Anti-Biofilm Activity of Laurel Essential Oil against Vibrio parahaemolyticus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Determination of Minimum Inhibitory Concentration (MIC)
2.3. Determination of Subinhibitory Concentration (SIC)
2.4. Biofilm Formation Assay
2.5. Biofilm Metabolic Activity
2.6. Visualization of Biofilm
2.7. Swimming Motility Assay
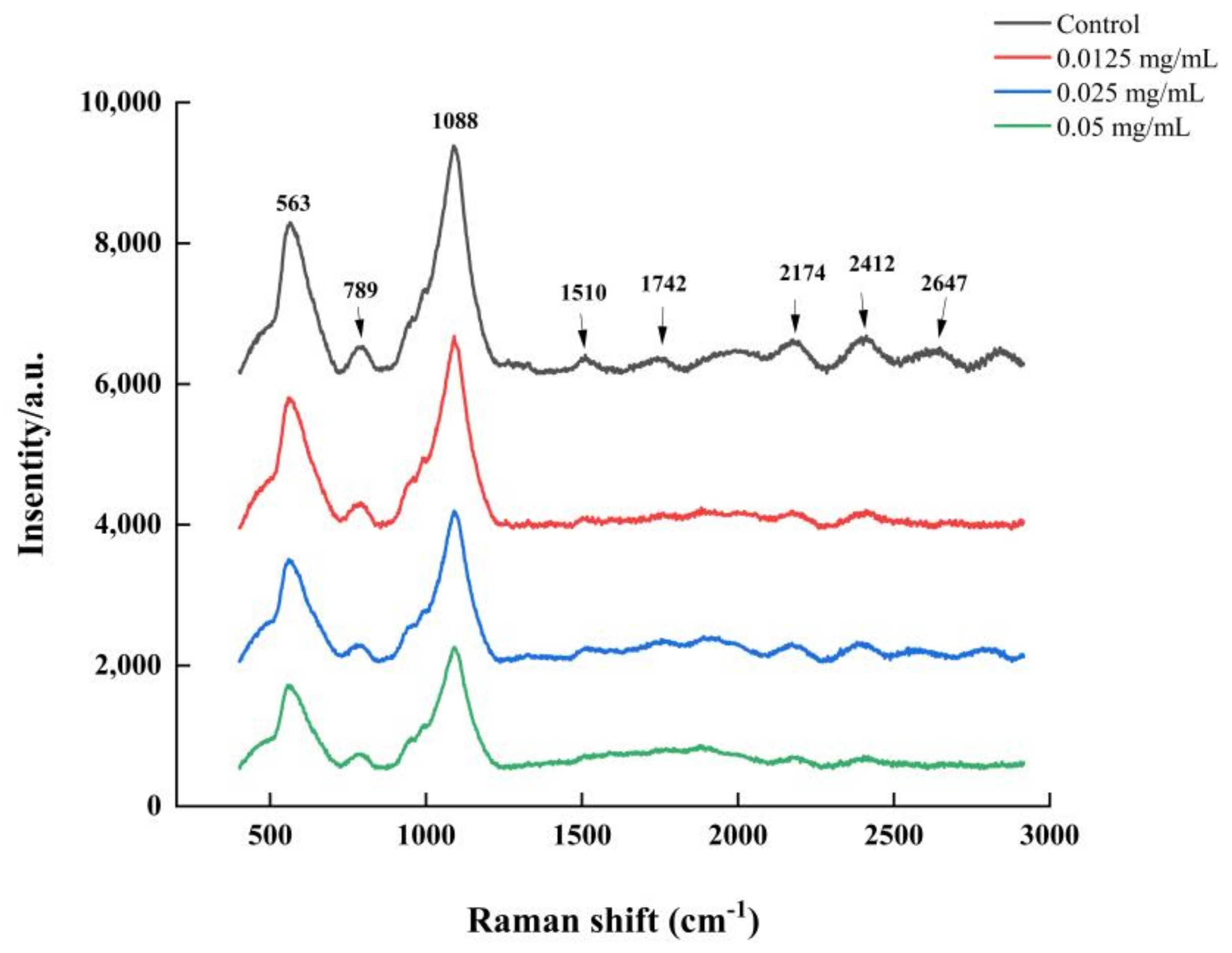
2.8. Raman Spectroscopy Analysis
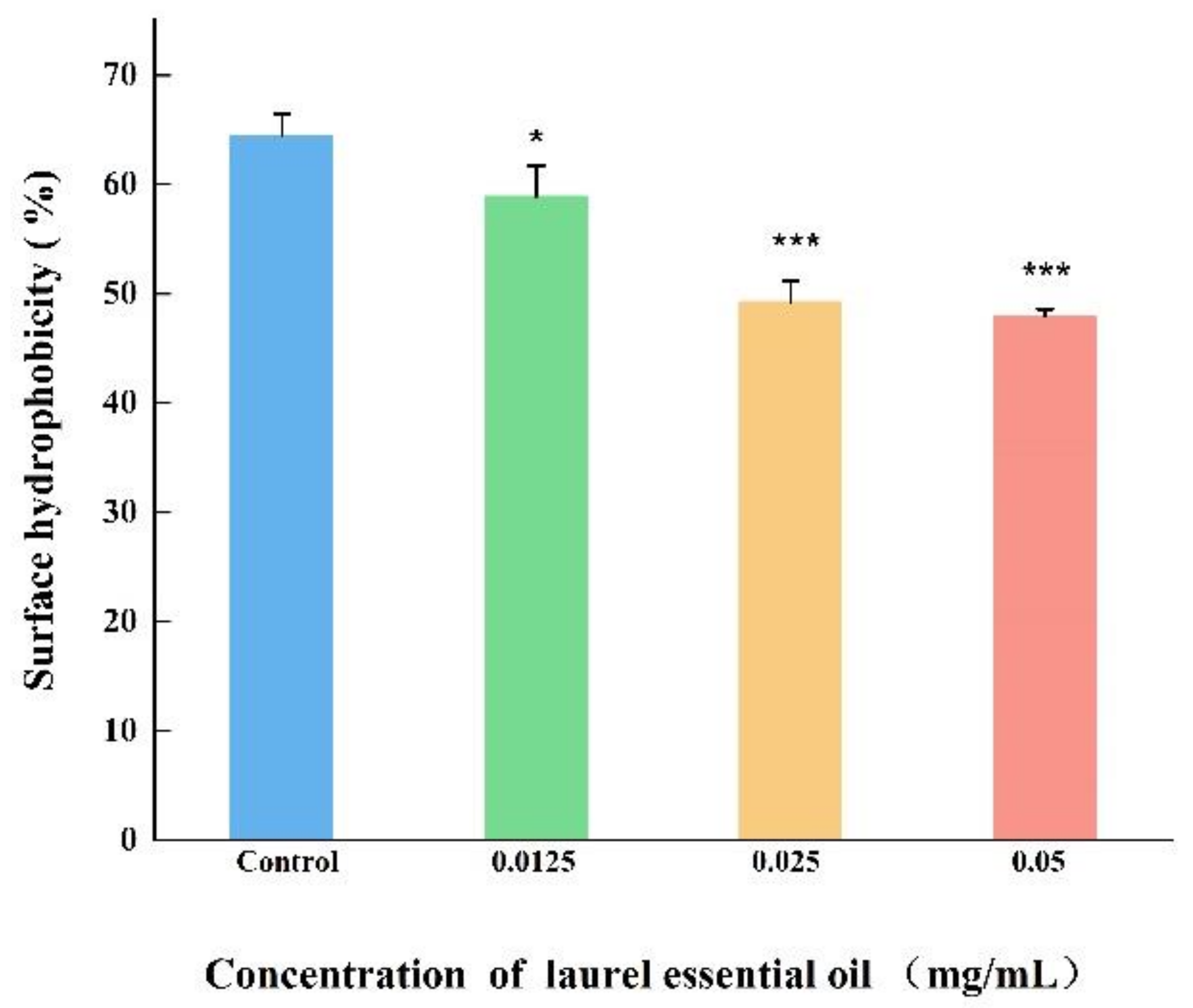
2.9. The Cell Surface Hydrophobicity (CSH) Assay
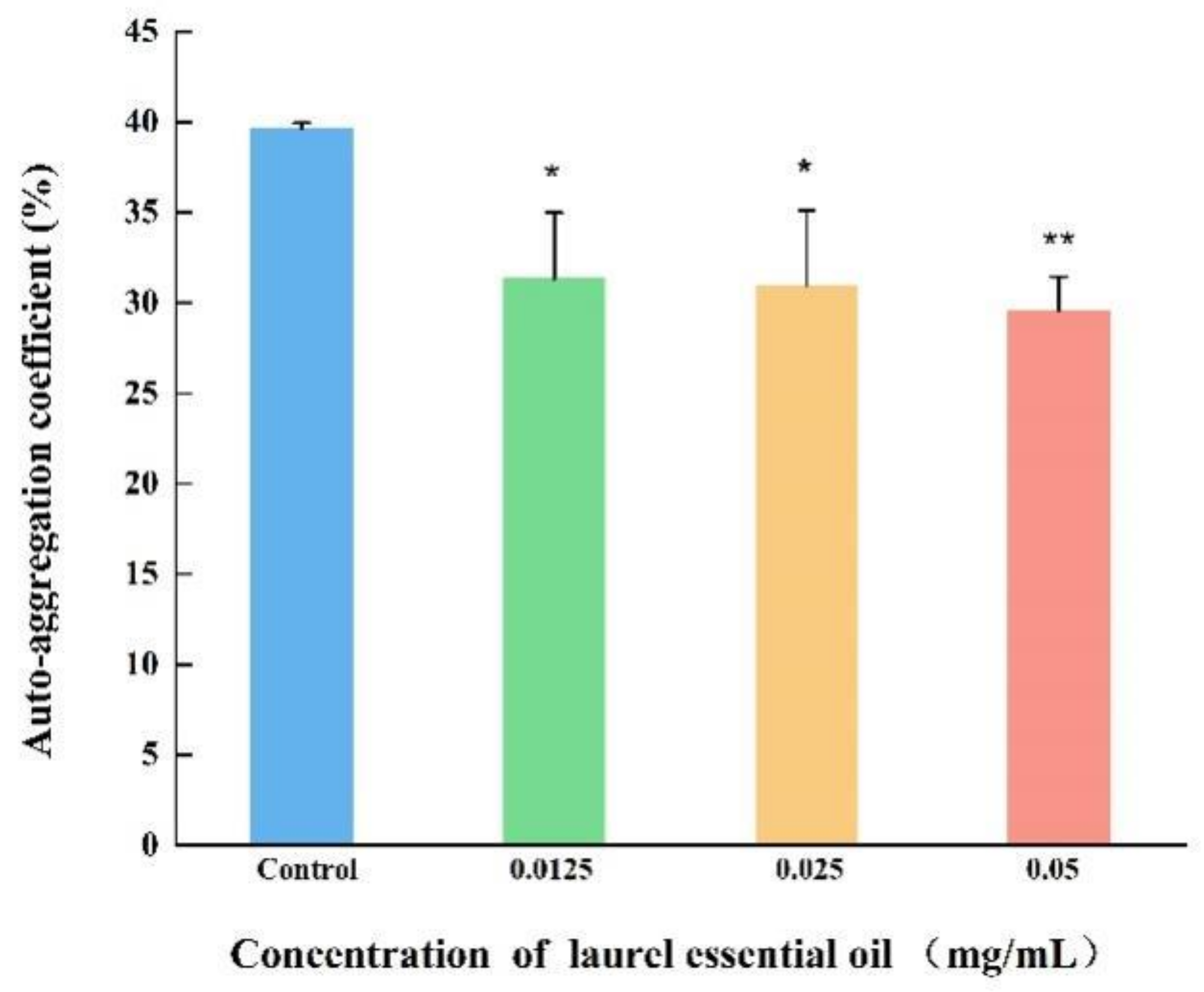
2.10. Auto-Aggregation Abilities Assays
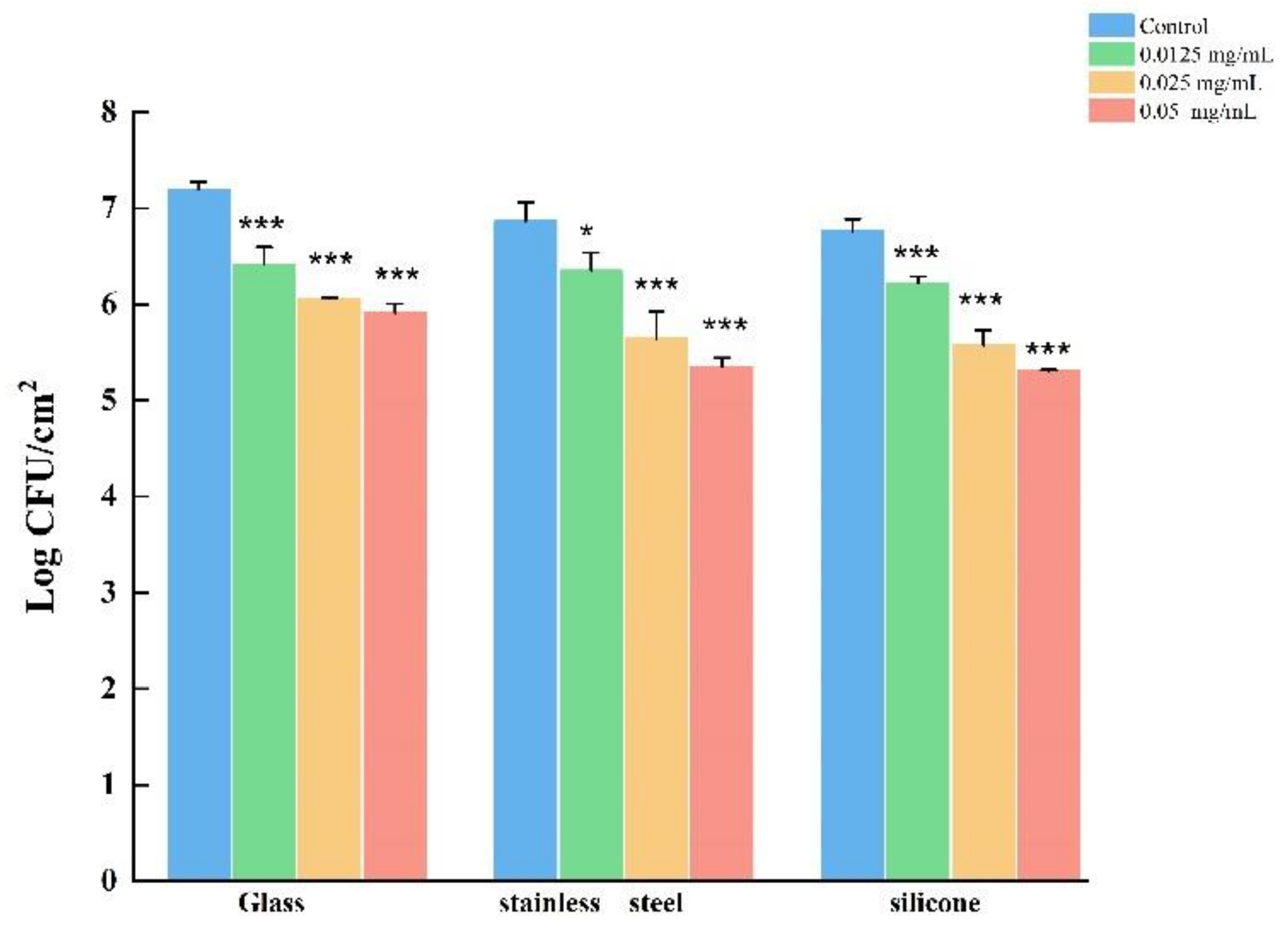
2.11. Biofilm Formation on Stainless-Steel (SS) Coupons, Glass and Food-Grade Silicone
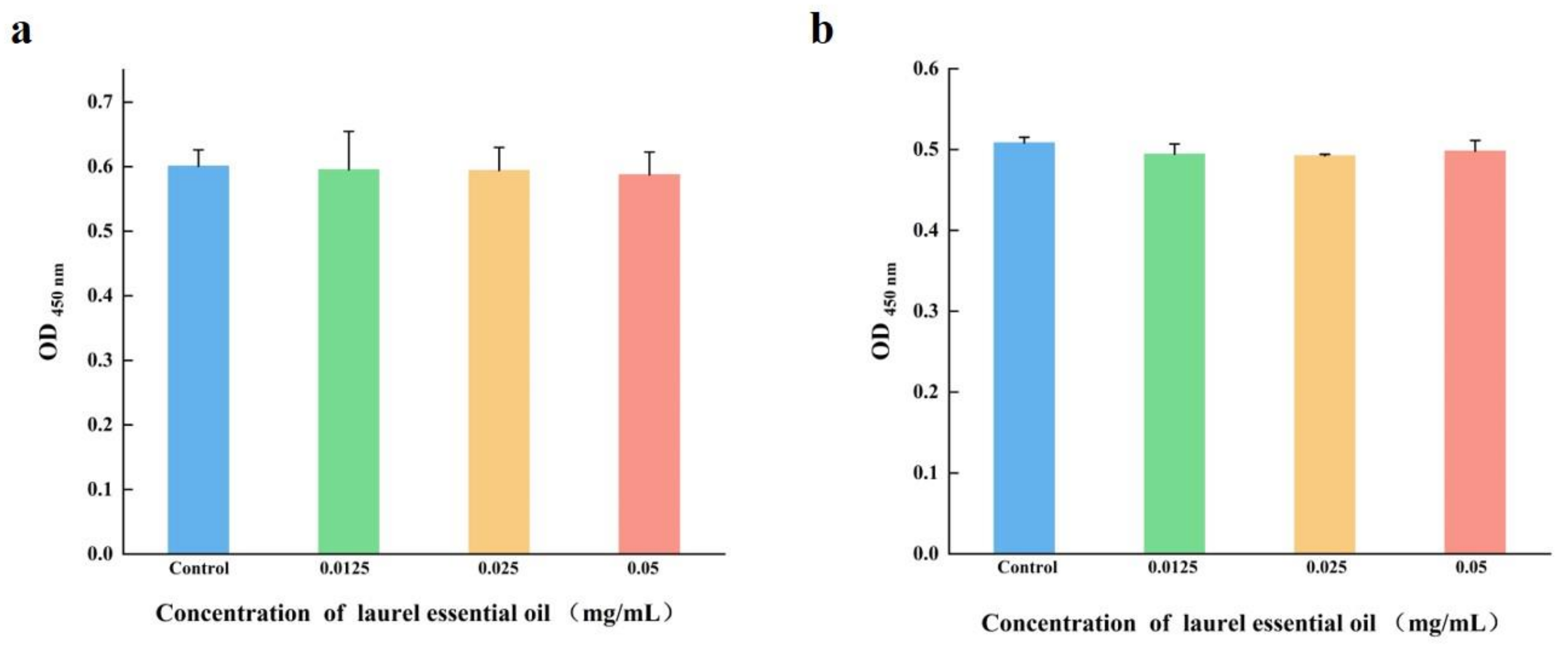
2.12. Cytotoxicity Analysis
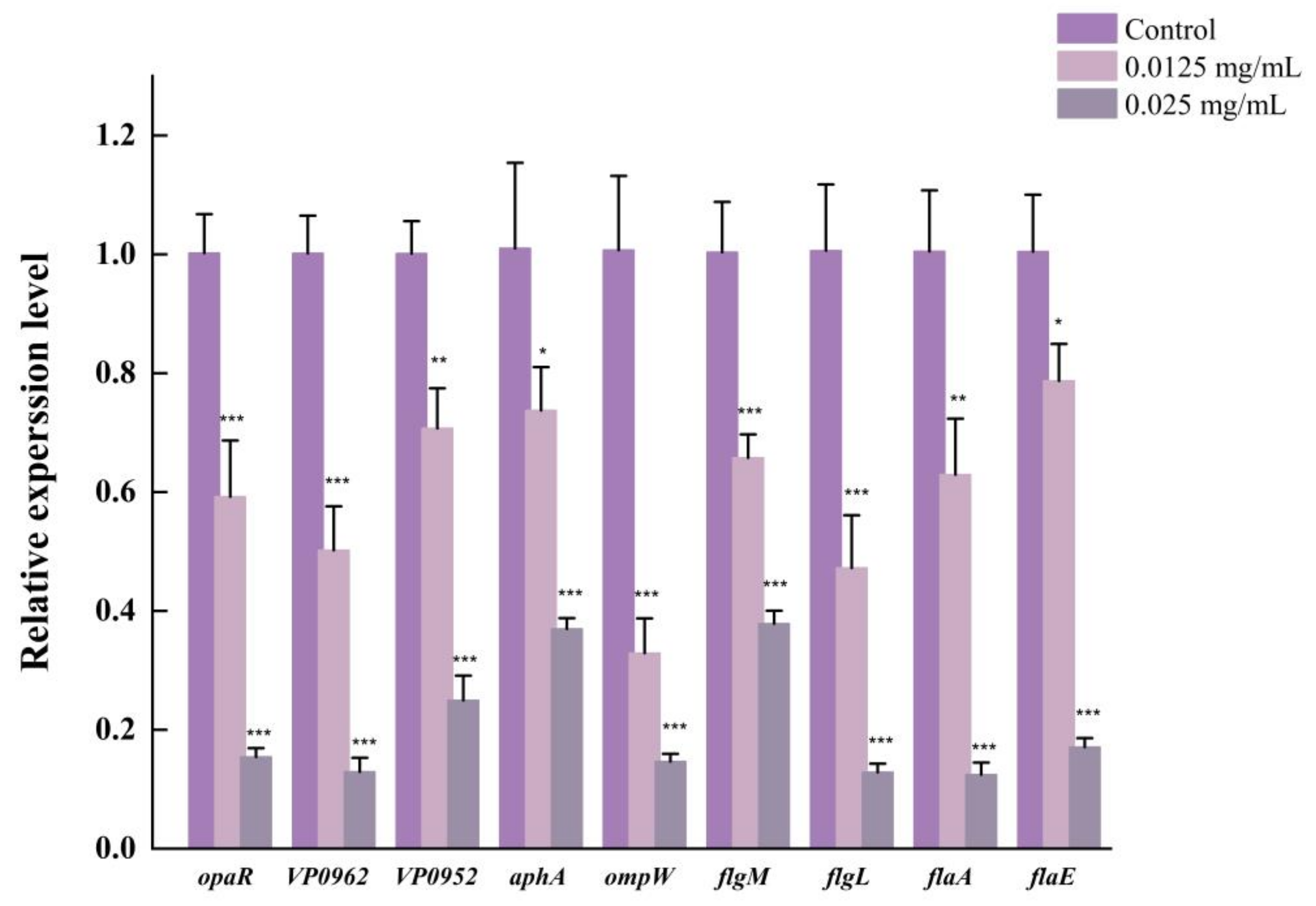
2.13. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Assay
2.14. Statistical Analysis
3. Results
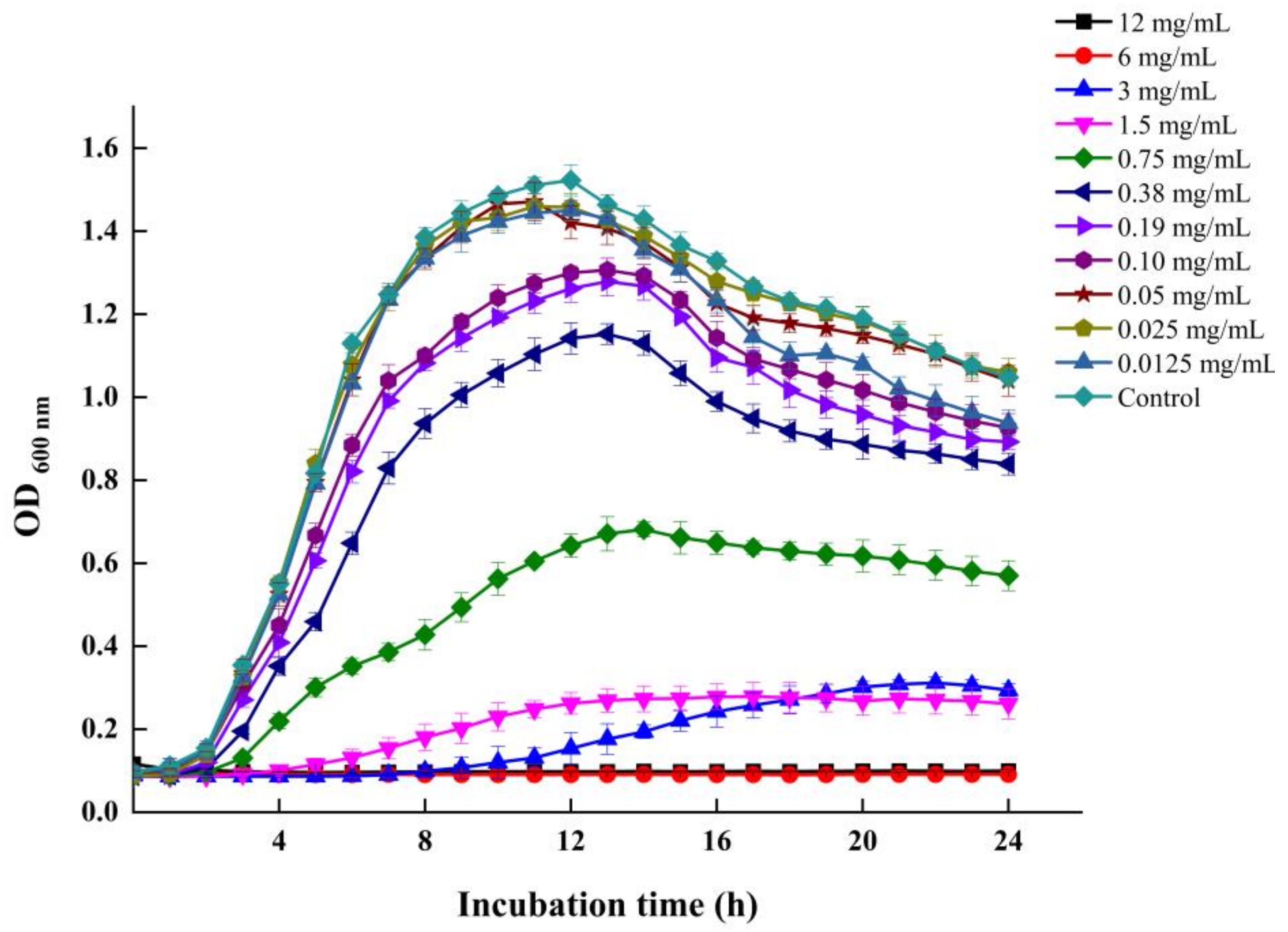
3.1. MICs and SICs of LEO against V. parahaemolyticus
3.2. LEO Inhibited Biofilm Formation on Microtiter Plates
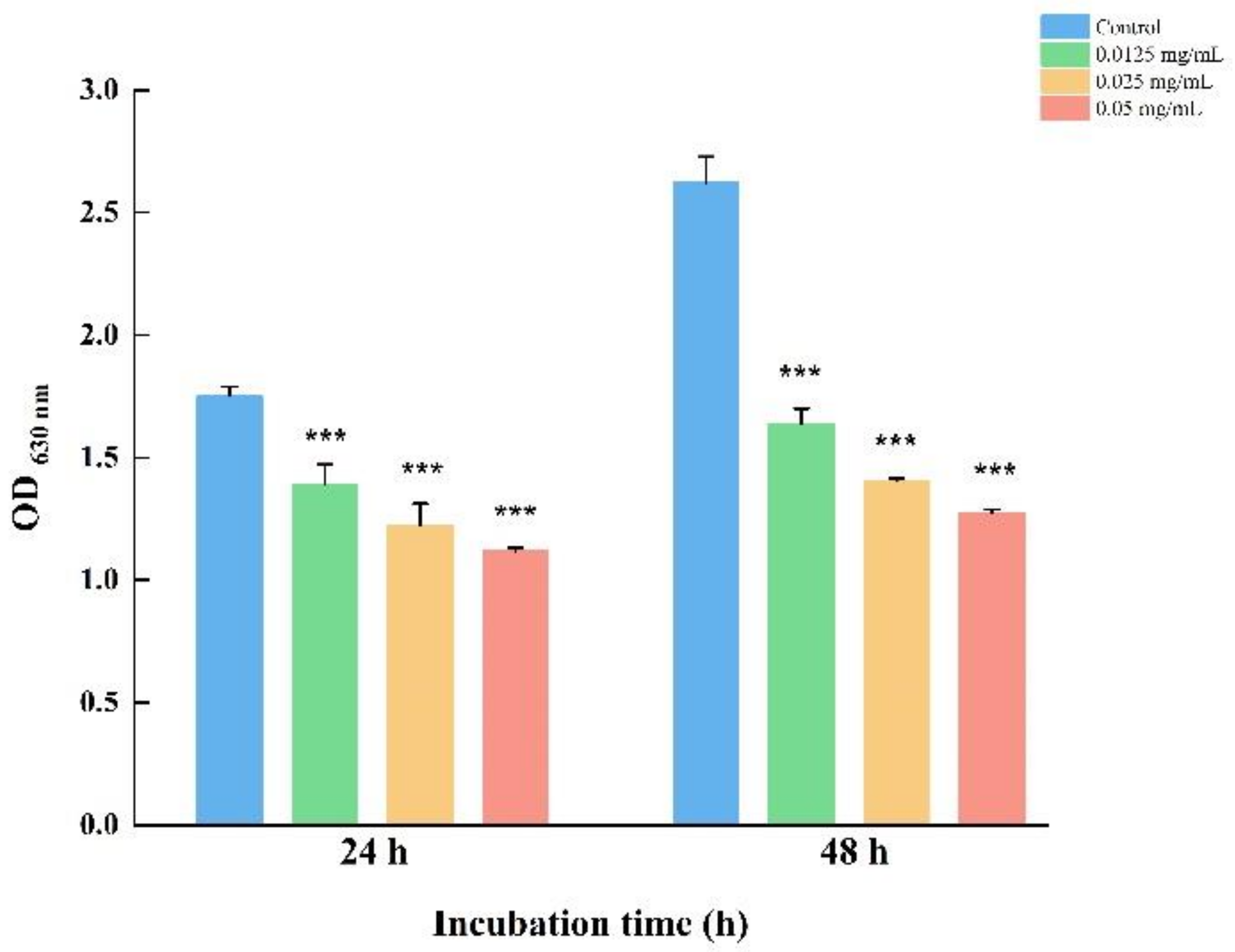
3.3. LEO Decreased Biofilm Metabolic Activity
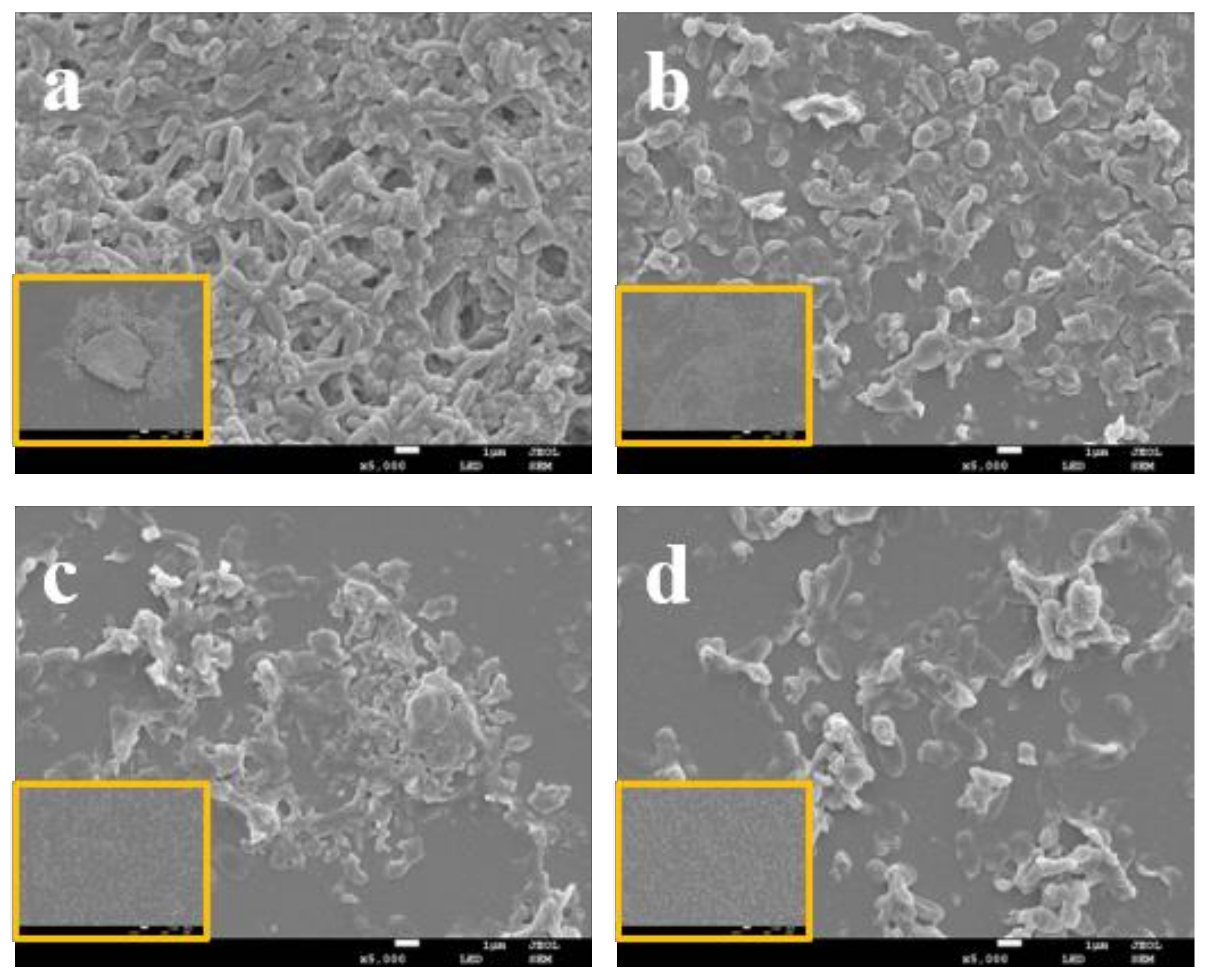
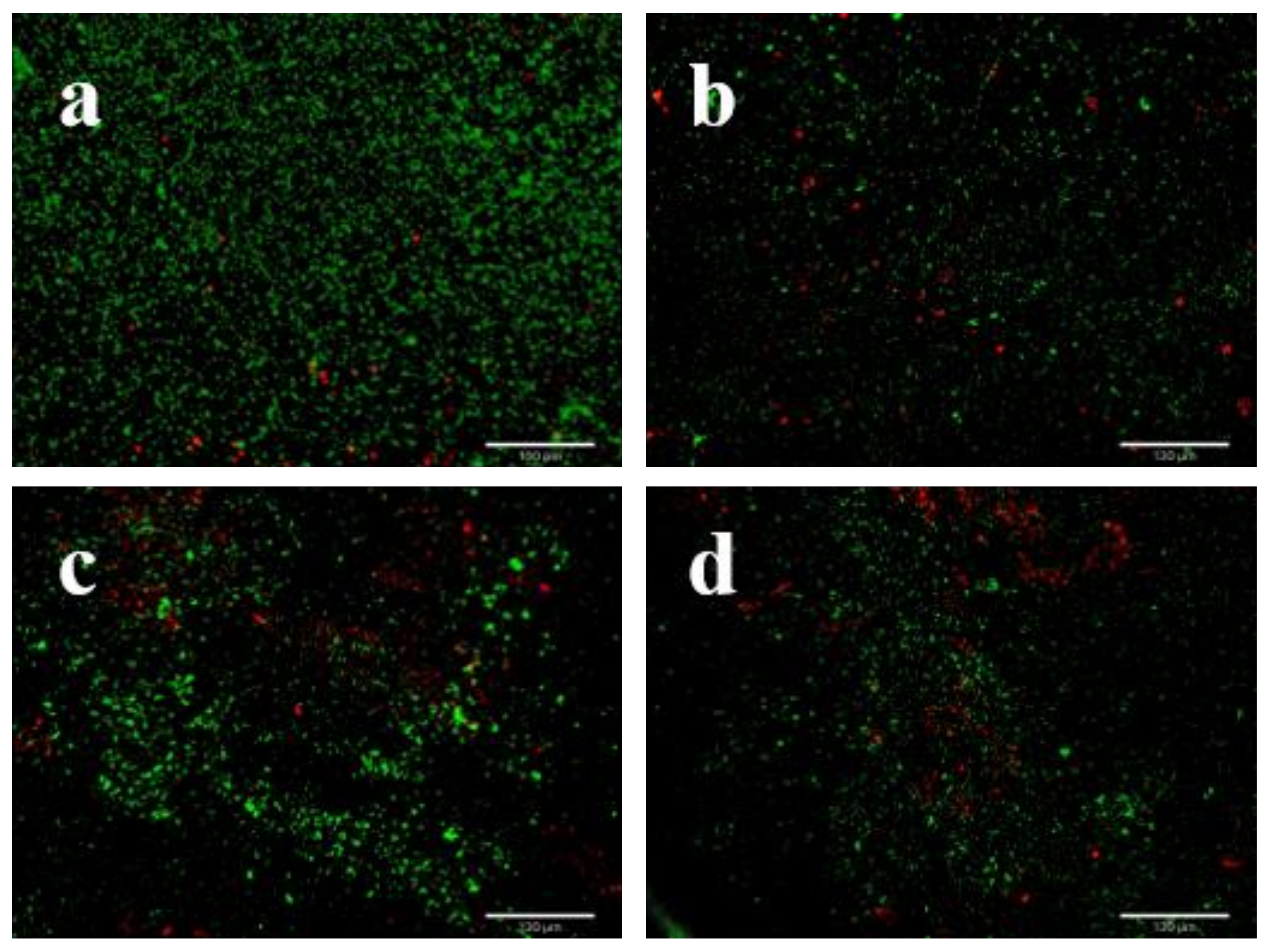
3.4. LEO Impaired V. parahaemolyticus Biofilm Structure
3.5. LEO Inhibited Bacterial Motility
3.6. LEO Modified V. parahaemolyticus Biofilm Composition
3.7. LEO Decreased CSH and Auto-Aggregation Abilities of V. parahaemolyticus
3.8. LEO Reduced Biofilm Cells on Food Contact Surfaces
3.9. LEO Was Non-Cytotoxic to Cells
