A Novel RAA Combined Test Strip Method Based on Dual Gene Targets for Pathogenic Vibrio vulnificus in Aquatic Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Culture and Extraction of Genomic DNA
2.2. Design and Synthesis of the Primers for RAA
2.3. Preparation of the MBGNP Probe
2.4. TS-DTL Preparation and Detection Procedure
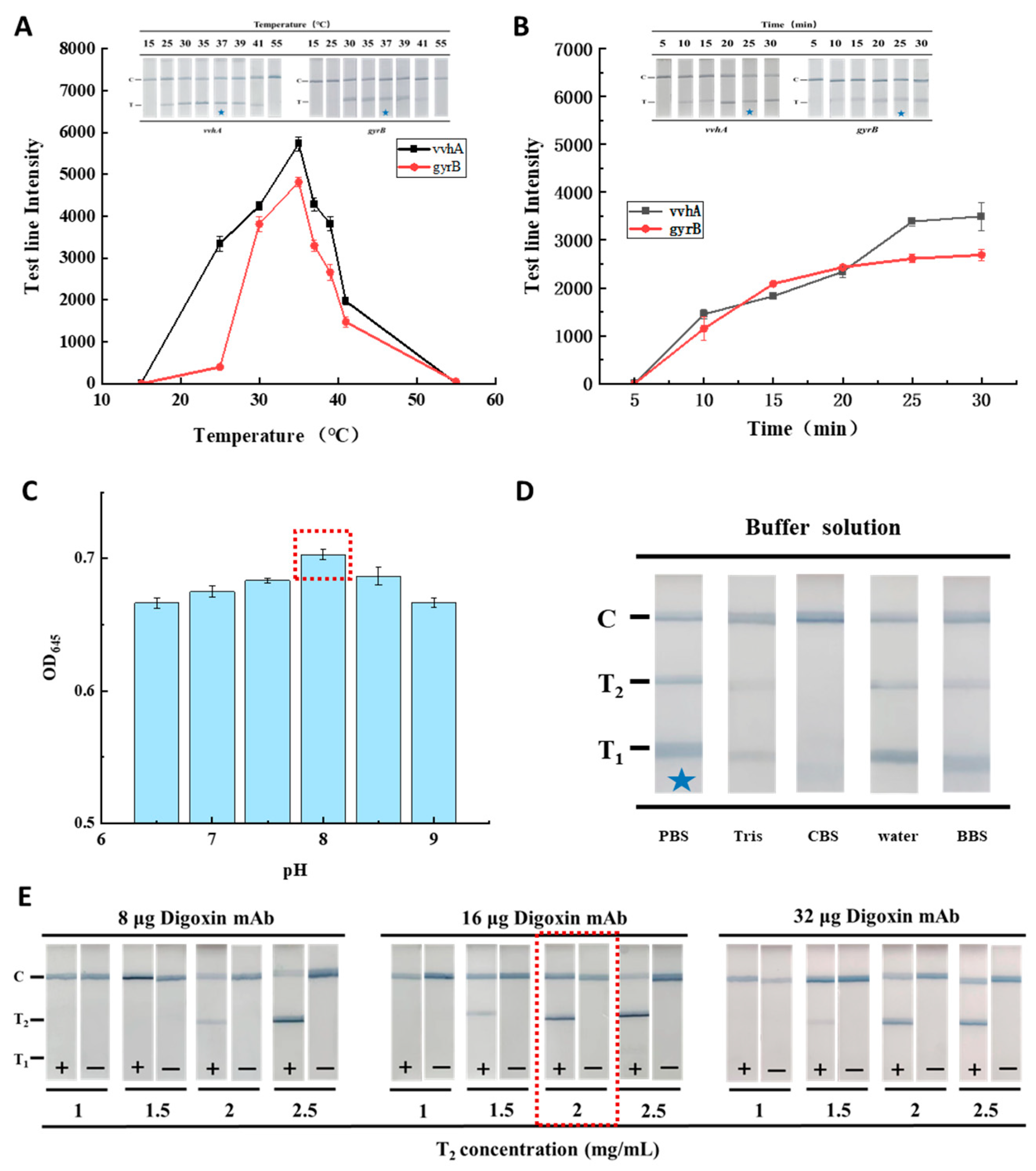
2.5. Optimization of the Reaction Conditions of RAA-TS-DTL
2.6. Specificity of RAA-TS-DTL for V. vulnificus
2.7. Sensitivity of RAA-TS-DTL for V. vulnificus in Oyster
2.8. Accuracy Evaluation of RAA-TS-DTL
3. Results
3.1. Principle of RAA-TS-DTL for the Detection of V. vulnificus
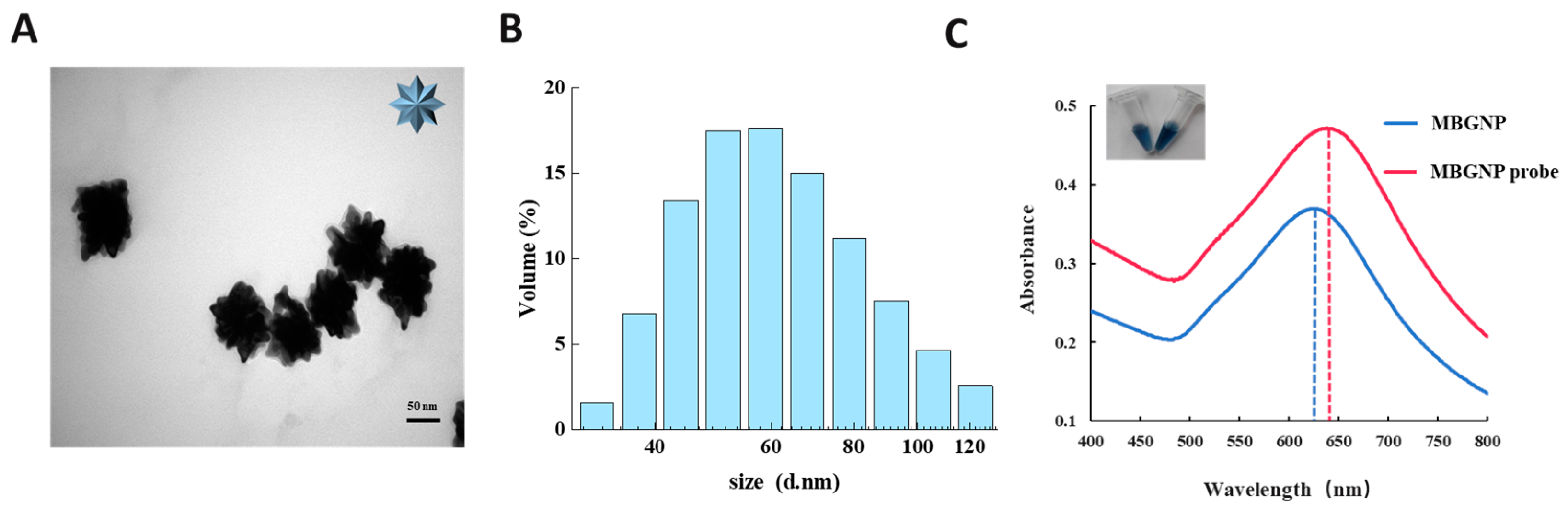
3.2. Characterization of the MBGNP and MBGNP Probe
3.3. Screening of Optimal Primer Pairs for RAA
3.4. Optimization of the RAA-TS-DTL System
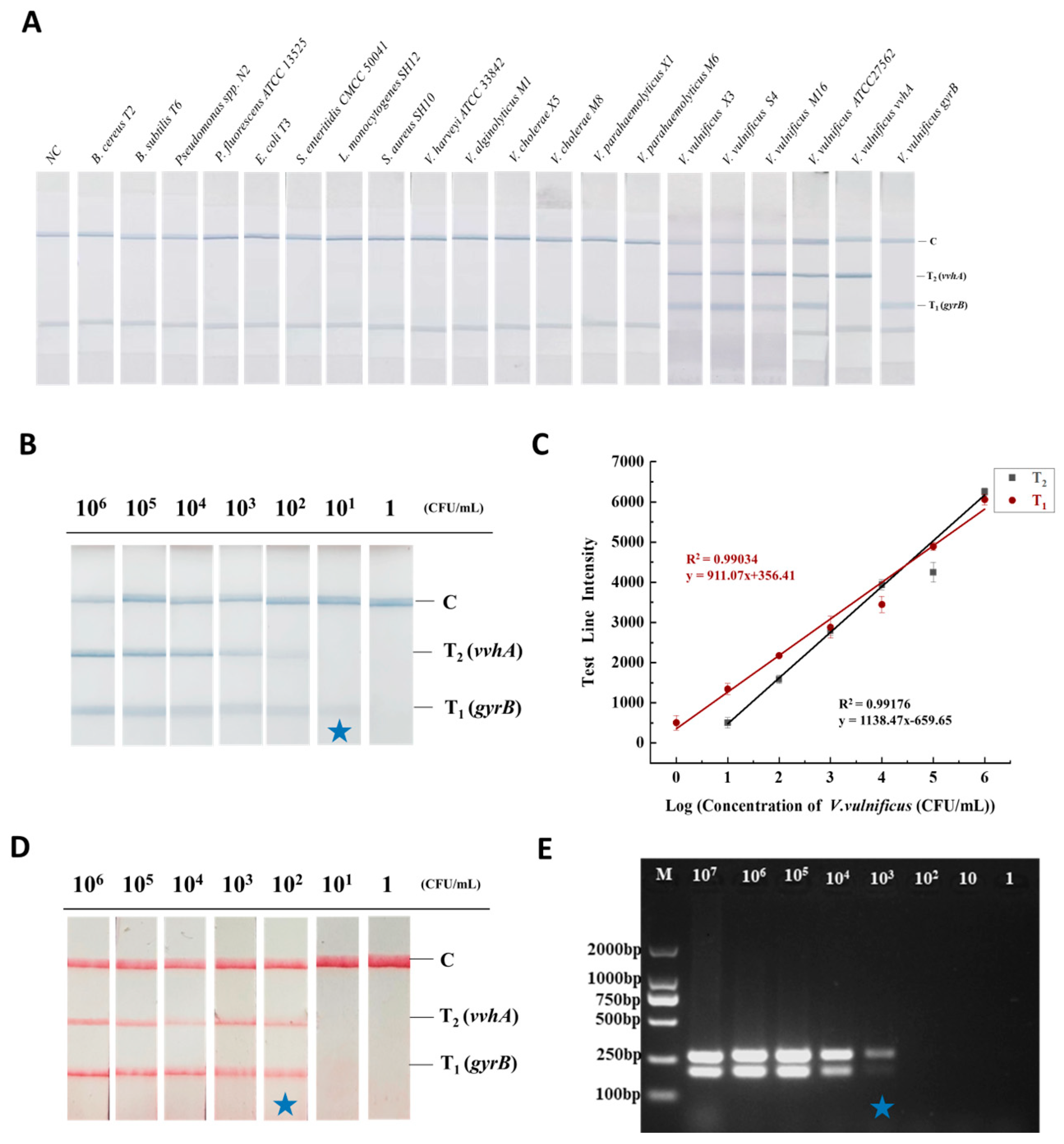
3.5. Detection Performance of the RAA-TS-DTL System
3.5.1. Specificity
3.5.2. Sensitivity of RAA-TS-DTL for V. vulnificus in Oyster
3.5.3. Accuracy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Baker-Austin, C.; Stockley, L.; Rangdale, R.; Martinez-Urtaza, J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective. Environ. Microbiol. Rep. 2010, 2, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D. The Biology of Vibrio vulnificus. Microbiol. Spectr. 2015, 3, 349. [Google Scholar] [CrossRef]
- Jones, M.K.; Oliver, J.D. Vibrio vulnificus: Disease and Pathogenesis. Infect. Immun. 2009, 77, 1723–1733. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D. Vibrio vulnificus: New insights into a deadly opportunistic pathogen. Environ. Microbiol. 2018, 20, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Archer, E.J.; Baker-Austin, C.; Osborn, T.J.; Jones, N.R.; Martinez-Urtaza, J.; Trinanes, J.; Oliver, J.D.; Gonzalez, F.J.C.; Lake, I.R. Climate warming and increasing Vibrio vulnificus infections in North America. Sci. Rep. 2023, 13, 3893. [Google Scholar] [CrossRef] [PubMed]
