Bacterial and Fungal Communities Are Specifically Modulated by the Cocoa Bean Fermentation Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Fermentation Process and Sample Collection
2.2. DNA Extraction
2.3. DNA Amplification
2.4. Sequence Data Preparation
2.5. PICRUST Analysis
3. Results and Discussion
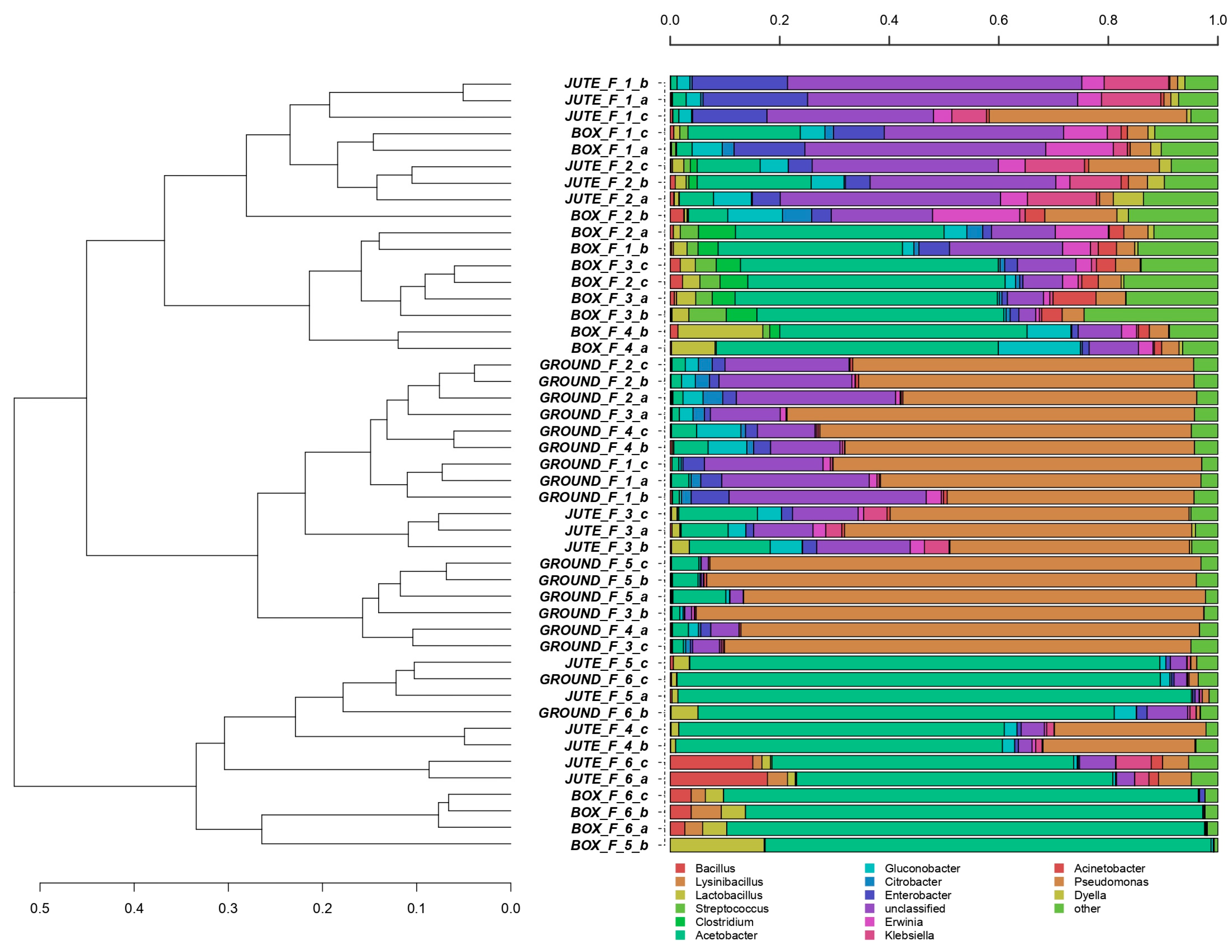
3.1. Overview of the Bacterial and Fungal Communities Involved in Different Fermentation Methods
3.2. Effects of the Method and the Days of Fermentation on the Abundance of OTUs
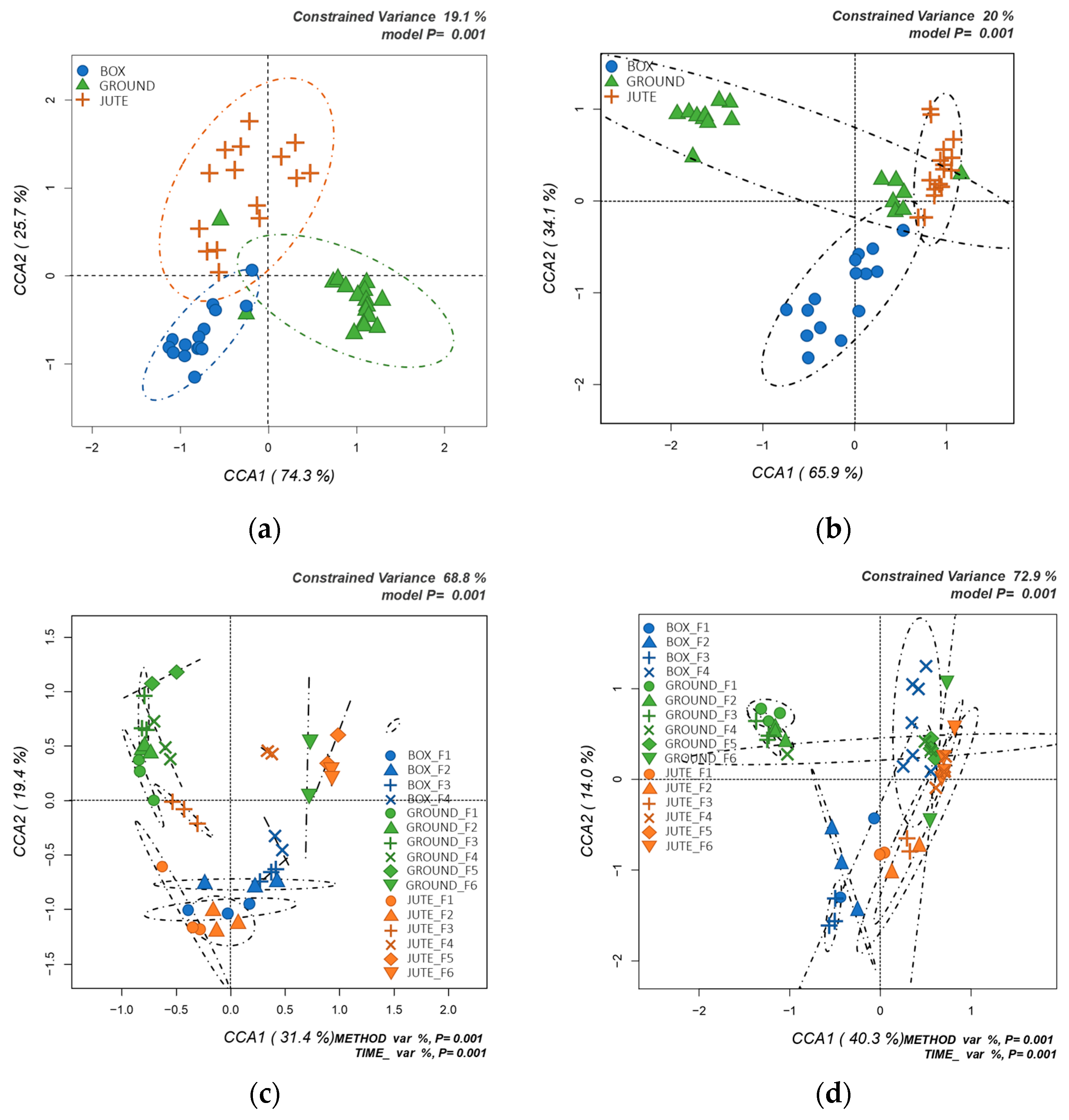
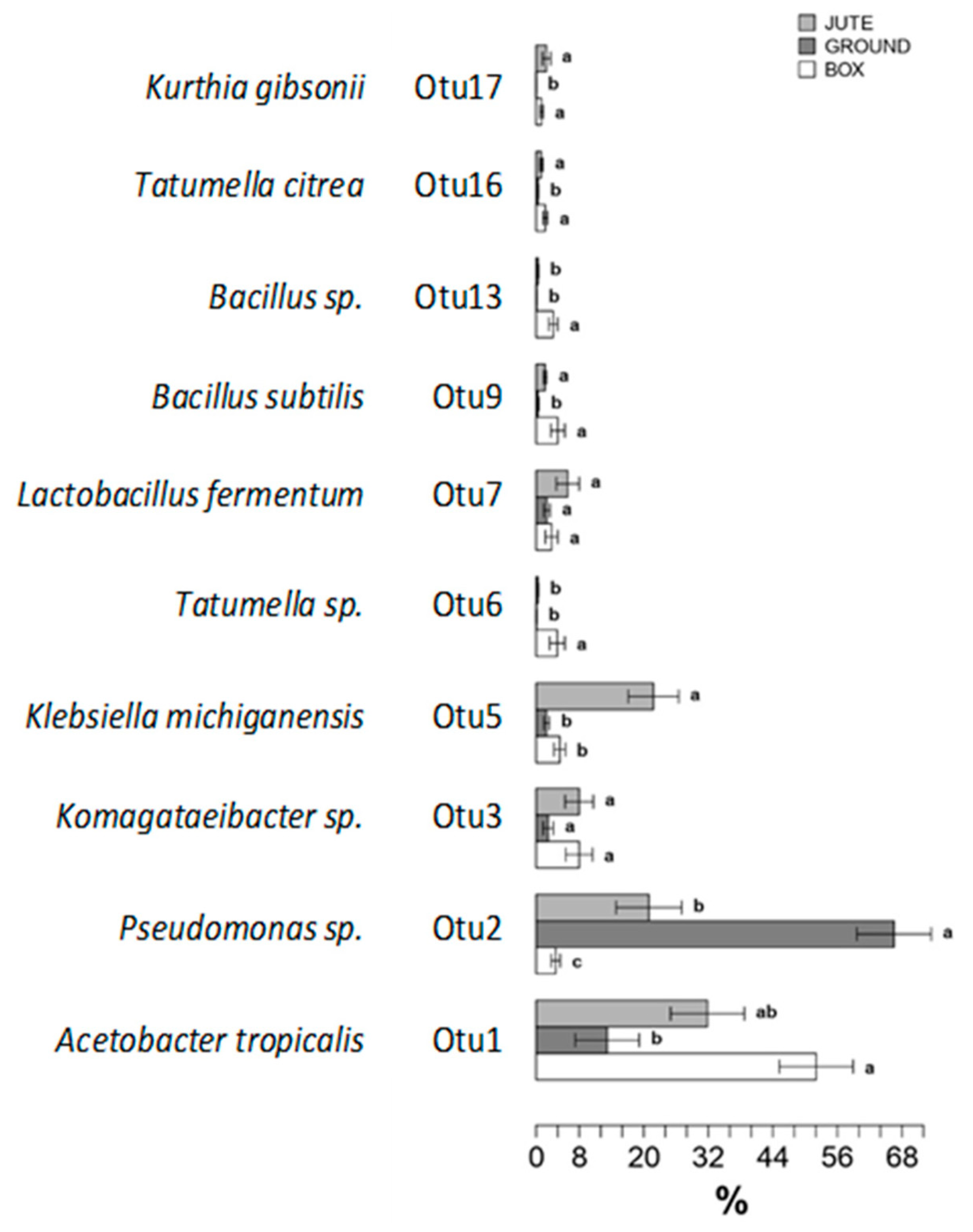
3.3. Influence of the Fermentation Method on the Bacterial Communities
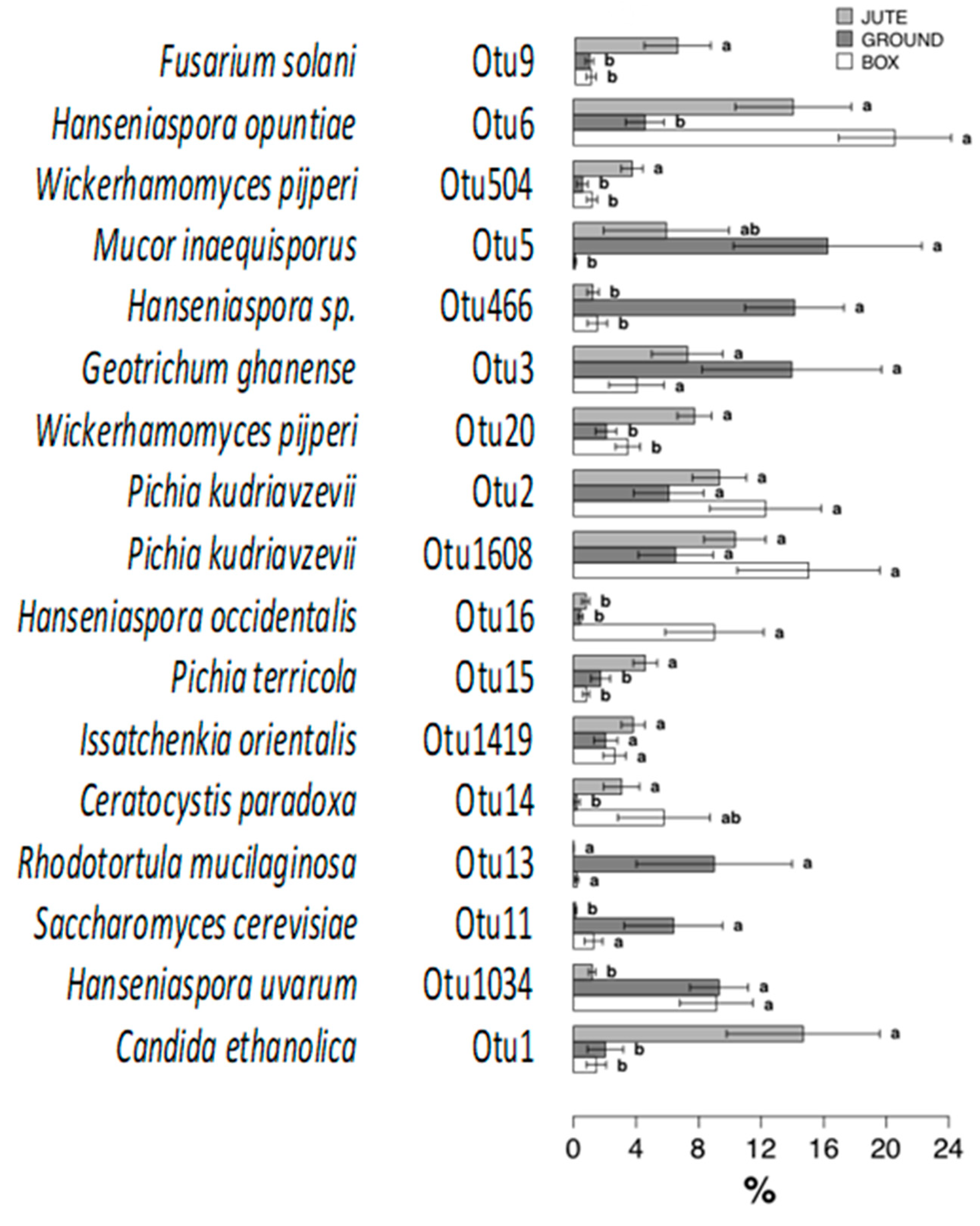
3.4. Influence of the Fermentation Method on the Fungal Communities
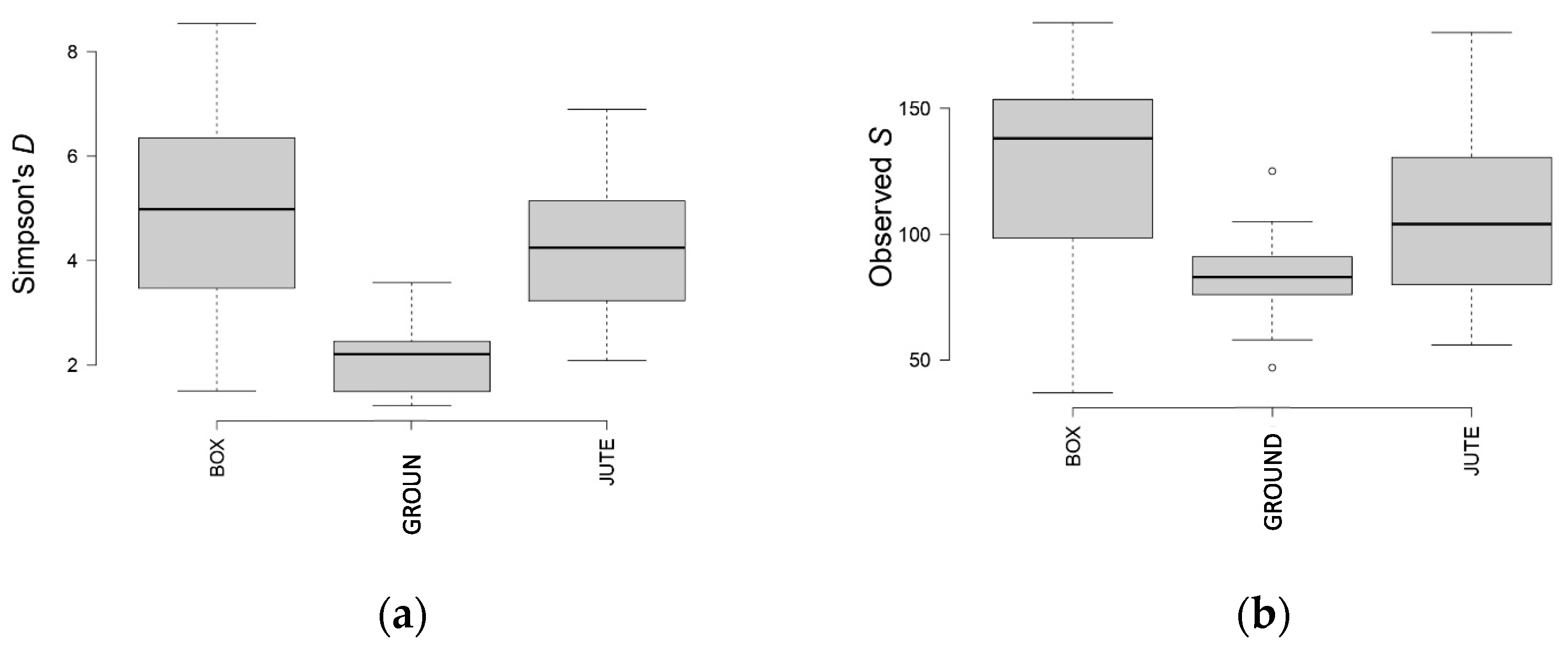
3.5. Biodiversity Indexes
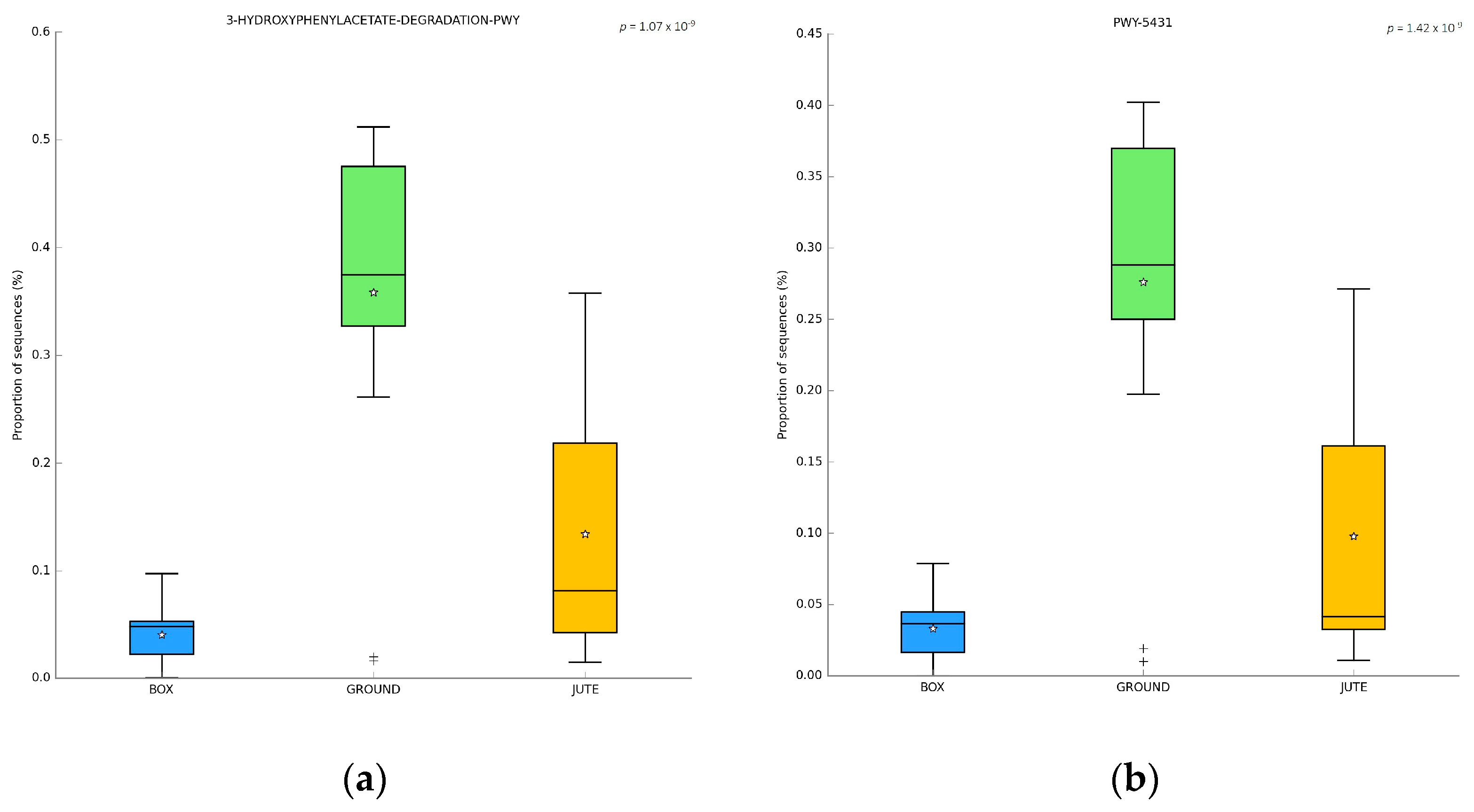
3.6. PICRUST Analysis
3.7. Evaluation of the Three Fermentation Methods
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lefeber, T.; Janssens, M.; Moens, F.; Gobert, W.; de Vuyst, L. Interesting starter culture strains for controlled cocoa bean fermentation revealed by simulated cocoa pulp fermentations of cocoa-specific lactic acid bacteria. Appl. Environ. Microbiol. 2011, 77, 6694–6698. [Google Scholar] [CrossRef]
- Cempaka, L.; Aliwarga, L.; Purwo, S.; Kresnowati, M.T.A.P. Dynamics of cocoa bean pulp degradation during cocoa bean fermentation: Effects of yeast starter culture addition. J. Math. Fundam. Sci. 2014, 46, 14–25. [Google Scholar] [CrossRef]