3.10. LEO Down-Regulated Expression of Genes Related to Biofilm Formation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, J.; Li, X.; Qiu, Y.; Xue, X.; Zhang, M.; Yang, W.; Zhang, Y. Quorum sensing regulates transcription of the pilin gene mshA1 of MSHA pilus in Vibrio parahaemolyticus. Gene 2022, 807, 145961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, L.; Osei-Adjei, G.; Zhang, Y.; Yang, W.; Yin, Z.; Zhou, D. Autoregulation of ToxR and its regulatory actions on major virulence gene loci in Vibrio parahaemolyticus. Front. Cell. Infect. Microbiol. 2018, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Robert-Pillot, A.; Copin, S.; Gay, M.; Malle, P.; Quilici, M.L. Total and pathogenic Vibrio parahaemolyticus in shrimp: Fast and reliable quantification by real-time PCR. Int. J. Food Microbiol. 2010, 143, 190–197. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, Y.; Qiu, Y.; Yang, H.; Yang, W.; Yin, Z.; Xia, P. H-NS is a repressor of major virulence gene loci in Vibrio parahaemolyticus. Front. Microbiol. 2014, 5, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Mizan, M.F.R.; Bang, H.J.; Sadekuzzaman, M.; Lee, N.; Kim, T.J.; Ha, S.D. Molecular characteristics, biofilm-forming abilities, and quorum sensing molecules in Vibrio parahaemolyticus strains isolated from marine and clinical environments in Korea. Biofouling 2017, 33, 369–378. [Google Scholar]
- Mizan, M.F.R.; Ashrafudoulla, M.; Sadekuzzaman, M.; Kang, I.; Ha, S.-D. Effects of NaCl, glucose, and their combinations on biofilm formation on black tiger shrimp (Penaeus monodon) surfaces by Vibrio parahaemolyticus. Food Control. 2018, 89, 203–209. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. J. 2011, 17, 7. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, R.; Qi, X.; Zhou, B.; Wang, J.; Chen, Y.; Zhang, H. Epidemiology of foodborne disease outbreaks caused by Vibrio parahaemolyticus during 2010–2014 in Zhejiang Province, China. Food Control. 2017, 77, 110–115. [Google Scholar] [CrossRef]