- Wangman, P.; Surasilp, T.; Pengsuk, C.; Sithigorngul, P.; Longyant, S. Development of a species-specific monoclonal antibody for rapid detection and identification of foodborne pathogen Vibrio vulnificus. J. Food Saf. 2021, 41, e12939. [Google Scholar] [CrossRef]
- Jadeja, R.; Janes, M.E.; Simonson, J.G. Development of rapid and sensitive antiflagellar monoclonal antibody based lateral flow device for the detection of Vibrio vulnificus from oyster homogenate. Food Control 2015, 56, 110–113. [Google Scholar] [CrossRef]
- D’Souza, C.; Kumar, B.K.; Rai, P.; Deekshit, V.K.; Karunasagar, I. Application of gyrB targeted SYBR green based qPCR assay for the specific and rapid detection of Vibrio vulnificus in seafood. J. Microbiol. Methods 2019, 166, 105747. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-D.; Xu, Y.-G.; Qiu, S.-P.; Wang, Y.; Gao, H.-J.; Gao, S.-Y. Establishment of a quantitative real-time PCR for detecting vvhA gene in Vibrio vulnificus. Food Mach. 2016, 31–70. [Google Scholar]
- Tian, Z.; Yang, L.; Qi, X.; Zheng, Q.; Shang, D.; Cao, J. Visual LAMP method for the detection of Vibrio vulnificus in aquatic products and environmental water. BMC Microbiol. 2022, 22, 256. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, P.; Dong, Y.; Chen, S.; Shen, H.; Jiang, G.; Zhu, H.; Dong, J.; Gao, S. An isothermal recombinase polymerase amplification and lateral flow strip combined method for rapid on-site detection of Vibrio vulnificus in raw seafood. Food Microbiol 2021, 98, 103664. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Cai, Y.; Zhu, L.; Wang, H.; Lu, Y. Rapid and highly sensitive detection of Toxigenic Vibrio cholerae based on recombinase-aided amplification combining with lateral flow assay. Food Anal. Methods 2021, 14, 687–696. [Google Scholar] [CrossRef]
