S.; Wang, K.; Zi, X.; Zhang, R.; Wang, G.; Kang, J.; Li, Z.; Dou, T.; Ge, C. Physicochemical, Nutritional Properties and Metabolomics Analysis Fat Deposition Mechanism of Chahua Chicken No. 2 and Yao Chicken. Genes 2022, 13, 1358. [Google Scholar]
- Shen, L.; Ma, J.; Zhou, H.; Chen, L.; Tang, J.; Zhang, K.; Zhao, Y.; Niu, L.; Zhang, S.; Jiang, A.; et al. Plasma Metabolomic Profiling Reveals Preliminary Biomarkers of Pork Quality Based on pH Value. Foods 2022, 11, 4005. [Google Scholar] [CrossRef]
- Gonzalez-Rivas, P.A.; Chauhan, S.S.; Ha, M.; Fegan, N.; Dunshea, F.R.; Warner, R.D. Effects of heat stress on animal physiology, metabolism, and meat quality: A review. Meat Sci. 2020, 162, 108025. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, M.; Jaspal, M.H.; Hayat, Z.; Yar, M.K.; Badar, I.H.; Ullah, S.; Hussain, Z.; Ali, S.; Farid, M.U.; Farooq, M.Z.; et al. Effect of animal age, postmortem chilling rate, and aging time on meat quality attributes of water buffalo and humped cattle bulls. Anim. Sci. J. 2020, 91, e13354. [Google Scholar] [CrossRef] [PubMed]
- Bowker, B.; Zhuang, H. Detection of razor shear force differences in broiler breast meat due to the woody breast condition depends on measurement technique and meat state1. Poult. Sci. 2019, 98, 6170–6176. [Google Scholar]
- Picard, B.; Gagaoua, M. Muscle Fiber Properties in Cattle and Their Relationships with Meat Qualities: An Overview. J. Agric. Food Chem. 2020, 68, 6021–6039. [Google Scholar]
- Hwang, Y.H.; Lee, S.J.; Lee, E.Y.; Joo, S.T. Effects of carcass weight increase on meat quality and sensory properties of pork loin. J. Anim. Sci. Technol. 2020, 62, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Grant, J.; Goldberg, E.M.; Ryland, D.; Aliani, M. Investigation of low molecular weight peptides (<1 kDa) in chicken meat and their contribution to meat flavor formation. J. Sci. Food Agric. 2019, 99, 1728–1739. [Google Scholar]
- Jin, Y.; Cui, H.; Yuan, X.; Liu, L.; Liu, X.; Wang, Y.; Ding, J.; Xiang, H.; Zhang, X.; Liu, J.; et al. Identification of the main aroma compounds in Chinese local chicken high-quality meat. Food Chem. 2021, 359, 129930. [Google Scholar] [PubMed]
- Xu, X.; Yang, H.; Xu, Z.; Li, X.; Leng, X. The comparison of largemouth bass (Micropterus salmoides) fed trash fish and formula feeds: Growth, flesh quality and metabolomics. Front. Nutr. 2022, 9, 966248. [Google Scholar] [CrossRef]
- Jung, S.; Bae, Y.S.; Kim, H.J.; Jayasena, D.D.; Lee, J.H.; Park, H.B.; Heo, K.N.; Jo, C. Carnosine, anserine, creatine, and inosine 5'-monophosphate contents in breast and thigh meats from 5 lines of Korean native chicken. Poult. Sci. 2013, 92, 3275–3282. [Google Scholar] [CrossRef]