- De Vuyst, L.; Weckx, S. The cocoa bean fermentation process: From ecosystem analysis to starter culture development. J. Appl. Microbiol. 2016, 121, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Ardhana, M.M.; Fleet, G.H. The microbial ecology of cocoa bean fermentations in Indonesia. Int. J. Food Microbiol. 2003, 86, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Hernández, C.; Mota-Gutierrez, J.; Ferrocino, I.; Hernández-Estrada, Z.J.; González-Ríos, O.; Cocolin, L.; Suárez-Quiroz, M.L. The challenges and perspectives of the selection of starter cultures for fermented cocoa beans. Int. J. Food Microbiol. 2019, 301, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Lefeber, T.; Janssens, M.; Camu, N.; de Vuyst, L. Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media toward development of a starter culture for cocoa bean fermentation. Appl. Environ. Microbiol. 2010, 76, 7708–7716. [Google Scholar] [CrossRef]
- Afoakwa, E.O. Chocolate Science and Technology; John Wiley & Sons: New York, NY, USA, 2016. [Google Scholar]
- Guehi, T.S.; Dadie, A.T.; Koffi, K.P.; Dabonne, S.; Ban-Koffi, L.; Kedjebo, K.D.; Nemlin, G.J. Performance of different fermentation methods and the effect of their duration on the quality of raw cocoa beans. Int. J. Food Sci. Technol. 2010, 45, 2508–2514. [Google Scholar] [CrossRef]
- Streule, S.; Leischtfeld, S.F.; Galler, M.; Schwenninger, S.M. Monitoring of cocoa post-harvest process practices on a small-farm level at five locations in Ecuador. Heliyon 2022, 8, e09628. [Google Scholar] [CrossRef]
- Nielsen, D.S.; Teniola, O.D.; Ban-Koffi, L.; Owusu, M.; Andersson, T.S.; Holzapfel, W.H. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int. J. Food Microbiol. 2007, 114, 168–186. [Google Scholar] [CrossRef]
- Papalexandratou, Z.; Vrancken, G.; de Bruyne, K.; Vandamme, P.; de Vuyst, L. Spontaneous organic cocoa bean box fermentations in Brazil are characterized by a restricted species diversity of lactic acid bacteria and acetic acid bacteria. Food Microbiol. 2011, 28, 1326–1338. [Google Scholar] [CrossRef]