- Xiao, X.; Lin, Z.; Huang, X.; Lu, J.; Zhou, Y.; Zheng, L.; Lou, Y. Rapid and Sensitive Detection of Vibrio vulnificus Using CRISPR/Cas12a Combined with a Recombinase-Aided Amplification Assay. Front. Microbiol. 2021, 12, 767315. [Google Scholar] [CrossRef]
- Lu, Y.-K.; Xu, D.; Liu, W.-Y.; Xie, J.; Lu, Y. A Rapid Tricolour Immunochromatographic Assay for Simultaneous Detection of Tricaine and Malachite Green. Biosensors 2022, 12, 456. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Li, J.; Huang, X.; Duan, H.; Ji, Y.; Xiong, Y. Effect of the tip length of multi-branched AuNFs on the detection performance of immunochromatographic assays. Anal. Methods 2016, 8, 3316–3324. [Google Scholar] [CrossRef]
- Sun, R.; Chen, J.; Wang, Y.; Zhang, Z.; Li, Y.; Li, F.; Ma, C.; Han, Q.; Shi, Y. Rapid, specific and sensitive detection of Vibrio parahaemolyticus in seafood by accelerated strand exchange amplification. Anal. Methods 2023, 15, 655–662. [Google Scholar] [CrossRef]
- Pedrosa de Macena, L.d.G.; de Oliveira Pereira, J.S.; da Silva, J.C.; Ferreira, F.C.; Maranhao, A.G.; Lanzarini, N.M.; Miagostovich, M.P. Quantification of infectious Human mastadenovirus in environmental matrices using PMAxx-qPCR. Braz. J. Microbiol. 2022, 53, 1465–1471. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Xue, C.-h.; Liu, Q. Changes in microbial flora of Pacific oysters (Crassostrea gigas) during refrigerated storage and its shelf-life extension by chitosan. Int. J. Food Microbiol. 2009, 131, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Froelich, B.A.; Noble, R.T. Factors Affecting the Uptake and Retention of Vibrio vulnificus in Oysters. Appl. Environ. Microbiol. 2014, 80, 7454–7459. [Google Scholar] [CrossRef]
