Evaluation of the Novel mTA10 Selective Broth, MSB, for the Co-Enrichment and Detection of Salmonella spp., Escherichia coli O157 and Listeria monocytogenes in Ready-to-Eat Salad Samples
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
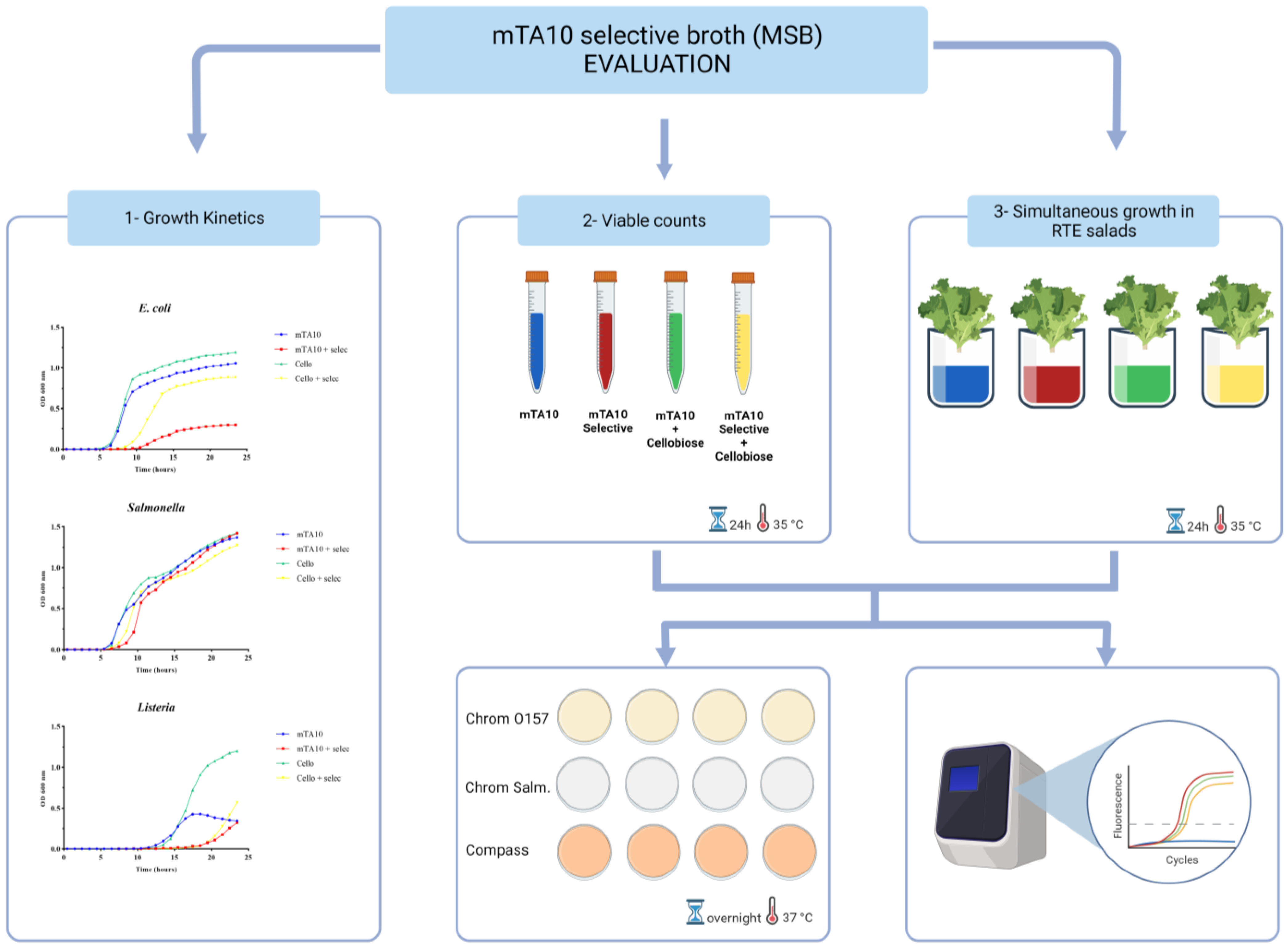
2.2. Selective Medium Formulation and Evaluation
2.2.1. Individual Bacterial Growth
2.2.2. Mixed Bacterial Cultures Growth
2.2.3. Evaluation of the Growth Capacity of Mixed Bacterial Cultures in Spiked RTE Salads
2.3. RTE Salad Sample Inoculation and Processing in the New Method Implementing MSB
2.4. DNA Extraction
2.5. Pathogen Detection by Multiplex qPCR
2.6. Fitness-for-Purpose
2.7. Data Representation
3. Results
3.1. MSB Evaluation
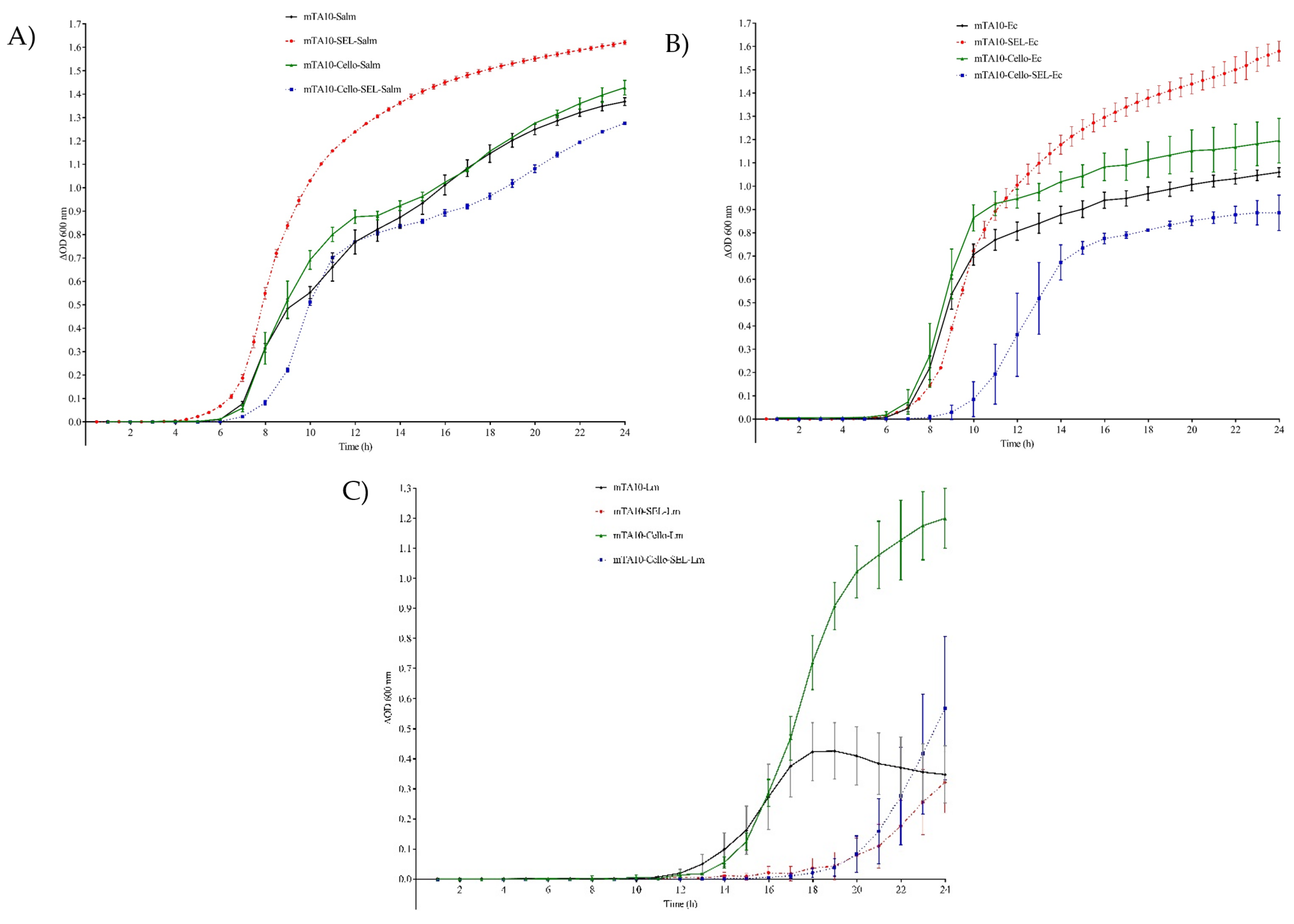
3.1.1. Individual Bacterial Growth
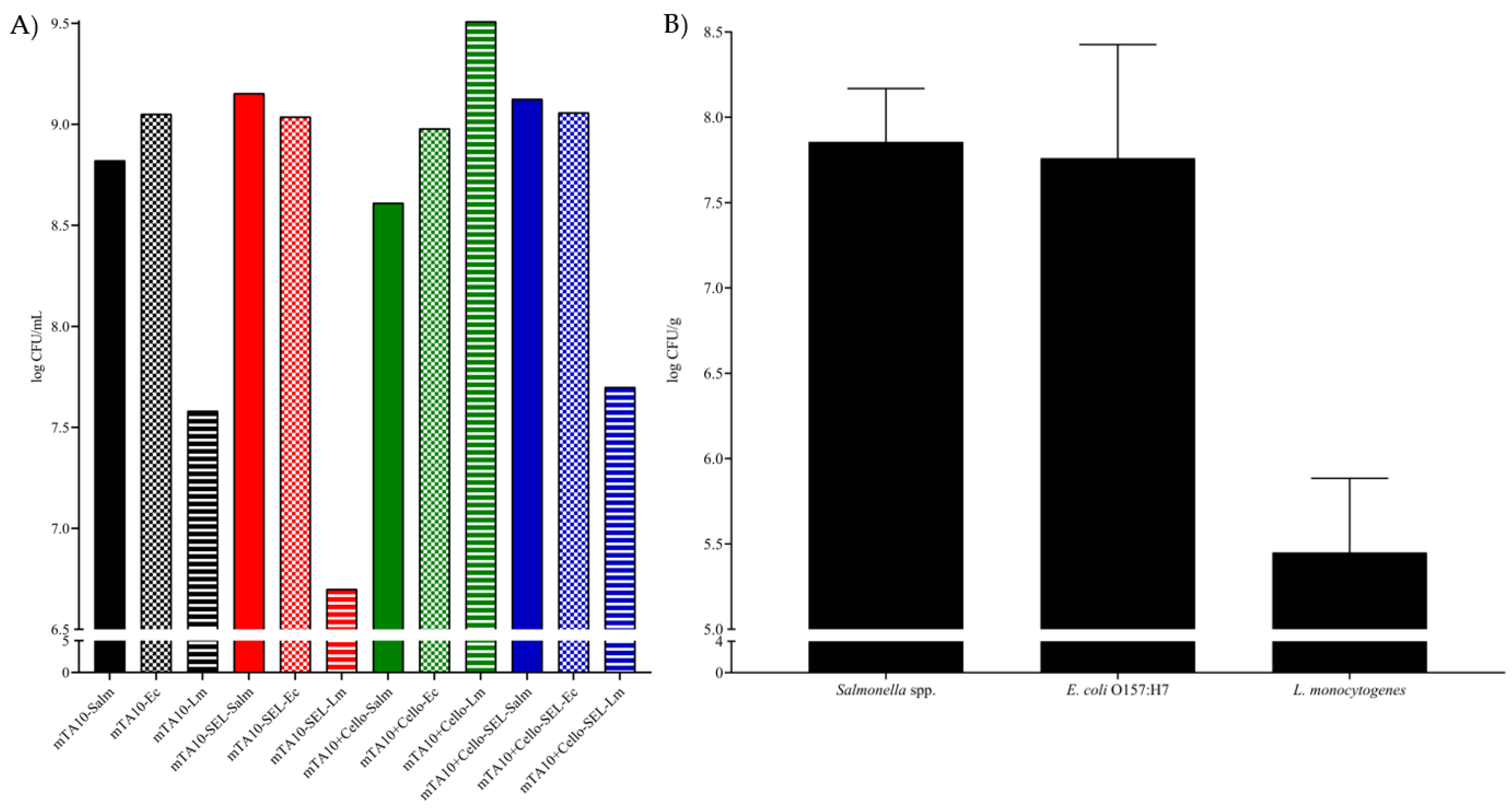
3.1.2. Mixed Bacterial Cultures Growth
3.1.3. Mixed Bacterial Culture Growth in MSB Spiked RTE Salads
3.2. Fitness-for-Purpose
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015.
- Panwar, S.; Duggirala, K.S.; Yadav, P.; Debnath, N.; Yadav, A.K.; Kumar, A. Advanced Diagnostic Methods for Identification of Bacterial Foodborne Pathogens: Contemporary and Upcoming Challenges. Crit. Rev. Biotechnol. 2022, 43, 982–1000. [Google Scholar] [CrossRef] [PubMed]
- Deng, K.; Wang, S.S.; Kiener, S.; Smith, E.; Chen, K.S.; Pamboukian, R.; Laasri, A.; Pelaez, C.; Ulaszek, J.; Kmet, M.; et al. Multi-Laboratory Validation Study of a Real-Time PCR Method for Detection of Salmonella in Baby Spinach. Food Microbiol. 2023, 114, 104299. [Google Scholar] [CrossRef] [PubMed]
- Brehm-Stecher, B.; Young, C.; Jaykus, L.-A.; Mary Lou, T. Sample Preparation: The Forgotten Beginning. J. Food Prot. 2009, 72, 1774–1789. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, M.; Diez-Valcarce, M.; Robles, S.; Losilla-Garcia, B.; Cook, N.; D’Agostino, M.; Diez-Valcarce, M.; Robles, S.; Losilla-Garcia, B.; Cook, N. A Loop-Mediated Isothermal Amplification-Based Method for Analysing Animal Feed for the Presence of Salmonella. Food Anal. Methods 2015, 8, 2409–2416. [Google Scholar] [CrossRef]