- Camu, N.; De Winter, T.; Verbrugghe, K.; Cleenwerck, I.; Vandamme, P.; Takrama, J.S.; Vancanneyt, M.; De Vuyst, L. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl. Environ. Microbiol. 2007, 73, 1809–1824. [Google Scholar] [CrossRef] [PubMed]
- Daniel, H.M.; Vrancken, G.; Takrama, J.F.; Camu, N.; De Vos, P.; De Vuyst, L. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res. 2009, 9, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Meersman, E.; Steensels, J.; Mathawan, M.; Wittocx, P.-J.; Saels, V.; Struyf, N.; Bernaert, H.; Vrancken, G.; Verstrepen, K.J. Detailed analysis of the microbial population in Malaysian spontaneous cocoa pulp fermentations reveals a core and variable microbiota. PLoS ONE 2013, 8, e81559. [Google Scholar] [CrossRef] [PubMed]
- Visintin, S.; Alessandria, V.; Valente, A.; Dolci, P.; Cocolin, L. Molecular identification and physiological characterization of yeasts, lactic acid bacteria and acetic acid bacteria isolated from heap and box cocoa bean fermentations in West Africa. Int. J. Food Microbiol. 2016, 216, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Papalexandratou, Z.; Camu, N.; Falony, G.; De Vuyst, L. Comparison of the bacterial species diversity of spontaneous cocoa bean fermentations carried out at selected farms in Ivory Coast and Brazil. Food Microbiol. 2011, 28, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Bortolini, C.; Patrone, V.; Puglisi, E.; Morelli, L. Detailed analyses of the bacterial populations in processed cocoa beans of different geographic origin, subject to varied fermentation conditions. Int. J. Food Microbiol. 2016, 236, 98–106. [Google Scholar] [CrossRef]