- Chen, B.; Huang, J.; Li, H.; Zeng, Q.-H.; Wang, J.J.; Liu, H.; Zhao, Y. Eradication of planktonic Vibrio parahaemolyticus and its sessile biofilm by curcumin-mediated photodynamic inactivation. Food Control. 2020, 113, 107181. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.J.; Qian, H.; Tan, L.; Zhang, Z.; Liu, H.; Zhao, Y. Insights into the role of extracellular DNA and extracellular proteins in biofilm formation of Vibrio parahaemolyticus. Front. Microbiol. 2020, 11, 813–826. [Google Scholar]
- Han, N.; Mizan, M.F.R.; Jahid, I.K.; Ha, S.-D. Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food Control. 2016, 70, 161–166. [Google Scholar]
- Hartmann, I.; Carranza, P.; Lehner, A.; Stephan, R.; Eberl, L.; Riedel, K. Genes involved in Cronobacter sakazakii biofilm formation. Appl. Environ. Microbiol. 2010, 76, 2251–2261. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tan, L.; Guo, L.; Zhang, P.; Malakar, P.K.; Ahmed, F.; Zhao, Y. Acidic electrolyzed water more effectively breaks down mature Vibrio parahaemolyticus biofilm than DNase I. Food Control. 2020, 117, 107312. [Google Scholar]
- Dobrinčić, A.; Brunović, B.; Čošić, Z.; Pelaić, Z.; Repajić, M.; Levaj, B. The influence of laurel essential oil treatment on the shelf-life and sensory characteristics of fresh-cut apples during storage Croatian Journal of Food Technology, Biotechnology and Nutrition. FTB Food Technol. Biotechnol. 2022, 17, 77–84. [Google Scholar]
- Ozogul, Y.; El Abed, N.; Ozogul, F. Antimicrobial effect of laurel essential oil nanoemulsion on food-borne pathogens and fish spoilage bacteria. Food Chem. 2022, 368, 130831. [Google Scholar] [CrossRef] [PubMed]
- Merghni, A.; Marzouki, H.; Hentati, H.; Aouni, M.; Mastouri, M. Antibacterial and antibiofilm activities of Laurus nobilis L. essential oil against Staphylococcus aureus strains associated with oral infections. Curr. Res. Transl. Med. 2016, 64, 29–34. [Google Scholar]