- Parichehr, M.; Mohammad, K.; Abbas, D.; Mehdi, K. Developing a Multiplex Real-Time PCR with a New Pre-Enrichment to Simultaneously Detect Four Foodborne Bacteria in Milk. Future Microbiol. 2019, 14, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.S.; Cox, N.A. Universal Preenrichment Broth for the Simultaneous Detection of Salmonella and Listeria in Foods. J. Food Prot. 1992, 55, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Kubota, J.; Fujihara, K.; Honjoh, K.; Masayoshi Iio, N.F.; Nakabe, M.; Oda, S.; Satoyama, T.; Takasu, K.; Nakanishi, H.; et al. Simultaneous Enrichment of Salmonella spp., Escherichia coli O157:H7, Vibrio parahaemolyticus, Staphylococcus aureus, Bacillus cereus, and Listeria monocytogenes by Single Broth and Screening of the Pathogens by Multiple. Food Sci. Technol. Res. 2009, 15, 427–438. [Google Scholar] [CrossRef]
- Kawasaki, S.; Horikoshi, N.; Okada, Y.; Takeshita, K.; Sameshima, T.; Kawamoto, S. Multiplex PCR for Simultaneous Detection of Salmonella spp., Listeria monocytogenes, and Escherichia coli O157: H7 in Meat Samples. J. Food Prot. 2005, 68, 551–556. [Google Scholar] [CrossRef]
- Kawasaki, S.; Fratamico, P.M.; Kamisaki-Horikoshi, N.; Okada, Y.; Takeshita, K.; Sameshima, T.; Kawamoto, S. Development of the Multiplex PCR Detection Kit for Salmonella spp., Listeria monocytogenes, and Escherichia coli O157:H7. Jarq-Japan Agric. Res. Q. 2011, 45, 77–81. [Google Scholar] [CrossRef]
- Omiccioli, E.; Amagliani, G.; Brandi, G.; Magnani, M. A New Platform for Real-Time PCR Detection of Salmonella spp., Listeria monocytogenes and Escherichia coli O157 in Milk. Food Microbiol. 2009, 26, 615–622. [Google Scholar] [CrossRef]
- Garrido, A.; Chapela, M.-J.; Román, B.; Fajardo, P.; Lago, J.; Vieites, J.M.J.M.; Cabado, A.G.A.G. A New Multiplex Real-Time PCR Developed Method for Salmonella spp. and Listeria monocytogenes Detection in Food and Environmental Samples. Food Control 2013, 30, 76–85. [Google Scholar] [CrossRef]
- Villamizar-Rodríguez, G.; Fernández, J.; Marín, L.; Muñiz, J.; González, I.; Lombó, F. Multiplex Detection of Nine Food-Borne Pathogens by MPCR and Capillary Electrophoresis after Using a Universal Pre-Enrichment Medium. Front. Microbiol. 2015, 6, 1194. [Google Scholar] [CrossRef] [PubMed]