- Vasileiadis, S.; Puglisi, E.; Trevisan, M.; Scheckel, K.G.; Langdon, K.A.; McLaughlin, M.J.; Lombi, E.; Donner, E. Changes in soil bacterial communities and diversity in response to long-term silver exposure. FEMS Microbiol. Ecol. 2015, 91, fiv114. [Google Scholar] [CrossRef]
- Bruns, T.D.; Lee, S.B.; Taylor, J.W.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics Hydraulic Lift and Common Mycorrhizal Networks View Project Evolution of Gene Expression View Project. 1990. Available online: https://www.researchgate.net/publication/223397588 (accessed on 20 January 2023).
- Berry, D.; Schawb, C.; Milinovich, G.; Reichert, J.; Mahfoudh, B.K.; Decker, T.; Engel, M.; Hai, B.; Hainzl, E.; Heider, S.; et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012, 6, 2091–2106. [Google Scholar] [CrossRef]
- Berry, D.; Mahfoudh, K.B.; Wagner, M.; Loy, A. Barcoded Primers Used in Multiplex Amplicon Pyrosequencing Bias Amplification. Appl. Environ. Microbiol. 2012, 78, 612. [Google Scholar] [CrossRef]
- Puglisi, E.; Romaniello, F.; Galletti, S.; Boccaleri, E.; Frache, A.; Cocconcelli, P.S. Selective bacterial colonization processes on polyethylene waste samples in an abandoned landfill site. Sci. Rep. 2019, 9, 14138. [Google Scholar] [CrossRef]
- Bartram, A.K.; Lynch, M.D.J.; Stearns, J.C.; Moreno-Hagelsieb, G.; Neufeld, J.D. Generation of Multimillion-Sequence 16S rRNA Gene Libraries from Complex Microbial Communities by Assembling Paired-End Illumina Reads. Appl. Environ. Microbiol. 2011, 77, 3846–3852. [Google Scholar] [CrossRef] [PubMed]
- Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; Sahl, J.W.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Papalexandratou, Z.; Falony, G.; Romanens, E.; Jimenez, J.C.; Amores, F.; Daniel, H.-M.; De Vuyst, L. Species Diversity, Community Dynamics, and Metabolite Kinetics of the Microbiota Associated with Traditional Ecuadorian Spontaneous Cocoa Bean Fermentations. Appl. Environ. Microbiol. 2011, 77, 7698–7714. [Google Scholar] [CrossRef]