- Bonnin-Jusserand, M.; Copin, S.; Le Bris, C.; Brauge, T.; Gay, M.; Brisabois, A.; Grard, T.; Midelet-Bourdin, G. Vibrio species involved in seafood-borne outbreaks (Vibrio cholerae, V-parahaemolyticus and V-vulnificus): Review of microbiological versus recent molecular detection methods in seafood products. Crit. Rev. Food Sci. Nutr. 2019, 59, 597–610. [Google Scholar] [CrossRef]
- Park, S.B.; Chang, S.K.C. Development of Recombinase Polymerase Amplification Combined with Lateral Flow Dipstick Assay to Detect Hemolysin Gene of Vibrio vulnificus in Oysters. J. Food Prot. 2022, 85, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Fu, Y.; Zhang, S.; Pan, Z.; Xia, J.; Zhu, P.; Guo, J. An Assay Combining Droplet Digital PCR With Propidium Monoazide Treatment for the Accurate Detection of Live Cells of Vibrio vulnificus in Plasma Samples. Front. Microbiol. 2022, 13, 927285. [Google Scholar] [CrossRef] [PubMed]
- Perez Roig, A.; Carmona-Salido, H.; Sanjuan, E.; Fouz, B.; Amaro, C. A multiplex PCR for the detection of Vibrio vulnificus hazardous to human and/or animal health from seafood. Int. J. Food Microbiol. 2022, 377, 109778. [Google Scholar] [CrossRef]
- Chun-Hua, R.; Chao-Qun, H.; Peng, L.; Qing-Bai, W. Sensitive and rapid identification of Vibrio vulnificus by loop-mediated isothermal amplification. Microbiol. Res. 2009, 164, 514–521. [Google Scholar]
- Zhang, L.; Wang, M.; Cong, D.; Ding, S.; Cong, R.; Yue, J.; Geng, J.; Hu, C. Rapid, specific and sensitive detection of Vibrio vulnificus by loop-mediated isothermal amplification targeted to vvhA gene. Acta Oceanol. Sin. 2018, 37, 83–88. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, C.; Yu, C.; Si, Z.; Huang, D.; Shen, P.; Fang, M.; Xu, Z. Integration of a CRISPR Cas12a-assisted multicolor biosensor and a micropipette tip enables visible point-of-care testing of foodborne Vibrio vulnificus. Analyst 2023, 148, 3509–3517. [Google Scholar] [CrossRef]