- Azinheiro, S.; Carvalho, J.; Prado, M.; Garrido-Maestu, A. Multiplex Detection of Salmonella spp., E. coli O157 and L. monocytogenes by qPCR Melt Curve Analysis in Spiked Infant Formula. Microorganisms 2020, 8, 1359. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.C.; Bhunia, A.K. SEL, a Selective Enrichment Broth for Simultaneous Growth of Salmonella enterica, Escherichia coli O157: H7, and Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 4853–4866. [Google Scholar] [CrossRef] [PubMed]
- Suo, Y.; Gao, S.; Xie, Y.; Liu, Y.; Qu, Y.; Lin, T.; Zhou, C. A Multipathogen Selective Enrichment Broth for Simultaneous Growth of Salmonella enteria, Escherichia coli O157:H7, and Shigella flexneri. J. Food Saf. 2018, 38, e12388. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Bhunia, A.K.; Tang, C.; Wang, C.; Shi, H. Development of a Multi-Pathogen Enrichment Broth for Simultaneous Growth of Five Common Foodborne Pathogens. J. Gen. Appl. Microbiol. 2015, 61, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Zhang, L.; Wu, H.; Yu, Y.; Tang, Y.; Liu, D.; Li, X. Simultaneous Detection of Salmonella, Listeria monocytogenes, and Staphylococcus aureus by Multiplex Real-Time PCR Assays Using High-Resolution Melting. Food Anal. Methods 2014, 7, 1960–1972. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Liu, J.; Cai, Z.; Bai, X. Development and Evaluation of a Multiplex PCR for Simultaneous Detection of Five Foodborne Pathogens. J. Appl. Microbiol. 2012, 112, 823–830. [Google Scholar] [CrossRef]
- Kim, J.; Shin, H.; Park, H.; Jung, H.; Kim, J.; Cho, S.; Ryu, S.; Jeon, B. Microbiota Analysis for the Optimization of Campylobacter isolation from Chicken Carcasses Using Selective Media. Front. Microbiol. 2019, 10, 1381. [Google Scholar] [CrossRef]
- Costa-Ribeiro, A.; Azinheiro, S.; Mota, S.; Prado, M.; Lamas, A.; Garrido-Maestu, A. Assessment of the Presence of Acinetobacter spp. Resistant to β-Lactams in Commercial Ready-to-Eat Salad Samples. Food Microbiol. 2024, 118, 104410. [Google Scholar] [CrossRef]
- Sant’Ana, A.S.; Franco, B.D.G.M.; Schaffner, D.W. Risk of Infection with Salmonella and Listeria monocytogenes Due to Consumption of Ready-to-Eat Leafy Vegetables in Brazil. Food Control 2014, 42, 1–8. [Google Scholar] [CrossRef]