- Hamdouche, Y.; Guehi, T.; Durand, N.; Kedjebo, K.B.D.; Montet, D.; Meile, J.C. Dynamics of microbial ecology during cocoa fermentation and drying: Towards the identification of molecular markers. Food Control 2015, 48, 117–122. [Google Scholar] [CrossRef]
- Camu, N.; De Winter, T.; Addo, S.K.; Takrama, J.S.; Bernaert, H.; De Vuyst, L. Fermentation of cocoa beans: Influence of microbial activities and polyphenol concentrations on the flavour of chocolate. J. Sci. Food Agric. 2008, 88, 2288–2297. [Google Scholar] [CrossRef]
- Agyirifo, D.S.; Wamalwa, M.; Otwe, E.P.; Galyuon, I.; Runo, S.; Takrama, J.; Ngeranwa, J. Metagenomics analysis of cocoa bean fermentation microbiome identifying species diversity and putative functional capabilities. Heliyon 2019, 5, e02170. [Google Scholar] [CrossRef]
- Serra, J.L.; Moura, F.G.; Pereira, G.V.D.M.; Soccol, C.R.; Rogez, H.; Darnet, S. Determination of the microbial community in Amazonian cocoa bean fermentation by Illumina-based metagenomic sequencing. LWT 2019, 106, 229–239. [Google Scholar] [CrossRef]
- Schwan, R.F.; Wheals, A.E. The Microbiology of Cocoa Fermentation and its Role in Chocolate Quality. Crit. Rev. Food Sci. Nutr. 2004, 44, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Rocha, F.; Cala, M.P.; Mejía, J.L.A.; Rodríguez-López, C.M.; Chica, M.J.; Olarte, H.H.; Fernández-Niño, M.; Barrios, A.F.G. Dissecting fine-flavor cocoa bean fermentation through metabolomics analysis to break down the current metabolic paradigm. Sci. Rep. 2021, 11, 21904. [Google Scholar] [CrossRef] [PubMed]