- Yan, J.; Liu, P.; Xu, L.; Huan, H.; Zhou, W.; Xu, X.; Shi, Z. Effects of exogenous inosine monophosphate on growth performance, flavor compounds, enzyme activity, and gene expression of muscle tissues in chicken. Poult. Sci. 2018, 97, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Dalle-Zotte, A.; Ricci, R.; Cullere, M.; Serva, L.; Tenti, S.; Marchesini, G. Research Note: Effect of chicken genotype and white striping-wooden breast condition on breast meat proximate composition and amino acid profile. Poult. Sci. 2020, 99, 1797–1803. [Google Scholar] [CrossRef] [PubMed]
- Boschetti, E.; Bordoni, A.; Meluzzi, A.; Castellini, C.; Dal-Bosco, A.; Sirri, F. Fatty acid composition of chicken breast meat is dependent on genotype-related variation of FADS1 and FADS2 gene expression and desaturating activity. Animals 2016, 10, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Ballester, M.; Ramayo-Caldas, Y.; Revilla, M.; Corominas, J.; Castelló, A.; Estellé, J.; Fernández, A.I.; Folch, J.M. Integration of liver gene co-expression networks and eGWAs analyses highlighted candidate regulators implicated in lipid metabolism in pigs. Sci. Rep. 2017, 7, 46539. [Google Scholar] [PubMed]
- Li, B.; Weng, Q.; Dong, C.; Zhang, Z.; Li, R.; Liu, J.; Jiang, A.; Li, Q.; Jia, C.; Wu, W.; et al. A Key Gene, PLIN1, Can Affect Porcine Intramuscular Fat Content Based on Transcriptome Analysis. Genes 2018, 9, 194. [Google Scholar] [CrossRef]
- Huang, C.Y.; Oka, S.I.; Xu, X.; Chen, C.F.; Tung, C.Y.; Chang, Y.Y.; Mourad, Y.; Vehra, O.; Ivessa, A.; Yehia, G.; et al. PERM1 regulates genes involved in fatty acid metabolism in the heart by interacting with PPARα and PGC-1α. Sci. Rep. 2022, 12, 14576. [Google Scholar] [PubMed]
- Su, X.; Tan, Q.S.; Parikh, B.H.; Tan, A.; Mehta, M.N.; Sia-Wey, Y.; Tun, S.B.; Li, L.J.; Han, X.Y.; Wong, T.Y.; et al. Characterization of Fatty Acid Binding Protein 7 (FABP7) in the Murine Retina. Invest. Ophthalmol. Vis. Sci. 2016, 57, 3397–3408. [Google Scholar] [PubMed]
- Busato, S.; Ford, H.R.; Abdelatty, A.M.; Estill, C.T.; Bionaz, M. Peroxisome Proliferator-Activated Receptor Activation in Precision-Cut Bovine Liver Slices Reveals Novel Putative PPAR Targets in Periparturient Dairy Cows. Front. Vet. Sci. 2022, 99, 31264. [Google Scholar]
- ALJohani, A.M.; Syed, D.N.; Ntambi, J.M. Insights into Stearoyl-CoA Desaturase-1 Regulation of Systemic Metabolism. Trends Endocrinol. Metab. 2017, 28, 831–842. [Google Scholar]
- Abu-Farha, M.; Al-Khairi, I.; Cherian, P.; Chandy, B.; Sriraman, D.; Alhubail, A.; Al-Refaei, F.; AlTerki, A.; Abubaker, J. Increased ANGPTL3, 4 and ANGPTL8/betatrophin expression levels in obesity and T2D. Lipids Health Dis. 2016, 15, 181. [Google Scholar]