- Abadias, M.; Usall, J.; Anguera, M.; Solsona, C.; Viñas, I. Microbiological Quality of Fresh, Minimally-Processed Fruit and Vegetables, and Sprouts from Retail Establishments. Int. J. Food Microbiol. 2008, 123, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Prado, M. Combination of Immunomagnetic Separation and Real-Time Recombinase Polymerase Amplification (IMS-QRPA) for Specific Detection of Listeria monocytogenes in Smoked Salmon Samples. J. Food Sci. 2019, 84, 1881–1887. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Bai, Y.; Liu, Y.; Zhou, C.; Zhou, X.; Zhang, D.; Shi, C.; Suo, Y. SSEL, a Selective Enrichment Broth for Simultaneous Growth of Salmonella enterica, Staphylococcus aureus, Escherichia coli O157: H7, and Listeria monocytogenes. J. Food Saf. 2020, 40, 4853–4866. [Google Scholar] [CrossRef]
- Yu, Y.-G.G.; Wu, H.; Liu, Y.-Y.Y.; Li, S.-L.L.; Yang, X.-Q.Q.; Xiao, X.-L.L. A Multipathogen Selective Enrichment Broth for Simultaneous Growth of Salmonella enterica Serovar Enteritidis, Staphylococcus aureus, and Listeria monocytogenes. Can. J. Microbiol. 2010, 56, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M. (Ed.) Handbook of Microbiological Media, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2004; ISBN 9780429129032. [Google Scholar]
- Zimbro, M.J.; Power, D.A.; Miller, S.M.; Wilson, G.E.; Johnson, J.A. (Eds.) Difco & BBL Manual: Manual of Microbiological Culture Media, 2nd ed.; Becton, Dickinson and Company: Sparks, MD, USA, 2009; ISBN 0972720715. [Google Scholar]
- Singh, T. Antimicrobial Efficacy of Octenidine Dihydrochloride and Artemisia Annua Plant Extract as Root Canal Irrigants—An In Vivo Study. J. Dent. Oral Sci. 2021, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Al-Kafaween, M.A.; Khan, R.S.; Hilmi, M.; Bakar, A.; Bouacha, M. Effect of Growth Media and Optical Density on Biofilm Formation by Staphylococcus epidermidis Soil Fertility and Soil Microbiology. Soil Water Analysis for Irrigation Purposes. View Project Antibacterial Activity View Project. EC Microbiol. 2019, 15, 277–282. [Google Scholar]
- Neidhardt, F.C.; Bloch, P.L.; Smith, D.F. Culture Medium for Enterobacteria. Microbiology 1974, 119, 736–747. [Google Scholar] [CrossRef]