- Romanens, E.; Näf, R.; Lobmaier, T.; Pedan, V.; Leischtfeld, S.F.; Meile, L.; Schwenninger, S.M. A lab-scale model system for cocoa bean fermentation. Appl. Microbiol. Biotechnol. 2018, 102, 3349–3362. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Armisen, T.; Papalexandratou, Z.; Hendryckx, H.; Camu, N.; Vrancken, G.; De Vuyst, L.; Cornelis, P. Diversity of the total bacterial community associated with Ghanaian and Brazilian cocoa bean fermentation samples as revealed by a 16 S rRNA gene clone library. Appl. Microbiol. Biotechnol. 2010, 87, 2281–2292. [Google Scholar] [CrossRef]
- Pereira, G.V.D.M.; Magalhães-Guedes, K.T.; Schwan, R.F. rDNA-based DGGE analysis and electron microscopic observation of cocoa beans to monitor microbial diversity and distribution during the fermentation process. Food Res. Int. 2013, 53, 482–486. [Google Scholar] [CrossRef]
- Soumahoro, S.; Ouattara, H.G.; Droux, M.; Nasser, W.; Niamke, S.L.; Reverchon, S. Acetic acid bacteria (AAB) involved in cocoa fermentation from Ivory Coast: Species diversity and performance in acetic acid production. J. Food Sci. Technol. 2020, 57, 1904–1916. [Google Scholar] [CrossRef]
- Pitiwittayakul, N.; Yukphan, P.; Sintuprapa, W.; Yamada, Y.; Theeragool, G. Identification of acetic acid bacteria isolated in Thailand and assigned to the genus Acetobacter by groEL gene sequence analysis. Ann. Microbiol. 2015, 65, 1557–1564. [Google Scholar] [CrossRef]
- Kawasaki, H.; Uchimura, T.; Lisdiyanti, K.K.P. Identification of Acetobacterstrains isolated from Indonesian sources, and proposals of Acetobacter syzygii sp. Nov., Acetobacter cibinongensis sp. Nov., and Acetobacter orientalis sp. Nov. J. Gen. Appl. Microbiol. 2001, 47, 119–131. [Google Scholar]