- Keles, U.; Ow, J.R.; Kuentzel, K.B.; Zhao, L.N.; Kaldis, P. Liver-derived metabolites as signaling molecules in fatty liver disease. Cell Mol. Life Sci. 2022, 80, 4. [Google Scholar] [PubMed]
- Xu, Y.; Chen, H.; Wan, K.; Tang, Z.; Sun, W.; Wu, L.; Ren, Z.; Ding, Q.; Liang, K.; Sun, Z. Effects of Long-Term Low-Protein Diets Supplemented with Sodium Dichloroacetate and Glucose on Metabolic Biomarkers and Intestinal Microbiota of Finishing Pigs. Animals 2022, 12, 2522. [Google Scholar] [CrossRef]
- Maurya, S.K.; Mishra, R. Co-Localization and Interaction of Pax5 with Iba1 in Brain of Mice. Cell Mol. Neurobiol. 2018, 38, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Huang, S.; Luo, S.; Liao, H.; Wang, Y.; Deng, X.; Ma, F.; Ma, C.W.; Zhou, L. Identification of genes underlying the enhancement of immunity by a formula of lentinan, pachymaran and tremelia polysaccharides in immunosuppressive mice. Sci. Rep. 2018, 8, 10082. [Google Scholar] [PubMed]
- Halperin, D.; Stavsky, A.; Kadir, R.; Drabkin, M.; Wormser, O.; Yogev, Y.; Dolgin, V.; Proskorovski-Ohayon, R.; Perez, Y.; Nudelman, H.; et al. CDH2 mutation affecting N-cadherin function causes attention-deficit hyperactivity disorder in humans and mice. Nat. Commun. 2021, 12, 6187. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, X.; Cui, H.; Liu, R.; Zhao, G.; Wen, J. Transcriptional insights into key genes and pathways controlling muscle lipid metabolism in broiler chickens. BMC Genom. 2019, 20, 863. [Google Scholar]
- Ishaq, M.; Bandara, N.; Morgan, S.; Nowell, C.; Mehdi, A.M.; Lyu, R.; McCarthy, D.; Anderson, D.; Creek, D.J.; Achen, M.G.; et al. Key signaling networks are dysregulated in patients with the adipose tissue disorder, lipedema. Int. J. Obes. 2022, 46, 502–514. [Google Scholar]
- Bandet, C.L.; Tan-Chen, S.; Bourron, O.; Le-Stunff, H.; Hajduch, E. Sphingolipid Metabolism: New Insight into Ceramide-Induced Lipotoxicity in Muscle Cells. Int. J. Mol. Sci. 2019, 20, 479. [Google Scholar] [CrossRef]
- Harjunpää, H.; Llort-Asens, M.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Palacionyte, J.; Januskevicius, A.; Vasyle, E.; Rimkunas, A.; Bajoriuniene, I.; Miliauskas, S.; Malakauskas, K. IL-5 and GM-CSF, but Not IL-3, Promote the Proliferative Properties of Inflammatory-like and Lung Resident-like Eosinophils in the Blood of Asthma Patients. Cells 2022, 11, 3804. [Google Scholar] [CrossRef]
- Zhang, M.; Zheng, D.; Peng, Z.; Zhu, Y.; Li, R.; Wu, Q.; Li, Y.; Li, H.; Xu, W.; Zhang, M.; et al. Identification of Differentially Expressed Genes and Lipid Metabolism Signaling Pathways between Muscle and Fat Tissues in Broiler Chickens. J. Poult. Sci. 2021, 58, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Yang, C.; Wang, S.; Ma, Y. Identifying Key Genes and Functionally Enriched Pathways of Diverse Adipose Tissue Types in Cattle. Front. Genet. 2022, 13, 790690. [Google Scholar] [CrossRef] [PubMed]