- Teo, A.Y.; Knabel, S.J. Development of a Simple Recovery-Enrichment System for Enhanced Detection of Heat-Injured Listeria monocytogenes in Pasteurized Milk. J. Food Prot. 2000, 63, 462–472. [Google Scholar] [CrossRef]
- Taylor, A.J.; Stasiewicz, M.J. Persistent and Sporadic Listeria monocytogenes Strains Do Not Differ When Growing at 37 °C, in Planktonic State, under Different Food Associated Stresses or Energy Sources. BMC Microbiol. 2019, 19, 257. [Google Scholar] [CrossRef]
- Morishige, Y.; Fujimori, K.; Amano, F. Differential Resuscitative Effect of Pyruvate and Its Analogues on VBNC (Viable But Non-Culturable) Salmonella. Microbes Environ. 2013, 28, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Cox, L.J.; Dooley, D.; Beumer, R. Effect of Lithium Chloride and Other Inhibitors on the Growth of Listeria spp. Food Microbiol. 1990, 7, 311–325. [Google Scholar] [CrossRef]
- Jacobsen, C.N. The Influence of Commonly Used Selective Agents on the Growth of Listeria monocytogenes. Int. J. Food Microbiol. 1999, 50, 221–226. [Google Scholar] [CrossRef]
- Kinner, J.A.; Moats, W.A. Selective Action of Sodium Cholate-MgCl2 Broth and Its Possible Use in Isolation of Salmonellae and Other Enteric Pathogens. J. Food Prot. 1978, 41, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Zanaroli, G.; Fedi, S.; Carnevali, M.; Fava, F.; Zannoni, D. Use of Potassium Tellurite for Testing the Survival and Viability of Pseudomonas pseudoalcaligenes KF707 in Soil Microcosms Contaminated with Polychlorinated Biphenyls. Res. Microbiol. 2002, 153, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Maestu, A.; Fuciños, P.; Azinheiro, S.; Carvalho, J.; Prado, M. Systematic Loop-Mediated Isothermal Amplification Assays for Rapid Detection and Characterization of Salmonella spp., Enteritidis and Typhimurium in Food Samples. Food Control 2017, 80, 297–306. [Google Scholar] [CrossRef]
- Sánchez, A.; Vázquez, J.A.; Quinteiro, J.; Sotelo, C.G. Modeling Real-Time PCR Kinetics: Richards Reparametrized Equation for Quantitative Estimation of European Hake (Merluccius Merluccius). J. Agric. Food Chem. 2013, 61, 3488–3493. [Google Scholar] [CrossRef] [PubMed]