- Pereira, G.V.D.M.; Magalhães, K.T.; de Almeida, E.G.; Coelho, I.D.S.; Schwan, R.F. Spontaneous cocoa bean fermentation carried out in a novel-design stainless steel tank: Influence on the dynamics of microbial populations and physical–chemical properties. Int. J. Food Microbiol. 2013, 161, 121–133. [Google Scholar] [CrossRef]
- Nielsen, D.S.; Hønholt, S.; Tano-Debrah, K.; Jespersen, L. Yeast populations associated with Ghanaian cocoa fermentations analysed using denaturing gradient gel electrophoresis (DGGE). Yeast 2005, 22, 271–284. [Google Scholar] [CrossRef]
- Douglass, A.P.; Offei, B.; Braun-Galleani, S.; Coughlan, A.Y.; Martos, A.A.R.; Ortiz-Merino, R.A.; Byrne, K.P.; Wolfe, K.H. Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: One species, four names. PLoS Pathog. 2018, 14, e1007138. [Google Scholar] [CrossRef]
- Papalexandratou, Z.; De Vuyst, L. Assessment of the yeast species composition of cocoa bean fermentations in different cocoa-producing regions using denaturing gradient gel electrophoresis. FEMS Yeast Res. 2011, 11, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Ho, V.T.T.; Zhao, J.; Fleet, G. Yeasts are essential for cocoa bean fermentation. Int. J. Food Microbiol. 2014, 174, 72–87. [Google Scholar] [CrossRef] [PubMed]
- Mozzi, F.; Raya, R.R.; Vignolo, G.M.; de Vuyst, L.; Weckx, S. Biotechnology of Lactic Acid Bacteria: Novel Applications, Second Edition. Edited the Functional Role of Lactic Acid Bacteria in Cocoa Bean Fermentation Chapter 16 16.1. Introduction. 2016. Available online: www.tomsekstra.dk (accessed on 20 January 2023).
- Herman, P.M.J. Development of Open Source Software for Ecological Modelling View Project Face-It: Functional Biodiversity in a Changing Sedimentary Environment: Implications for Biogeochemistry and Food Webs in a Managerial Setting. View Project. Available online: https://www.researchgate.net/publication/237139172 (accessed on 15 February 2023).