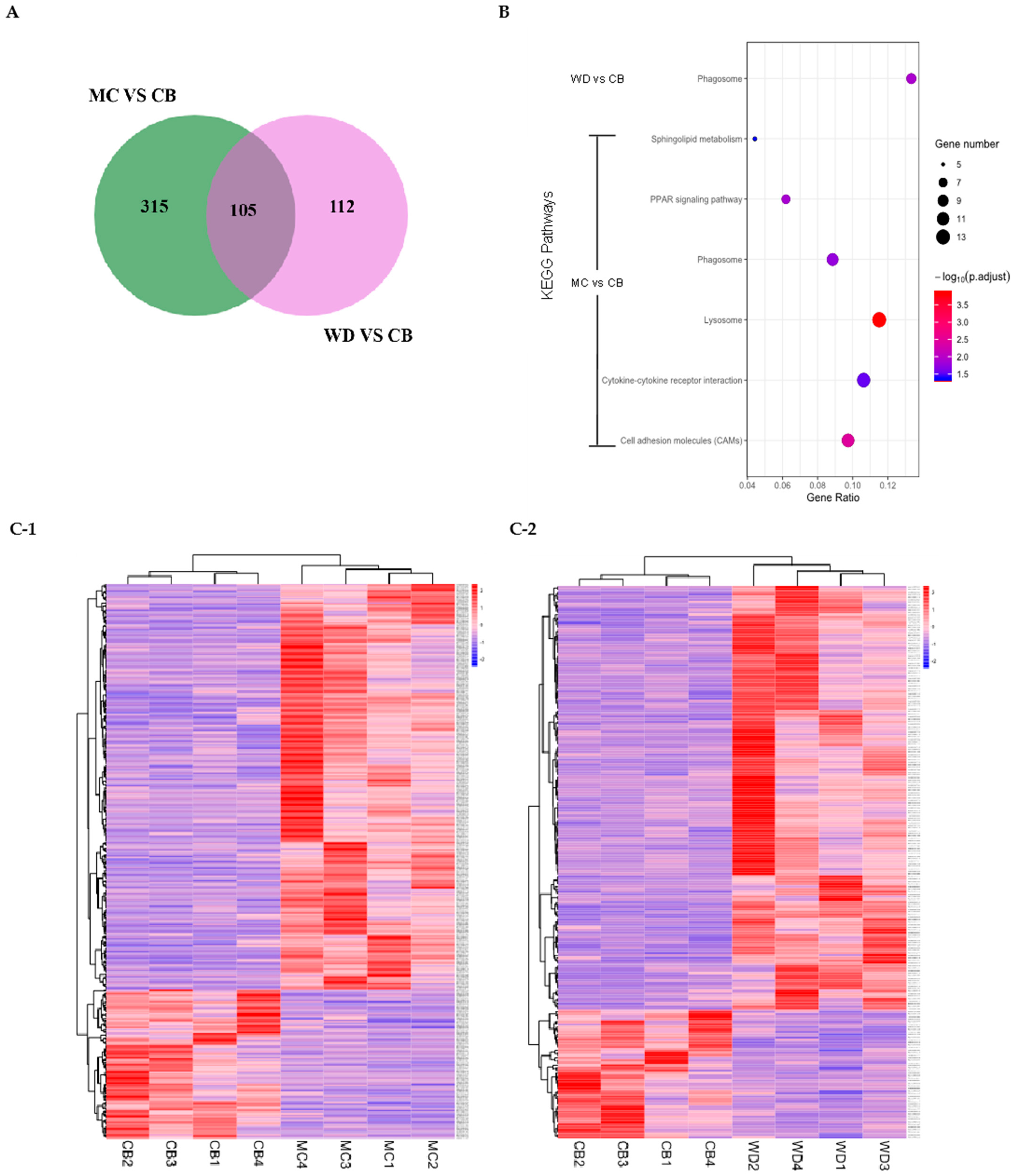
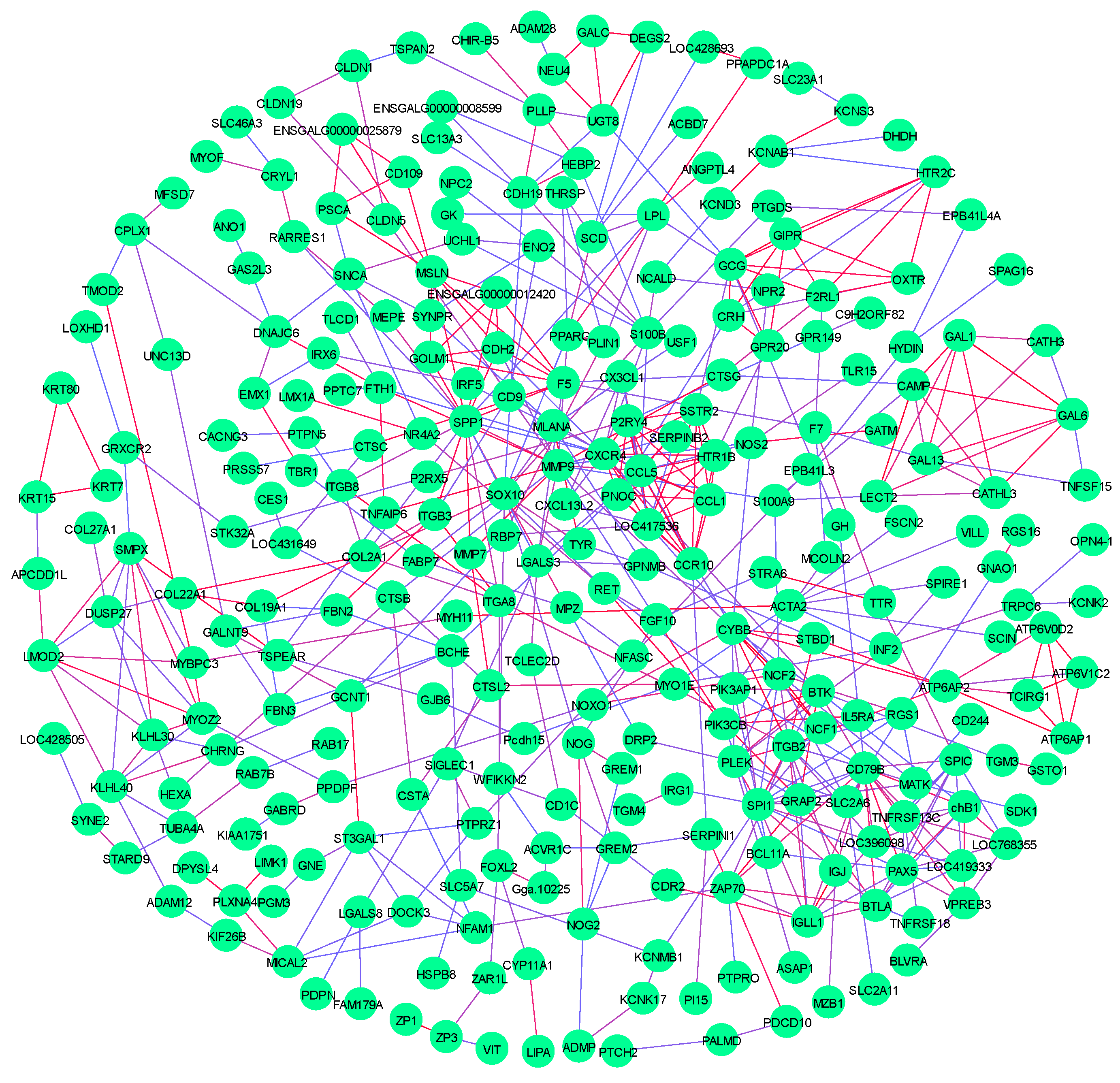
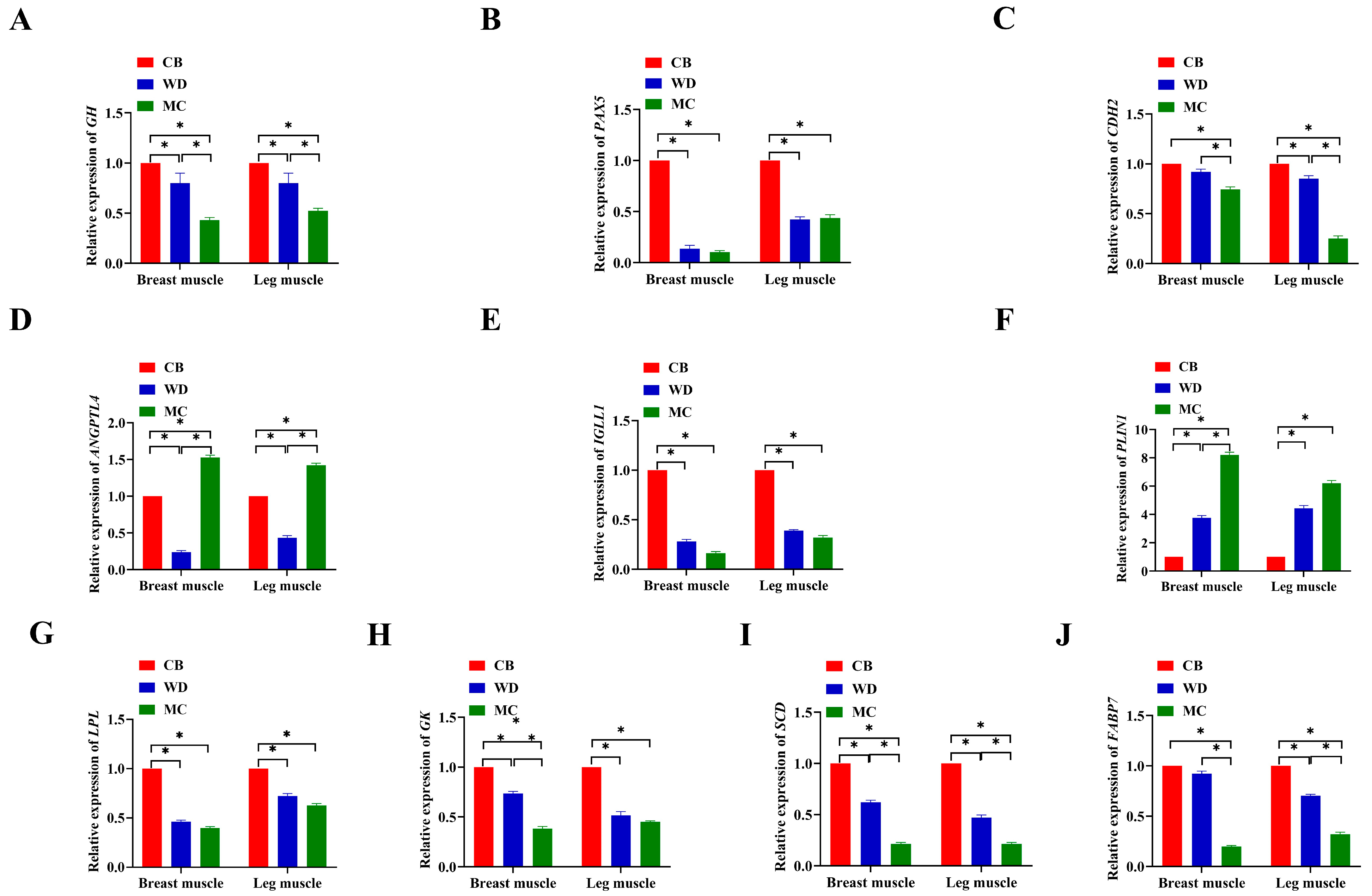
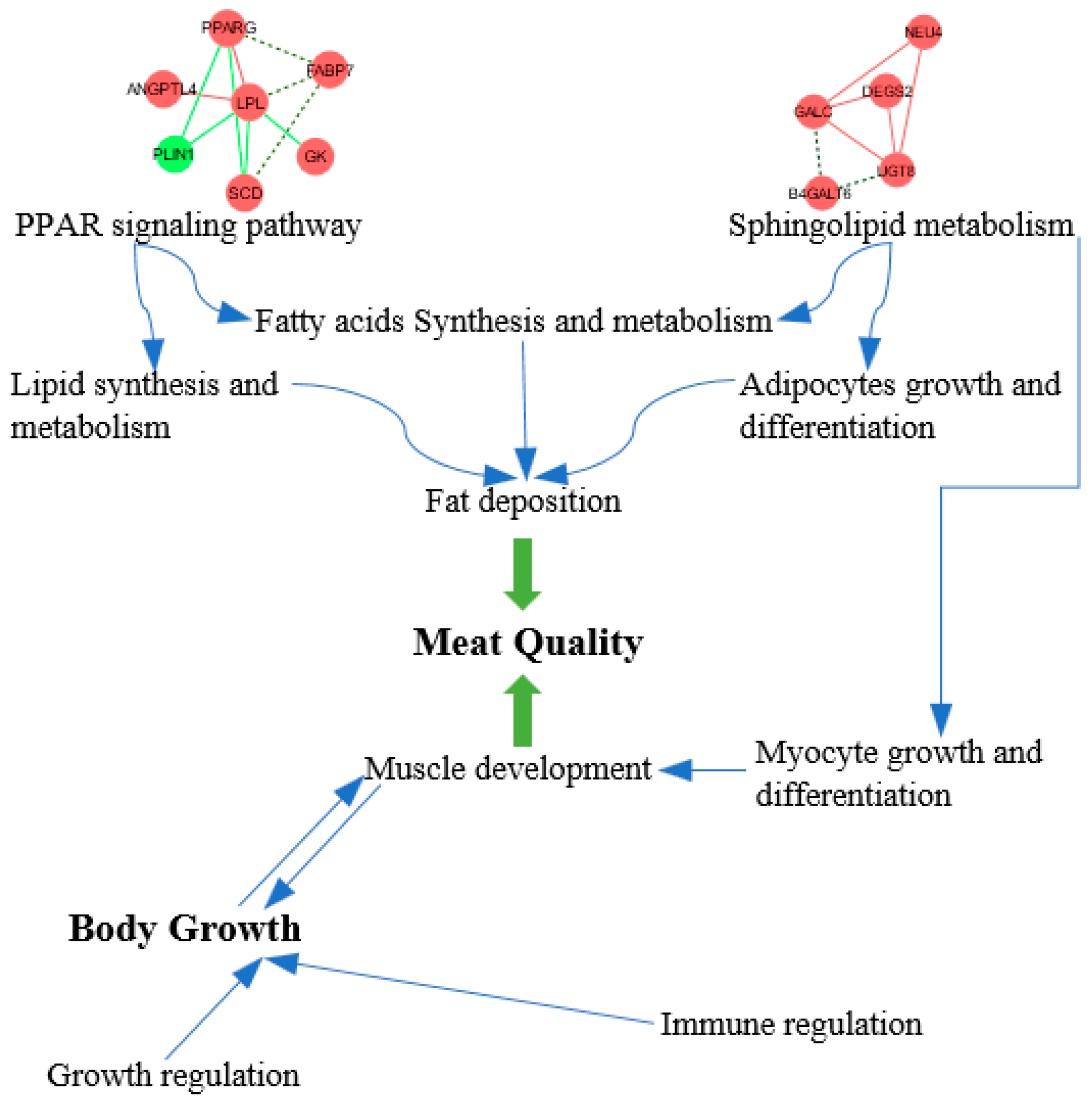
| Compositions of Diets % | Phase I | Phase II |
|---|---|---|
| Corn | 58.85 | 61.25 |
| Soy protein | 25.29 | 22.44 |
| Wheat bran | 8.90 | 9.50 |
| Fish meal | 3.00 | 3.00 |
| Calcium hydrogen phosphate | 1.47 | 1.41 |
| Limestone | 1.10 | 1.00 |
| Lys | 0 | 0.02 |
| Met | 0.12 | 0.11 |
| Sodium Chloride | 0.27 | 0.27 |
| Minerals and vitamins | 1.00 | 1.00 |
| Total | 100 | 100 |
| Nutrients levels | ||
| Metabolism energy (MJ·kg−1) | 12.00 | 12.10 |
| Crude protein | 18.50 | 17.00 |
| Calcium | 0.95 | 0.95 |
| Phosphorus | 0.68 | 0.65 |
| Lys | 0.96 | 0.95 |
| Met | 0.40 | 0.38 |
| Age (Day) | Vaccine | Way |
|---|---|---|
| 1 | Marek’s disease | Subcutaneous injection |
| 3 | Newcastle | Oral vaccination |
| 12 | Gumboro | Subcutaneous injection |
| 20 | Newcastle | Oral vaccination |
| 42 | Fowl cholera | Subcutaneous injection |
| Age (Day) | Density (Birds/m2) | Temperature (°C) | Relative Humidity (%) | Light Intensity |
|---|---|---|---|---|
| 1–3 | 15 | 33–35 | 65–70 | 25 Lx, 24 h |
| 4–7 | 15 | 30–33 | 65–70 | 10 Lx, 23 h |
| 8–14 | 15 | 28–30 | 60–65 | 10 Lx, 23 h |
| 15–21 | 6 | 26–28 | 55–60 | 8 Lx, 18 h |
| 22–28 | 6 | 24–26 | 55–60 | 8 Lx, 18 h |
| 29–35 | 3 | 21–24 | 55 | 8 Lx, 18 h |
| 36–84 | 3 | 18–21 | 55 | 5 Lx, 18 h |
| Items | Measurement Methods |
|---|---|
| Breast muscle weight | The weight of the breast without skin and adherent fat |
| Breast muscle rate | Percentage of breast muscle weight in body weight |
| Leg muscle weight | The weight of the two legs without skin and adherent fat |
| Leg muscle rate | Percentage of leg muscle weight in body weight |
| Gene | Primer Sequence (5′-3′) | Annealing Temperature (°C) | Product Size (bp) |
|---|---|---|---|
| β-actin | F: tggactcctacaaccaacgg R: catcctccttgaactcgcag | 58.8 | 258 |
| PLIN1 | F: atggtgagaggcagagcatt R: cttcttcacgctggagatgc | 56.6 | 185 |
| LPL | F: ggttcctggacagatggaca R: caacatcctttcccaccagc | 58.8 | 239 |
| FABP7 | F: tgacgaatacatgaaggcgc R: catcaaattcctcgccgagt | 58.3 | 167 |
| SCD | F: caagttctccgagacgcatg R: gggcttgtagtatctccgct | 56.6 | 178 |
| ANGPTL4 | F: tggaagactgggagggaaac R: gtttgtgtccgctttgaggt | 57.6 | 185 |
| GK | F: cgggaacttcttatggctgc R: aatggtatcccgcagtcctt | 58.3 | 202 |
| PAX5 | F: atcagcaagtcccagtctcc R: gtctccacgcatctgtttcc | 57.6 | 239 |
| IGLL1 | F: accaacagaccctcgaacat R: ttgtcccggccccaaatata | 58.8 | 157 |
| CDH2 | F: caaaactttcggaccctgca R: gtggtggcttcttttgggtt | 58.8 | 232 |
| GH | F: agctgcttcggttttcactg R: atcgtaggtgggtctgagga | 57.6 | 209 |