- Yayun, J.; Chaochuan, Z.; Ming, J.; Ruolan, Z.; Qiaoli, W.; Fuyuan, H.; Yongliang, L.; Laibao, Z. An ultrasensitive colorimetric foodborne pathogenic detection method using a CRISPR/Cas12A mediated strand displacement/hybridization chain reaction. J. Agric. Food Chem. 2023, 71, 4193–4200. [Google Scholar] [CrossRef]
- Lantz, P.G.; Hahn-Haegerdal, B.; Radstroem, P. Sample preparation methods in PCR-based detection of food pathogens. Trends Food Sci. Technol. 1994, 5, 384–389. [Google Scholar] [CrossRef]
- Panicker, G.; Bej, A.K. Real-time PCR detection of Vibrio vulnificus in oysters: Comparison of oligonucleotide primers and probes targeting vvhA. Appl. Environ. Microbiol. 2005, 71, 5702–5709. [Google Scholar] [CrossRef]


| Number | Bacterial Strains | Species and Strain | Source |
|---|---|---|---|
| 1 | Vibrio vulnificus | Vibrio vulnificus ATCC27562 | GDMCC |
| 2 | Vibrio vulnificus X3 | Isolated from shrimp | |
| 3 | Vibrio vulnificus S4 | Isolated from scallop | |
| 4 | Vibrio vulnificus M16 | Isolated from oysters | |
| 5 | Other Vibrio spp. | Vibrio parahaemolyticus M6 | Isolated from oysters |
| 6 | Vibrio parahaemolyticus X1 | Isolated from shrimp | |
| 7 | Vibrio cholerae M8 | Isolated from oysters | |
| 8 | Vibrio cholerae X5 | Isolated from shrimp | |
| 9 | Vibrio alginolyticus M1 | Isolated from oysters | |
| 10 | Vibrio harveyi ATCC 33842 | GDMCC | |
| 11 | Other bacterial strains | Staphylococcus aureus SH10 | Preserved in our laboratory |
| 12 | Listeria monocytogenes SH12 | ||
| 13 | Salmonella enteritidis CMCC 50041 | CMCC | |
| 14 | Escherichia coli T3 | Isolated from aquaculture water | |