- Caputo, L.; Nazzaro, F.; Souza, L.F.; Aliberti, L.; De Martino, L.; Fratianni, F.; De Feo, V. Laurus nobilis: Composition of essential oil and its biological activities. Molecules 2017, 22, 930–940. [Google Scholar] [PubMed]
- Vilela, J.; Martins, D.; Monteiro-Silva, F.; González-Aguilar, G.; de Almeida, J.M.M.M.; Saraiva, C. Antimicrobial effect of essential oils of Laurus nobilis L. and Rosmarinus officinallis L. on shelf-life of minced “Maronesa” beef stored under different packaging conditions. Food Packag. Shelf Life 2016, 8, 71–80. [Google Scholar]
- Dadalioǧlu, I.; Evrendilek, G.A. Chemical Compositions and Antibacterial Effects of Essential Oils of Turkish Oregano (Origanum minutiflorum), Bay Laurel (Laurus nobilis), Spanish Lavender (Lavandula stoechas L.), and Fennel (Foeniculum vulgare) on Common Foodborne Pathogens. J. Agric. Food Chem. 2004, 52, 8255–8260. [Google Scholar]
- Miao, X.; Liu, H.; Zheng, Y.; Guo, D.; Shi, C.; Xu, Y.; Xia, X. Inhibitory Effect of thymoquinone on Listeria monocytogenes ATCC 19115 biofilm formation and virulence attributes critical for human infection. Front. Cell. Infect. Microbiol. 2019, 9, 304. [Google Scholar]
- Shi, C.; Song, K.; Zhang, X.; Sun, Y.; Sui, Y.; Chen, Y.; Xia, X. Antimicrobial Activity and Possible Mechanism of Action of Citral against Cronobacter sakazakii. PLoS ONE. 2016, 11, e0159006. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Yan, J.; Ding, Z.; Xie, J. The HD-GYP domain protein of Shewanella putrefaciens YZ08 regulates biofilm formation and spoilage activities. Food Res. Int. 2022, 157, 111466. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, W.; Qin, N.; Ren, X.; Xia, X. Propionate and butyrate inhibit biofilm formation of Salmonella typhimurium grown in laboratory media and food models. Foods 2022, 11, 3493. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Gao, J.; Liu, H.; Liu, J.; Jin, T.; Qin, N.; Xia, X. Antibiofilm effect of sodium butyrate against Vibrio parahaemolyticus. Food Control. 2022, 131, 108422. [Google Scholar] [CrossRef]
- Guo, D.; Yang, Z.; Zheng, X.; Kang, S.; Yang, Z.; Xu, Y.; Xia, X. Thymoquinone Inhibits Biofilm Formation and Attachment-Invasion in Host Cells of Vibrio parahaemolyticus. Foodborne Pathog. Dis. 2019, 16, 671–678. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, W.; Cao, Y.; Gao, J.; Jin, T.; Qin, N.; Xia, X. Punicalagin inhibits biofilm formation and virulence gene expression of Vibrio parahaemolyticus. Food Control. 2022, 139, 109045. [Google Scholar] [CrossRef]
- Ling, N.; Zhang, J.; Li, C.; Zeng, H.; He, W.; Ye, Y.; Wu, Q. The glutaredoxin gene, grxB, affects acid tolerance, surface hydrophobicity, auto-aggregation, and biofilm formation in Cronobacter sakazakii. Front. Microbiol. 2018, 9, 133. [Google Scholar] [CrossRef]
- Hossain, M.I.; Mizan, M.F.R.; Ashrafudoulla, M.; Nahar, S.; Joo, H.-J.; Jahid, I.K.; Ha, S.-D. Inhibitory effects of probiotic potential lactic acid bacteria isolated from kimchi against Listeria monocytogenes biofilm on lettuce, stainless-steel surfaces, and MBEC™ biofilm device. LWT 2020, 118, 108864. [Google Scholar] [CrossRef]
- Han, Q.; Song, X.; Zhang, Z.; Fu, J.; Wang, X.; Malakar, P.K.; Zhao, Y. Removal of foodborne pathogen biofilms by acidic electrolyzed water. Front. Microbiol. 2017, 8, 988. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Shakeel, M.; Majeed, M.I.; Nawaz, H.; Rashid, N.; Ali, A.; Haque, A.; Saleem, M. Surface-enhanced Raman spectroscopy for the characterization of pellets of biofilm forming bacterial strains of Staphylococcus epidermidis. Photodiagn. Photodyn. Ther. 2022, 40, 103145. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Hosaka, S.; Kanematsu, H.; Yoshitake, M. Marine biofilm model comprising a loop-Type biofilm reactor and a halomonas strain HIG FST4 1, an active biofilm-forming Bacterium. Coatings 2022, 12, 1605. [Google Scholar] [CrossRef]
- Lei, T.; Jiang, F.; He, M.; Zhang, J.; Zeng, H.; Chen, M.; Wu, Q. Prevalence, virulence, antimicrobial resistance, and molecular characterization of fluoroquinolone resistance of Vibrio parahaemolyticus from different types of food samples in China. Int. J. Food Microbiol. 2020, 317, 108461. [Google Scholar] [CrossRef]
- Wang, D.; Fletcher, G.C.; Gagic, D.; On, S.L.W.; Palmer, J.S.; Flint, S.H. Comparative genome identification of accessory genes associated with strong biofilm formation in Vibrio parahaemolyticus. Food Res. Int. 2023, 166, 112605. [Google Scholar] [CrossRef]
- Mohammadzadeh, M.; Mahmoudi, R.; Ghajarbeygi, P. Evaluation of Chemical Composition and Antibacterial Properties of Froriepia subpinnta Essential Oils from Guilan Region: Before and After Flowering. J. Essent. Oil Bear. Plants 2018, 21, 1119–1127. [Google Scholar] [CrossRef]
- Pilar Santamarina, M.; Roselló, J.; Giménez, S.; Amparo Blázquez, M. Commercial Laurus nobilis L. and Syzygium aromaticum L. Merr. & Perry essential oils against post-harvest phytopathogenic fungi on rice. LWT Food Sci. Technol. 2016, 65, 325–332. [Google Scholar]
- Santhakumari, S.; Nilofernisha, N.M.; Ponraj, J.G.; Pandian, S.K.; Ravi, A.V. In vitro and in vivo exploration of palmitic acid from Synechococcus elongatus as an antibiofilm agent on the survival of Artemia franciscana against virulent vibrios. J. Invertebr. Pathol. 