- Good, I.J. The Population Frequencies of Species and the Estimation of Population Parameters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Sparnins, V.L.; Chapman, P.J.; Dagley, S. Bacterial Degradation of 4-Hydroxyphenylacetic Acid and Homoprotocatechuic Acid. J. Bacteriol. 1974, 120, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Gibello, A.; Fernandez, J.; Ferrer, E.; Garrido-Pertierra, A. Catabolism of 3- and 4-hydroxyphenylacetic acid by Klebsiella pneumoniae. J. Gen. Microbiol. 1991, 137, 621–628. [Google Scholar] [CrossRef]
- Harwood, C.S.; Parales, R.E. The Β-Ketoadipate Pathway and The Biology of Self-Identity. Annu. Rev. Microbiol. 1996, 50, 553–590. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Sleeper, B.P.; Tsuchida, M.; Macdonald, D.L. The Bacterial Oxidation of Aromatic Compounds Iii. J. Bacteriol. 1950, 59, 137–151. [Google Scholar] [CrossRef]
- MacLean, A.M.; MacPherson, G.; Aneja, P.; Finan, T.M. Characterization of the β-Ketoadipate Pathway in Sinorhizobium meliloti. Appl. Environ. Microbiol. 2006, 72, 5403–5413. [Google Scholar] [CrossRef]
- Parke, D.; Ornston, L.N. Enzymes of the beta-ketoadipate pathway are inducible in Rhizobium and Agrobacterium spp. and constitutive in Bradyrhizobium spp. J. Bacteriol. 1986, 165, 288–292. [Google Scholar] [CrossRef]
- Illeghems, K.; Weckx, S.; De Vuyst, L. Applying meta-pathway analyses through metagenomics to identify the functional properties of the major bacterial communities of a single spontaneous cocoa bean fermentation process sample. Food Microbiol. 2015, 50, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Crafack, M.; Keul, H.; Eskildsen, C.E.; Petersen, M.A.; Saerens, S.; Blennow, A.; Skovmand-Larsen, M.; Swiegers, J.H.; Petersen, G.B.; Heimdal, H.; et al. Impact of starter cultures and fermentation techniques on the volatile aroma and sensory profile of chocolate. Food Res. Int. 2014, 63, 306–316. [Google Scholar] [CrossRef]
- Visintin, S.; Ramos, L.; Batista, N.; Dolci, P.; Schwan, F.; Cocolin, L. Impact of Saccharomyces cerevisiae and Torulaspora delbrueckii starter cultures on cocoa beans fermentation. Int. J. Food Microbiol. 2017, 257, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Batista, N.N.; Ramos, C.L.; Dias, D.R.; Pinheiro, A.C.M.; Schwan, R.F. The impact of yeast starter cultures on the microbial communities and volatile compounds in cocoa fermentation and the resulting sensory attributes of chocolate. J. Food Sci. Technol. 2016, 53, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J.; Henschke, P.A. The ‘buttery’ attribute of wine—Diacetyl—Desirability, spoilage and beyond. Int. J. Food Microbiol. 2004, 96, 235–252. [Google Scholar] [CrossRef]
- Castro-Alayo, E.M.; Idrogo-Vásquez, G.; Siche, R.; Cardenas-Toro, F.P. Formation of aromatic compounds precursors during fermentation of Criollo and Forastero cocoa. Heliyon 2019, 5, e01157. [Google Scholar] [CrossRef]
- Moreira, I.M.d.V.; Vilela, L.D.F.; Miguel, M.G.D.C.P.; Santos, C.; Lima, N.; Schwan, R.F. Impact of a Microbial Cocktail Used as a Starter Culture on Cocoa Fermentation and Chocolate Flavor. Molecules 2017, 22, 766. [Google Scholar] [CrossRef]
- Cocolin, L.; Dolci, P.; Rantsiou, K. Molecular Methods for Identification of Microorganisms in Traditional Meat Products. In Meat Biotechnology; Springer: New York, NY, USA, 2008; pp. 91–127. [Google Scholar] [CrossRef]
- Illeghems, K.; De Vuyst, L.; Papalexandratou, Z.; Weckx, S. Phylogenetic Analysis of a Spontaneous Cocoa Bean Fermentation Metagenome Reveals New Insights into Its Bacterial and Fungal Community Diversity. PLoS ONE 2012, 7, e38040. [Google Scholar] [CrossRef]
- Połka, J.; Rebecchi, A.; Pisacane, V.; Morelli, L.; Puglisi, E. Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 2015, 46, 342–356. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghisolfi, R.; Bandini, F.; Vaccari, F.; Bellotti, G.; Bortolini, C.; Patrone, V.; Puglisi, E.; Morelli, L. Bacterial and Fungal Communities Are Specifically Modulated by the Cocoa Bean Fermentation Method. Foods 2023, 12, 2024. https://doi.org/10.3390/foods12102024
Ghisolfi R, Bandini F, Vaccari F, Bellotti G, Bortolini C, Patrone V, Puglisi E, Morelli L. Bacterial and Fungal Communities Are Specifically Modulated by the Cocoa Bean Fermentation Method. Foods. 2023; 12(10):2024. https://doi.org/10.3390/foods12102024
Chicago/Turabian StyleGhisolfi, Rebecca, Francesca Bandini, Filippo Vaccari, Gabriele Bellotti, Cristian Bortolini, Vania Patrone, Edoardo Puglisi, and Lorenzo Morelli. 2023. "Bacterial and Fungal Communities Are Specifically Modulated by the Cocoa Bean Fermentation Method" Foods 12, no. 10: 2024. https://doi.org/10.3390/foods12102024
APA StyleGhisolfi, R., Bandini, F., Vaccari, F., Bellotti, G., Bortolini, C., Patrone, V., Puglisi, E., & Morelli, L. (2023). Bacterial and Fungal Communities Are Specifically Modulated by the Cocoa Bean Fermentation Method. Foods, 12(10), 2024. https://doi.org/10.3390/foods12102024