| 15 | Pseudomonas fluorescens ATCC 13525 | GDMCC | |
| 16 | Pseudomonas spp. N2 | Isolated from milk | |
| 17 | Bacillus subtilis T6 | Isolated from aquaculture water | |
| 18 | Bacillus cereus T2 | Isolated from aquaculture water |
| Assay | Number | Name | Sequence (5′–3′) and Modification | Amplicon Size (bp) |
|---|---|---|---|---|
| Basic RAA(vvhA) | 1 | F1 | GATACTTACGGTTACTCCATCGGTATTAAC | 300 |
| R1 | GATTGGGTTGAACTTCGTCTTATCAAATAC | |||
| 2 | F2 | GCGGAAGTGAACAAAGACGGCCCGAAAGT | 215 | |
| R2 | CAGTGAGCGGCGGTGAAATAGCATCCAAGC | |||
| 3 | F3 | GAAGTCAGTGGTCATTTACAACTACTC | 205 | |
| R3 | CGTCATAGTTCGGTTTGAAGTTGGAATAAGAG | |||
| 4 | F4 | ACTTACATTGGCCCATTCGCCAGCAGTTAT | 278 | |
| R4 | GATGAGCGGTTGTTGATGCGATAGTCTTTT | |||
| 5 | F5 | CTCATTTACTTACAACTACTCGAAAACCTTG | 238 | |
| R5 | ATAGTTCGGTTTGAAGTTGGAATAAGAGATTG | |||
| RAA-TS-DTL(vvhA) | F2-FAM | FAM-GCGGAAGTGAACAAAGACGGCCCGAAAGTG | 215 | |
| R2-Dig | Digoxin-CAGTGAGCGGCGGTGAAATAGCATCCAAGC | |||
| PCR | P-F1 | TTCCAACTTCAAACCGAACTATGA | 205 | |
| P-R1 | ATTCCAGTCGATGCGAATACGTTG |
| Assay | Number | Name | Sequence (5′–3′) and Modification | Amplicon Size (bp) |
|---|---|---|---|---|
| Basic RAA (gyrB) | 6 | F6 | CCGTAAGAACCAAGCAATCCTACCGCTAAA | 140 |
| R6 | TGTACTCGTCACGACCGATACCACAACCTA | |||
| 7 | F7 | ACAGCTACATGGACAAAGAAGGCTACTCGA | 157 | |
| R7 | TTCACTTCACTAGAAACCAGTTTGTCTTTA | |||
| 8 | F8 | GAAACCTTCACCAACATCGAATTTCATTAT | 426 | |
| R8 | CAGTGAGCGGCGGTGAAATAGCATCCAAGC | |||
| 9 | F9 | GAAACCTTCACCAACATCGAATTTCATTAT | 138 | |
| R9 | TTCATACATGAAGTGATCTTTCTTATCTTCTT | |||
| 10 | F10 | GCCAAACCAAAGACAAACTGGTTTCTAGTG | 360 | |
| R10 | CTACGTTTAGAATCTTACCTTTTAGCGGTAGG | |||
| RAA-TS-DTL (gyrB) | F10- Bio | Biotin-GCCAAACTAAAGACAAACTGGTTTCTAGTG | 360 | |
| R10-Dig | Digoxin-CTACGTTTAGAATCTTACCTTTTAGCGGTAGG | |||
| PCR | P-F2 | GTCCGCAGTGGAATCCTTCA | 285 | |