- Commission Regulation (EC) No 2073/2005 Microbiological Criteria for Foodstuffs 2005, 2073/2005. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:02005R2073-20200308 (accessed on 1 November 2023).
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Prado, M. Rapid and Sensitive Detection of Viable Listeria monocytogenes in Food Products by a Filtration-Based Protocol and QPCR. Food Microbiol. 2018, 73, 254–263. [Google Scholar] [CrossRef]
- Roumani, F.; Azinheiro, S.; Carvalho, J.; Prado, M.; Garrido-Maestu, A. Loop-Mediated Isothermal Amplification Combined with Immunomagnetic Separation and Propidium Monoazide for the Specific Detection of Viable Listeria monocytogenes in Milk Products, with an Internal Amplification Control. Food Control 2021, 125, 107975. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Fuciños, P.; Prado, M. Optimized Sample Treatment, Combined with Real-Time PCR, for Same-Day Detection of E. coli O157 in Ground Beef and Leafy Greens. Food Control 2020, 108, 106790. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Fuciños, P.; Prado, M. Development and Evaluation of Loop-Mediated Isothermal Amplification, and Recombinase Polymerase Amplification Methodologies, for the Detection of Listeria monocytogenes in Ready-to-Eat Food Samples. Food Control 2018, 86, 27–34. [Google Scholar] [CrossRef]
- Anderson, A.; Pietsch, K.; Zucker, R.; Mayr, A.; Müller-Hohe, E.; Messelhäusser, U.; Sing, A.; Busch, U.; Huber, I.; Muller-Hohe, E.; et al. Validation of a Duplex Real-Time PCR for the Detection of Salmonella spp. in Different Food Products. Food Anal. Methods 2011, 4, 259–267. [Google Scholar] [CrossRef]
- Tomás, D.; Rodrigo, A.; Hernández, M.; Ferrús, M.A. Validation of Real-Time PCR and Enzyme-Linked Fluorescent Assay-Based Methods for Detection of Salmonella spp. in Chicken Feces Samples. Food Anal. Methods 2009, 2, 180–189. [Google Scholar] [CrossRef]
- The Interagency Food Safety Analytics Collaboration (IFSAC). Foodborne Illness Source Attribution Estimates for Salmonella, Escherichia coli O157 (E. coli O157), Listeria monocytogenes (Lm), and Campylobacter; 2023. Available online: https://www.fda.gov/food/cfsan-constituent-updates/ifsac-releases-annual-report-2021-sources-foodborne-illness (accessed on 1 November 2023).