2017, 150, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Gao, Z.; Li, G.; Fu, F.; Liang, Z.; Zhu, H.; Shan, Y. Antimicrobial and antibiofilm efficacy and mechanism of essential oil from Citrus Changshan-huyou Y. B. chang against Listeria monocytogenes. Food Control. 2019, 105, 256–264. [Google Scholar] [CrossRef]
- Bai, J.-R.; Zhong, K.; Wu, Y.-P.; Elena, G.; Gao, H. Antibiofilm activity of shikimic acid against Staphylococcus aureus. Food Control. 2018, 95, 327–333. [Google Scholar] [CrossRef]
- Zheng, X.; Guo, J.; Rao, H.; Guo, D.; Huang, Y.; Xu, Y.; Shi, C. Antibacterial and antibiofilm activity of coenzyme Q0 against Vibrio parahaemolyticus. Food Control. 2020, 109, 106955. [Google Scholar]
- Liu, F.; Sun, Z.; Wang, F.; Liu, Y.; Zhu, Y.; Du, L.; Xu, W. Inhibition of biofilm formation and exopolysaccharide synthesis of Enterococcus faecalis by phenyllactic acid. Food Microbiol. 2020, 86, 103344. [Google Scholar]
- Cao, J.; Liu, H.; Wang, Y.; He, X.; Jiang, H.; Yao, J.; Chen, X. Antimicrobial and antivirulence efficacies of citral against foodborne pathogen Vibrio parahaemolyticus RIMD2210633. Food Control. 2021, 120, 107507. [Google Scholar]
- Yildiz, F.H.; Visick, K.L. Vibrio biofilms: So much the same yet so different. Trends Microbiol. 2009, 17, 109–118. [Google Scholar] [PubMed]
- Merino, L.; Trejo, F.M.; De Antoni, G.; Golowczyc, M.A. Lactobacillus strains inhibit biofilm formation of Salmonella sp. isolates from poultry. Food Res. Int. 2019, 123, 258–265. [Google Scholar] [PubMed]
- Jacobs, A.; Chenia, H.Y. Biofilm-forming capacity, surface hydrophobicity and aggregation characteristics of Myroides odoratus isolated from South African Oreochromis mossambicus fish. J. Appl. Microbiol. 2009, 107, 1957–1966. [Google Scholar] [CrossRef] [PubMed]


| Target Gene | Sequence of Primers (5′–3′) |
|---|---|