| P-R2 | TGGTTCTTACGGTTACGGCC |
| Sample Type | Number of Spiked Samples | Number of Negative Samples | Number of Samples Detected (+/−) | ||
|---|---|---|---|---|---|
| Traditional Culture | PCR-AGE | RAA-TS-DTL | |||
| Fish | 18 | 2 | 18/2 | 16/2 | 17/2 |
| Shrimp | 25 | 5 | 24/5 | 23/5 | 24/5 |
| Oyster | 7 | 3 | 6/3 | 6/3 | 6/3 |
| Method | Target Gene | Sensitivity | Sample | References |
|---|---|---|---|---|
| ICTS | / | 10 CFU/mL | oyster | [7] |
| qPCR | gyrB | 100 CFU/mL | clam meat | [8] |
| Multiple PCR | vvhA | 10 CFU /mL | cultured shrimps | [23] |
| ddPCR | vvhA | 15.4 CFU/mL | plasma | [22] |
| LAMP | gyrB | 10 fg/μL | culture solution | [10] |
| RPA-LFD | vvhA | 30 CFU/mL | oyster | [21] |
| RAA-CRISPR | vvhA | 20 CFU/mL | Seafood | [26] |
| RAA-TS-DTL | vvhA gyrB | 23 CFU/mL 6 CFU/mL | oyster | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Zhang, G.; Xu, D.; Ye, J.; Lu, Y. A Novel RAA Combined Test Strip Method Based on Dual Gene Targets for Pathogenic Vibrio vulnificus in Aquatic Products. Foods 2023, 12, 3605. https://doi.org/10.3390/foods12193605
Liu W, Zhang G, Xu D, Ye J, Lu Y. A Novel RAA Combined Test Strip Method Based on Dual Gene Targets for Pathogenic Vibrio vulnificus in Aquatic Products. Foods. 2023; 12(19):3605. https://doi.org/10.3390/foods12193605
Chicago/Turabian StyleLiu, Wenyue, Guangying Zhang, Di Xu, Jingqin Ye, and Ying Lu. 2023. "A Novel RAA Combined Test Strip Method Based on Dual Gene Targets for Pathogenic Vibrio vulnificus in Aquatic Products" Foods 12, no. 19: 3605. https://doi.org/10.3390/foods12193605
APA StyleLiu, W., Zhang, G., Xu, D., Ye, J., & Lu, Y. (2023). A Novel RAA Combined Test Strip Method Based on Dual Gene Targets for Pathogenic Vibrio vulnificus in Aquatic Products. Foods, 12(19), 3605. https://doi.org/10.3390/foods12193605