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [CrossRef]
- Castro-Ibáñez, I.; Gil, M.I.; Allende, A. Ready-to-Eat Vegetables: Current Problems and Potential Solutions to Reduce Microbial Risk in the Production Chain. LWT Food Sci. Technol. 2017, 85, 284–292. [Google Scholar] [CrossRef]
- Thomas, G.A.; Gil, T.P.; Müller, C.T.; Rogers, H.J.; Berger, C.N. From Field to Plate: How Do Bacterial Enteric Pathogens Interact with Ready-to-Eat Fruit and Vegetables, Causing Disease Outbreaks? Food Microbiol. 2023, 117, 104389. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Suo, Y.; Zhang, Z.; Liu, D.; Ye, X.; Chen, S.; Zhao, Y. A Multiplex RT-PCR Assay for S. aureus, L. monocytogenes, and Salmonella spp. Detection in Raw Milk with Pre-Enrichment. Front. Microbiol. 2017, 8, 989. [Google Scholar] [CrossRef] [PubMed]
- Scheusner, D.L.; Busta, F.F.; Speck, M.L. Inhibition of Injured Escherichia coli by Several Selective Agents. Appl. Microbiol. 1971, 21, 46–49. [Google Scholar] [CrossRef]
- Myers, J.A.; Curtis, B.S.; Curtis, W.R. Improving Accuracy of Cell and Chromophore Concentration Measurements Using Optical Density. BMC Biophys. 2013, 6, 4. [Google Scholar] [CrossRef]
- Xiao, X.L.; Zhai, J.X.; Wu, H.; Liu, D.; Yu, Y.G.; Li, X.F. Development and Evaluation of a Selective Enrichment Broth for Simultaneous Growth of Salmonella enterica Serovar Enteritidis, Shigella dysenteriae and Staphylococcus aureus. Ann. Microbiol. 2014, 64, 1543–1551. [Google Scholar] [CrossRef]
- Germini, A.; Masola, A.; Carnevali, P.; Marchelli, R. Simultaneous Detection of Escherichia coli O157:H7, Salmonella spp., and Listeria monocytogenes by Multiplex PCR. Food Control 2009, 20, 733–738. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Van Giau, V.; Vo, T.K. Multiplex PCR for Simultaneous Identification of E. coli O157:H7, Salmonella spp. and L. monocytogenes in Food. 3 Biotech 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Osman, F.; Hodzic, E.; Omanska-Klusek, A.; Olineka, T.; Rowhani, A. Development and Validation of a Multiplex Quantitative PCR Assay for the Rapid Detection of Grapevine Virus A, B and D. J. Virol. Methods 2013, 194, 138–145. [Google Scholar] [CrossRef]
- Compston, L.I.; Sarkobie, F.; Li, C.; Candotti, D.; Opare-Sem, O.; Allain, J.P. Multiplex Real-Time PCR for the Detection and Quantification of Latent and Persistent Viral Genomes in Cellular or Plasma Blood Fractions. J. Virol. Methods 2008, 151, 47–54. [Google Scholar] [CrossRef]
| Component | g/L | Function | Reference |
|---|---|---|---|
| Tryptose | 10.0 | Provides amino acids, especially essential amino acids, large peptides and other nitrogenous substances. | [27,28] |
| Beef extract | 5.0 | Provides peptides, amino acids, nucleotides, organic acids, minerals and vitamins. | |
| Yeast extract | 5.0 | Source of amino acids, peptides and water-soluble vitamins such as B12 complex and carbohydrates. | |
| NaCl | 5.0 | Maintain osmotic balance. | [29,30] |
| MOPS | 8.5 | Buffering agents. | [31,32] |
| MOPS-Na | 13.7 | ||
| Cellobiose | 5.0 | Reducing sugar, which can be uptake by Listeria monocytogenes and used as a source of energy. | [33] |
| NaPyr | 1.1 | Used as an additional source of energy; bacterial growth inducer; free radical scavenger and reactive oxygen species (ROS)-quencher. | [34] |
| Lithium chloride | 1.0 | Broad-range inhibition of Gram-positive and Gram-negative bacteria. | [17,35,36] |
| Nalidixic acid | 0.0025 | Inhibition of competitive microbiota (able to inhibit microbiota that grow in the presence of LiCl). | [35] |
| Sodium cholate | 1.0 | Water-soluble bile salt that acts as a selective inhibitor. This substance interferes with the growth and incorporation of glucose and also inhibits flagellum formation in Gram-negative bacteria. | [37] |