| Items | CB | WD | MC | |||
|---|---|---|---|---|---|---|
| Breast Muscle | Leg Muscle | Breast Muscle | Leg Muscle | Breast Muscle | Leg Muscle | |
| pH 45 min | 6.40 ± 0.37 A | 6.55 ± 0.17 A | 5.95 ± 0.12 B | 6.41 ± 0.26 B | 5.89 ± 0.26 B | 6.17 ± 0.10 B |
| pH 24 h | 5.92 ± 0.35 | 6.55 ± 0.22 ** | 5.69 ± 0.21 | 6.40 ± 0.62 ** | 5.66 ± 0.10 | 6.09 ± 0.15 |
| L* | 50.16 ± 4.21 | 52.04 ± 3.63 A | 46.91 ± 3.78 | 42.22 ± 2.8 B | 50.82 ± 2.06 | 50.43 ± 2.26 A |
| a* | 3.47 ± 2.29 c | 7.64 ± 2.46 Bb* | 4.26 ± 1.46 b | 14.27 ± 4.79 Aa** | 5.65 ± 1.08 a | 9.36 ± 2.68 b |
| b* | 6.37 ± 1.67 | 5.97 ± 3.20 | 6.13 ± 3.12 | 5.35 ± 1.73 | 6.26 ± 1.79 | 5.58 ± 0.80 |
| water loss rate (%) | 13.49 ± 2.62 | 19.34 ± 4.54 A* | 9.77 ± 1.75 | 9.38 ± 2.01 B | 11.52 ± 3.74 | 11.34 ± 2.91 AB |
| shearing force (kg/f) | 4.71 ± 1.32 | 3.70 ± 2.52 | 4.73 ± 1.46 | 4.37 ± 1.19 | 4.46 ± 1.02 | 5.21 ± 0.97 |
| Items | CB | WD | MC | |||
|---|---|---|---|---|---|---|
| Breast Muscle | Leg Muscle | Breast Muscle | Leg Muscle | Breast Muscle | Leg Muscle | |
| Crude ash (%) | 1.33 ± 0.09 B | 1.27 ± 0.02 B | 1.72 ± 0.14 A** | 1.35 ± 0.03 B | 1.72 ± 0.03 A** | 1.59 ± 0.07 A |
| CP (%) | 24.26 ± 0.40 C | 24.46 ± 0.57 B | 27.30 ± 0.52 B** | 23.68 ± 0.18 C | 28.37 ± 0.51 A | 28.23 ± 0.13 A |
| CF (%) | 0.46 ± 0.05 C | 0.85 ± 0.20 B** | 0.86 ± 0.051 A | 2.08 ± 0.36 A** | 0.67 ± 0.02 B | 1.90 ± 0.05 A** |
| Water (%) | 73.95 ± 0.45 A | 73.43 ± 0.77 A | 70.12 ± 0.70 Ba | 72.89 ± 0.54 A | 69.23 ± 0.53 Bb | 68.29 ± 0.12 B |
| Inosine monophosphate (%) | 1.01 ± 0.15 Bb | 1.04 ± 0.14 B | 1.28 ± 0.11 A | 1.35 ± 0.04 A | 1.23 ± 0.09 a | 1.30 ± 0.07 A |
| Items | CB | WD | MC | ||||
|---|---|---|---|---|---|---|---|
| Breast Muscle | Leg Muscle | Breast Muscle | Leg Muscle | Breast Muscle | Leg Muscle | ||
| EAA | Thr | 2.81 ± 0.23 B | 2.83 ± 0.12 B | 4.25 ± 0.12 A | 4.26 ± 0.25 A | 2.64 ± 0.10 B | 2.63 ± 0.15 B |
| Val | 3.11 ± 0.18 B | 2.98 ± 0.17 B | 4.78 ± 0.07 A | 4.45 ± 0.24 A | 2.97 ± 0.16 B | 2.80 ± 0.13 B | |
| Met | 1.53 ± 0.04 B | 1.57 ± 0.10 B | 2.37 ± 0.13 A | 2.37 ± 0.12 A | 1.50 ± 0.07 B | 1.46 ± 0.11 B | |
| Ile | 2.99 ± 0.20 B | 2.94 ± 0.17 B | 4.41 ± 0.11 A | 4.29 ± 0.23 A | 2.79 ± 0.15 B | 2.75 ± 0.09 B | |
| Leu | 4.94 ± 0.32 B | 4.91 ± 0.24 B | 7.54 ± 0.20 A | 7.42 ± 0.37 A | 4.68 ± 0.20 B | 4.60 ± 0.20 B | |
| Phe | 1.56 ± 0.11 B | 1.68 ± 0.10 B | 2.89 ± 0.52 A | 3.08 ± 0.60 A | 1.51 ± 0.06 B | 1.59 ± 0.09 B | |
| Lys | 5.50 ± 0.33 B | 5.53 ± 0.26 B | 8.27 ± 0.24 A | 8.18 ± 0.52 A | 5.16 ± 0.16 B | 5.05 ± 0.32 B | |
| NEAA | Asp | 5.80 ± 0.43 B | 5.77 ± 0.32 B | 8.59 ± 0.24 A | 8.34 ± 0.46 A | 5.48 ± 0.30 B | 5.38 ± 0.27 B |
| Ser | 2.39 ± 0.18 B | 2.52 ± 0.11 B | 3.70 ± 0.17 A | 3.67 ± 0.26 A | 2.29 ± 0.06 B | 2.38 ± 0.15 B | |