| 16S rRNA | F: GCCTTCGGGAACTCTGAGACAG |
| R: GCTCGTTGCGGGACTTAACCCAA | |
| opaR | F: AGGGCATCGTTACCCAATC |
| R: TAAGTCAACATAGTCCGCATC | |
| VP0962 | F: GACCAAGACCCAGTGAGA |
| R: GGTAAAGCCAGCAAAGTT | |
| VP0952 | F: TATGATGGTGTTTGGTGC |
| R: TGTTTTTCTGAGCGTTTC | |
| aphA | F: ACACCCAACCGTTCGTGATG |
| R: GTTGAAGGCGTTGCGTAGTAAG | |
| ompW | F: TCGTGTCACCAAGTGTTTTCG |
| R: CGTGGCTGAATGGTGTTGC | |
| flgM | F: AGAAATGAAATCGCTACCAGT |
| R: GGAATTGCATAATTGCCTTA | |
| flgL | F: CGTCAGCGTCCACCACTT |
| R: GCGGCTCTGACTTACTGCTA | |
| flaA | F: CGGACTAAACCGTATCGCTGAAA |
| R: GGCTGCCCATAGAAAGCATTACA | |
| flaE | F: ACAGTGCGGATAGCCAGTA |
| R: CTTTGAGTAGCGTCTCGTTT |
| Strain | Origin | MICs(mg/mL) |
|---|---|---|
| ATCC 17802 | Shirasu food poisoning | 6 |
| RIMD 2210633Sm | Pandemic strain | 6 |
| VP 13 | Seafood | 3 |
| VP 48 | Seafood | 12 |
| VP 55 | Seafood | 6 |
| VP 481 | Seafood | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Liu, J.; Zou, Y.; Li, S.; Zhao, D.; Wang, H.; Xia, X. Anti-Biofilm Activity of Laurel Essential Oil against Vibrio parahaemolyticus. Foods 2023, 12, 3658. https://doi.org/10.3390/foods12193658
Zhu W, Liu J, Zou Y, Li S, Zhao D, Wang H, Xia X. Anti-Biofilm Activity of Laurel Essential Oil against Vibrio parahaemolyticus. Foods. 2023; 12(19):3658. https://doi.org/10.3390/foods12193658
Chicago/Turabian StyleZhu, Wenxiu, Jiaxiu Liu, Yue Zou, Shugang Li, Dongyun Zhao, Haisong Wang, and Xiaodong Xia. 2023. "Anti-Biofilm Activity of Laurel Essential Oil against Vibrio parahaemolyticus" Foods 12, no. 19: 3658. https://doi.org/10.3390/foods12193658
APA StyleZhu, W., Liu, J., Zou, Y., Li, S., Zhao, D., Wang, H., & Xia, X. (2023). Anti-Biofilm Activity of Laurel Essential Oil against Vibrio parahaemolyticus. Foods, 12(19), 3658. https://doi.org/10.3390/foods12193658