| Potassium tellurite | 0.0001 | Inhibition of competitive microbiota due to its oxidative capacity (inhibits Gram-negative bacteria and some Gram-positive bacteria unable to use). | [38] |
| Microorganism | Primer | Sequence 5′ → 3′ | Concentration (nM) | Modifications | Reference |
|---|---|---|---|---|---|
| Salmonella spp. | ttr-P3F | GGC TAA TTT AAC CCG TCG TCA G | 100 | [39] | |
| ttr-P3R | GTT TCG CCA CAT CAC GGT AGC | 100 | |||
| ttr-P3P | AAG TCG GTC TCG CCG TCG GTG | 100 | NED/MGBNFQ | ||
| L. monocytogenes | hly-P3F | CGC AAC AAA CTG AAG CAA AGG A | 200 | [42,43] | |
| hly-P3R | CGA TTG GCG TCT TAG GAC TTG C | 200 | |||
| hly-P3P | CAT GGC ACC//ACC AGC ATC TCC G | 150 | FAM/ZEN/IABkFQ | ||
| E. coli O157 | O157-rfbE-F | TCA ACA GTC TTG TAC AAG TCC AC | 800 | - | [44] |
| O157-rfbE-R | ACT GGC CTT GTT TCG ATG AG | 800 | - | ||
| O157-rfbE-P | AC TAG GAC CGC AGA GGA AAG AGA GGA A | 400 | Cy5/IAbRQSp | ||
| - | NC-IAC-F | AGT TGC ACA CAG TTA GTT CGA G | 100 | - | [24] |
| NC-IAC-R | TGG AGT GCT GGA CGA TTT GAA G | 100 | - | ||
| IAC-P | AGT GGC GGT//GAC ACT GTT GAC CT | 100 | YY/ZEN/IABkFQ | [45] |
| Salmonella spp. | E. coli O157:H7 | L. monocytogenes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mTA10 | mTA10 + Cello | mTA10 | mTA10 + Cello | mTA10 | mTA10 + Cello | |||||||
| N | S | N | S | N | S | N | S | N | S | N | S | |
| OD600max | 1.39 ± 0.02 | 1.55 ± 0.01 | 1.37 ± 0.06 | 1.17 ± 0.01 | 0.99 ± 0.02 | 1.48 ± 0.04 | 1.12 ± 0.09 | 0.87 ± 0.04 | 0.39 ± 0.10 | 0.32 ± 0.10 | 1.23 ± 0.13 | 0.57 ± 0.24 |
| µmax | 0.12 ± 0.01 | 0.25 ± 0.00 | 0.13 ± 0.01 | 0.13 ± 0.00 | 0.20 ± 0.02 | 0.21 ± 0.01 | 0.27 ± 0.05 | 0.18 ± 0.01 | 0.13 ± 0.01 | 0.07 ± 0.00 | 0.23 ± 0.02 | 0.16 ± 0.04 |
| λ | 5.71 ± 0.07 | 5.92 ± 0.09 | 5.58 ± 0.16 | 6.73 ± 0.02 | 6.67 ± 0.05 | 7.04 ± 0.03 | 6.79 ± 0.64 | 10.01 ± 0.90 | 13.56 ± 0.41 | 19.25 ± 1.34 | 14.76 ± 0.36 | 20.18 ± 0.90 |
| Microorganism | Inoculation Level * | N | PA | NA | PD | ND | SE | SP | AC |
|---|---|---|---|---|---|---|---|---|---|
| Salmonella spp. | 19 | 3 | 3 | 93 | 100 | 94 | |||
| 10 | 3 | 3 | |||||||
| 5 | 5 | 5 | |||||||
| 3.5 | 4 | 3 | 1 | ||||||
| 0 | 2 | 2 | 0 | ||||||
| E. coli O157:H7 | 9.6 | 3 | 3 | 86 | 100 | 88 | |||
| 8.6 | 5 | 3 | 2 | ||||||
| 6.4 | 4 | 4 | |||||||
| 3.8 | 3 | 3 | |||||||
| 0 | 2 | 2 | 0 | ||||||
| L. monocytogenes | 46 | 3 | 3 | 93 | 100 | 94 | |||
| 31 | 5 | 5 | |||||||
| 9.4 | 4 | 3 | 1 | ||||||
| 3.7 | 3 | 3 | |||||||
| 0 | 2 | 2 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa-Ribeiro, A.; Lamas, A.; Prado, M.; Garrido-Maestu, A. Evaluation of the Novel mTA10 Selective Broth, MSB, for the Co-Enrichment and Detection of Salmonella spp., Escherichia coli O157 and Listeria monocytogenes in Ready-to-Eat Salad Samples. Foods 2024, 13, 63. https://doi.org/10.3390/foods13010063
Costa-Ribeiro A, Lamas A, Prado M, Garrido-Maestu A. Evaluation of the Novel mTA10 Selective Broth, MSB, for the Co-Enrichment and Detection of Salmonella spp., Escherichia coli O157 and Listeria monocytogenes in Ready-to-Eat Salad Samples. Foods. 2024; 13(1):63. https://doi.org/10.3390/foods13010063
Chicago/Turabian StyleCosta-Ribeiro, Ana, Alexandre Lamas, Marta Prado, and Alejandro Garrido-Maestu. 2024. "Evaluation of the Novel mTA10 Selective Broth, MSB, for the Co-Enrichment and Detection of Salmonella spp., Escherichia coli O157 and Listeria monocytogenes in Ready-to-Eat Salad Samples" Foods 13, no. 1: 63. https://doi.org/10.3390/foods13010063
APA StyleCosta-Ribeiro, A., Lamas, A., Prado, M., & Garrido-Maestu, A. (2024). Evaluation of the Novel mTA10 Selective Broth, MSB, for the Co-Enrichment and Detection of Salmonella spp., Escherichia coli O157 and Listeria monocytogenes in Ready-to-Eat Salad Samples. Foods, 13(1), 63. https://doi.org/10.3390/foods13010063