| Glu | 9.32 ± 0.65 B | 9.83 ± 0.83 B | 13.82 ± 0.56 A | 14.52 ± 0.76 A | 8.57 ± 0.39 B | 8.88 ± 0.42 B | |
| Gly | 2.92 ± 0.33 B | 3.24 ± 0.48 B | 4.13 ± 0.23 A | 4.61 ± 0.39 A | 2.81 ± 0.30 B | 3.24 ± 0.32 B | |
| Ala | 3.79 ± 0.28 B | 3.82 ± 0.33 B | 5.32 ± 0.49 A | 5.52 ± 0.42 A | 3.57 ± 0.17 B | 3.54 ± 0.18 B | |
| Cys | 0.62 ± 0.06 a | 0.42 ± 0.03 | 0.55 ± 0.07 a | 0.39 ± 0.10 | 0.46 ± 0.10 b | 0.59 ± 0.15 | |
| Tyr | 1.78 ± 0.13 B | 1.79 ± 0.09 B | 2.98 ± 0.22 A | 2.84 ± 0.14 A | 1.67 ± 0.08 B | 1.65 ± 0.10 B | |
| His | 2.48 ± 0.34 B | 2.24 ± 0.48 B | 4.24 ± 0.13 A | 3.98 ± 0.31 A | 2.32 ± 0.32 B | 2.17 ± 0.25 B | |
| Arg | 3.78 ± 0.26 B | 3.86 ± 0.26 B | 6.29 ± 0.18 A | 6.25 ± 0.32 A | 3.63 ± 0.20 B | 3.64 ± 0.18 B | |
| Pro | 1.13 ± 0.14 C | 1.74 ± 0.35 B | 2.66 ± 0.04 A | 3.02 ± 0.22 A | 1.50 ± 0.14 B | 1.71 ± 0.11 B | |
| TAA | 56.43 ± 2.13 B | 57.66 ± 2.20 B | 86.78 ± 3.10 A | 87.28 ± 3.20 A | 53.54 ± 1.95 B | 54.06 ± 1.97 B | |
| Items | CB | WD | MC | |||
|---|---|---|---|---|---|---|
| Breast Muscle | Leg Muscle | Breast Muscle | Leg Muscle | Breast Muscle | Leg Muscle | |
| C12:0 | 0.018 ± 0.003 b | 0.050 ± 0.005 a* | 0.025 ± 0.003 a | 0.028 ± 0.010 b | 0.000 ± 0.000 c | 0.000 ± 0.000 c |
| C14:0 | 0.305 ± 0.027 B | 1.285 ± 0.13 A** | 0.665 ± 0.077 A | 0.875 ± 0.321 B | 0.125 ± 0.014 C | 0.243 ± 0.015 C* |
| C15:0 | 0.040 ± 0.004 b | 0.183 ± 0.018 a** | 0.068 ± 0.008 a | 0.133 ± 0.049 a* | 0.018 ± 0.002 c | 0.040 ± 0.002 b* |
| C16:0 | 15.088 ± 1.355 B | 50.978 ± 5.165 A** | 24.720 ± 2.869 A | 31.813 ± 11.67 B | 6.375 ± 0.712 C | 10.950 ± 0.684 C* |
| C17:0 | 0.063 ± 0.006 b | 0.323 ± 0.033 a** | 0.128 ± 0.015 a | 0.265 ± 0.097 a* | 0.060 ± 0.007 b | 0.120 ± 0.007 b* |
| C18:0 | 7.210 ± 0.648 B | 18.308 ± 1.855 A** | 9.393 ± 1.09 A | 11.758 ± 4.316 B | 3.490 ± 0.39 C | 7.685 ± 0.48 B* |
| C20:0 | 0.060 ± 0.005 b | 0.260 ± 0.026 a** | 0.115 ± 0.013 a | 0.443 ± 0.162 a* | 0.025 ± 0.003 c | 0.070 ± 0.004 b* |
| C22:0 | 0.073 ± 0.007 a | 0.078 ± 0.008 | 0.040 ± 0.005 b | 0.058 ± 0.021 | 0.028 ± 0.003 c | 0.058 ± 0.004 * |
| C24:0 | 0.545 ± 0.049 a | 0.690 ± 0.07 * | 0.480 ± 0.056 a | 0.573 ± 0.21 | 0.333 ± 0.037 b | 0.510 ± 0.032 * |
| C14:1n5 | 0.095 ± 0.009 b | 0.493 ± 0.05 A** | 0.130 ± 0.015 a | 0.220 ± 0.081 B | 0.015 ± 0.002 c | 0.055 ± 0.003 C* |
| C15:1n5 | 0.118 ± 0.011 a* | 0.053 ± 0.005 a | 0.128 ± 0.015 a** | 0.020 ± 0.007 b | 0.000 ± 0.000 b | 0.010 ± 0.001 b* |
| C16:1n7 | 3.018 ± 0.271 B* | 1.598 ± 0.162 B | 5.078 ± 0.589 A | 7.273 ± 2.669 A | 0.750 ± 0.084 C | 1.845 ± 0.115 B* |
| C17:1n7 | 0.065 ± 0.006 b | 0.185 ± 0.019 a* | 0.155 ± 0.018 a | 0.190 ± 0.07 a | 0.018 ± 0.002 c | 0.058 ± 0.004 b* |
| C18:1n9c | 17.918 ± 1.609 B | 72.878 ± 7.384 A** | 39.915 ± 4.632 A | 51.053 ± 18.73 A | 9.583 ± 1.07 C | 16.600 ± 1.037 B* |
| C20:1n | 0.220 ± 0.020 b | 1.118 ± 0.113 a** | 0.823 ± 0.095 a | 1.065 ± 0.391 a | 0.158 ± 0.018 b | 0.320 ± 0.02 b* |
| C18:2n6c | 10.498 ± 0.943 B | 35.703 ± 3.618 A** | 20.665 ± 2.398 A | 30.825 ± 11.310 A | 6.69 ± 0.747 C | 12.983 ± 0.811 B* |
| C18:3n6 | 0.093 ± 0.008 b | 0.383 ± 0.039 a** | 0.143 ± 0.017 a | 0.253 ± 0.093 b | 0.088 ± 0.01 b | 0.165 ± 0.01 b* |
| C18:3n3 | 0.298 ± 0.027 b | 1.360 ± 0.138 a** | 0.645 ± 0.075 a | 1.058 ± 0.388 a | 0.220 ± 0.025 b | 0.413 ± 0.026 b* |
| C20:2n6 | 0.185 ± 0.017 a | 0.410 ± 0.042 a* | 0.223 ± 0.026 a | 0.408 ± 0.15 a | 0.123 ± 0.014 b | 0.233 ± 0.015 b* |
| C20:4n6 | 5.638 ± 0.506 a | 7.925 ± 0.803 A* | 4.350 ± 0.505 b | 5.413 ± 1.987 B | 3.140 ± 0.351 c | 5.530 ± 0.346 B* |
| C20:5n3 | 0.070 ± 0.006 | 0.125 ± 0.013 | 0.073 ± 0.008 | 0.115 ± 0.042 | 0.075 ± 0.008 | 0.083 ± 0.005 |
| C24:6n3 | 0.598 ± 0.054 a | 0.778 ± 0.079 a* | 0.398 ± 0.046 b | 0.538 ± 0.197 b | 0.390 ± 0.044 b | 0.773 ± 0.048 a* |
| SFA | 23.400 ± 2.102 B | 72.153 ± 7.311 A** | 35.633 ± 4.135 A | 45.943 ± 16.86 B | 10.453 ± 1.167 C | 19.675 ± 1.23 C* |
| MUFA | 21.433 ± 1.925 B | 76.323 ± 7.733 A** | 46.228 ± 5.365 A | 59.820 ± 21.957 A | 10.523 ± 1.175 C | 18.888 ± 1.18 B* |
| PUFA | 17.378 ± 1.561 B | 46.683 ± 4.73 A** | 26.495 ± 3.075 A | 38.608 ± 14.17 A | 10.725 ± 1.197 C | 20.178 ± 1.261 B* |
| USFA | 38.810 ± 3.486 B | 123.005 ± 12.46 A** | 72.723 ± 8.439 A | 98.428 ± 36.12 A | 21.248 ± 2.372 C | 39.065 ± 2.441 B* |
| EFA | 16.525 ± 1.484 b | 45.370 ± 4.597 A** | 25.803 ± 2.994 A | 37.548 ± 13.78 A | 10.138 ± 1.132 c | 19.090 ± 1.193 B* |
| TFA | 62.210 ± 5.587 B | 195.158 ± 19.77 A** | 108.355 ± 12.57 A | 144.370 ± 52.99 A | 31.700 ± 3.539 C | 58.740 ± 3.671 B* |
| Sample | Raw Data | Clean Data | Clean Data Ratio | ||||
|---|---|---|---|---|---|---|---|
| Sequences | Bases | Sequences | Bases | Sequences | Bases | ||
| CB1 | read1 | 44157511 | 6554832118 | 43990235 | 6444213183 | 99.62% | 98.31% |
| read2 | 6554832118 | 6445842898 | 98.34% | ||||
| CB2 | read1 | 48373644 | 7130323676 | 48212083 | 7021232185 | 99.67% | 98.47% |
| read2 | 7130323676 | 7024719635 | 98.52% | ||||
| CB3 | read1 | 50417672 | 7455132971 | 50360438 | 7407469730 | 99.89% | 99.36% |
| read2 | 7455132971 | 7407611791 | 99.36% | ||||
| CB4 | read1 | 50343815 | 7448999742 | 50260894 | 7382587236 | 99.84% | 99.11% |
| read2 | 7448999742 | 7386118122 | 99.16% | ||||
| CB5 | read1 | 47428679 | 7021075183 | 47242571 | 6906712188 | 99.61% | 98.37% |
| read2 | 7021075183 | 6904694579 | 98.34% | ||||
| WD1 | read1 | 41193762 | 6095723172 | 41151375 | 6058668197 | 99.90% | 99.39% |
| read2 | 6095723172 | 6058855779 | 99.40% | ||||
| WD2 | read1 | 46889983 | 6973814175 | 46789831 | 6906779152 | 99.79% | 99.04% |
| read2 | 6973814175 | 6909960586 | 99.08% | ||||
| WD3 | read1 | 54172288 | 8043390125 | 54046615 | 7963234169 | 99.77% | 99.00% |
| read2 | 8043390125 | 7966400727 | 99.04% | ||||
| MC1 | read1 | 43635413 | 6496325561 | 43576854 | 6455586208 | 99.87% | 99.37% |
| read2 | 6496325561 | 6453106964 | 99.33% | ||||
| MC2 | read1 | 46462679 | 6897145314 | 46389506 | 6836428320 | 99.84% | 99.12% |
| read2 | 6897145314 | 6842573604 | 99.21% | ||||
| MC3 | read1 | 45892152 | 6813870731 | 45820588 | 6758336835 | 99.84% | 99.18% |
| read2 | 6813870731 | 6760233491 | 99.21% | ||||
| MC4 | read1 | 44347624 | 6564043642 | 44210356 | 6469829352 | 99.69% | 98.56% |
| read2 | 6564043642 | 6470469881 | 98.57% | ||||
| Sample | Clean Reads | Mapped Reads | Mapped Ratio |
|---|---|---|---|
| CB1 | 43990235 | 33107050 | 75.26% |
| CB2 | 48212083 | 36578507 | 75.87% |
| CB3 | 50360438 | 39402006 | 78.24% |
| CB4 | 50260894 | 37509705 | 74.63% |
| WD1 | 47242571 | 34699668 | 73.45% |
| WD2 | 41151375 | 33949884 | 82.50% |
| WD3 | 46789831 | 35087694 | 74.99% |
| WD4 | 54046615 | 39697238 | 73.45% |
| MC1 | 43576854 | 32656494 | 74.94% |
| MC2 | 46389506 | 36346177 | 78.35% |
| MC3 | 45820588 | 34837393 | 76.03% |
| MC4 | 44210356 | 30483040 | 68.95% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhang, X.; Wang, K.; Li, Q.; Yan, S.; Shi, H.; Liu, L.; Liang, S.; Yang, M.; Su, Z.; et al. RNA-Seq Reveals Pathways Responsible for Meat Quality Characteristic Differences between Two Yunnan Indigenous Chicken Breeds and Commercial Broilers. Foods 2024, 13, 2008. https://doi.org/10.3390/foods13132008
Liu Y, Zhang X, Wang K, Li Q, Yan S, Shi H, Liu L, Liang S, Yang M, Su Z, et al. RNA-Seq Reveals Pathways Responsible for Meat Quality Characteristic Differences between Two Yunnan Indigenous Chicken Breeds and Commercial Broilers. Foods. 2024; 13(13):2008. https://doi.org/10.3390/foods13132008
Chicago/Turabian StyleLiu, Yong, Xia Zhang, Kun Wang, Qihua Li, Shixiong Yan, Hongmei Shi, Lixian Liu, Shuangmin Liang, Min Yang, Zhengchang Su, and et al. 2024. "RNA-Seq Reveals Pathways Responsible for Meat Quality Characteristic Differences between Two Yunnan Indigenous Chicken Breeds and Commercial Broilers" Foods 13, no. 13: 2008. https://doi.org/10.3390/foods13132008
APA StyleLiu, Y., Zhang, X., Wang, K., Li, Q., Yan, S., Shi, H., Liu, L., Liang, S., Yang, M., Su, Z., Ge, C., Jia, J., Xu, Z., & Dou, T. (2024). RNA-Seq Reveals Pathways Responsible for Meat Quality Characteristic Differences between Two Yunnan Indigenous Chicken Breeds and Commercial Broilers. Foods, 13(13), 2008. https://doi.org/10.3390/foods13132008






