Metabolic Regulation Effect and Potential Metabolic Biomarkers of Pre-Treated Delphinidin on Oxidative Damage Induced by Paraquat in A549 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Cell Culture and Sample Preparation
2.3. Cell Viability and ROS Assay
2.4. Metabolite Analysis
2.5. Metabolite Profiling Analysis
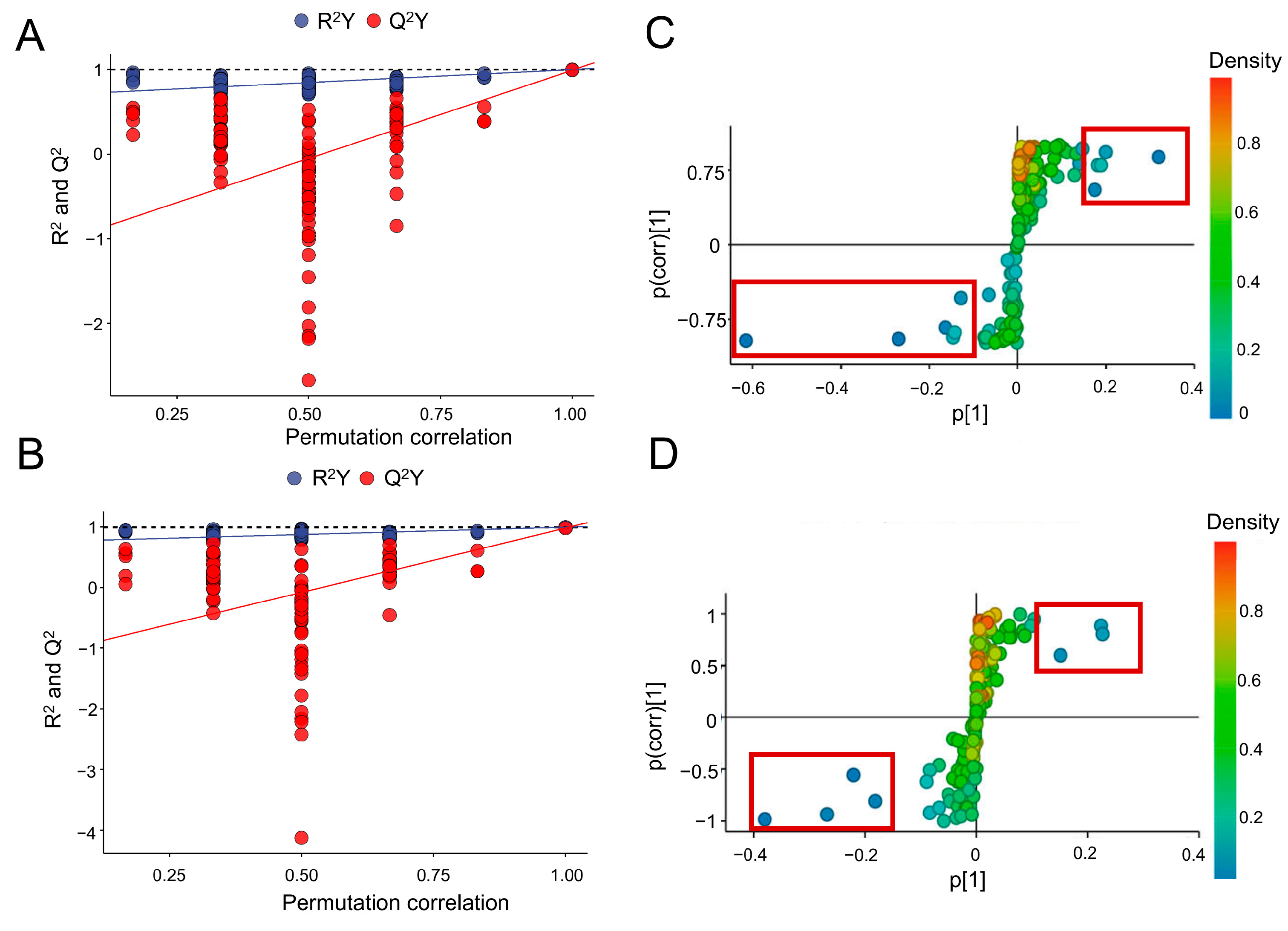
2.6. Multivariate Analysis
3. Results and Discussion
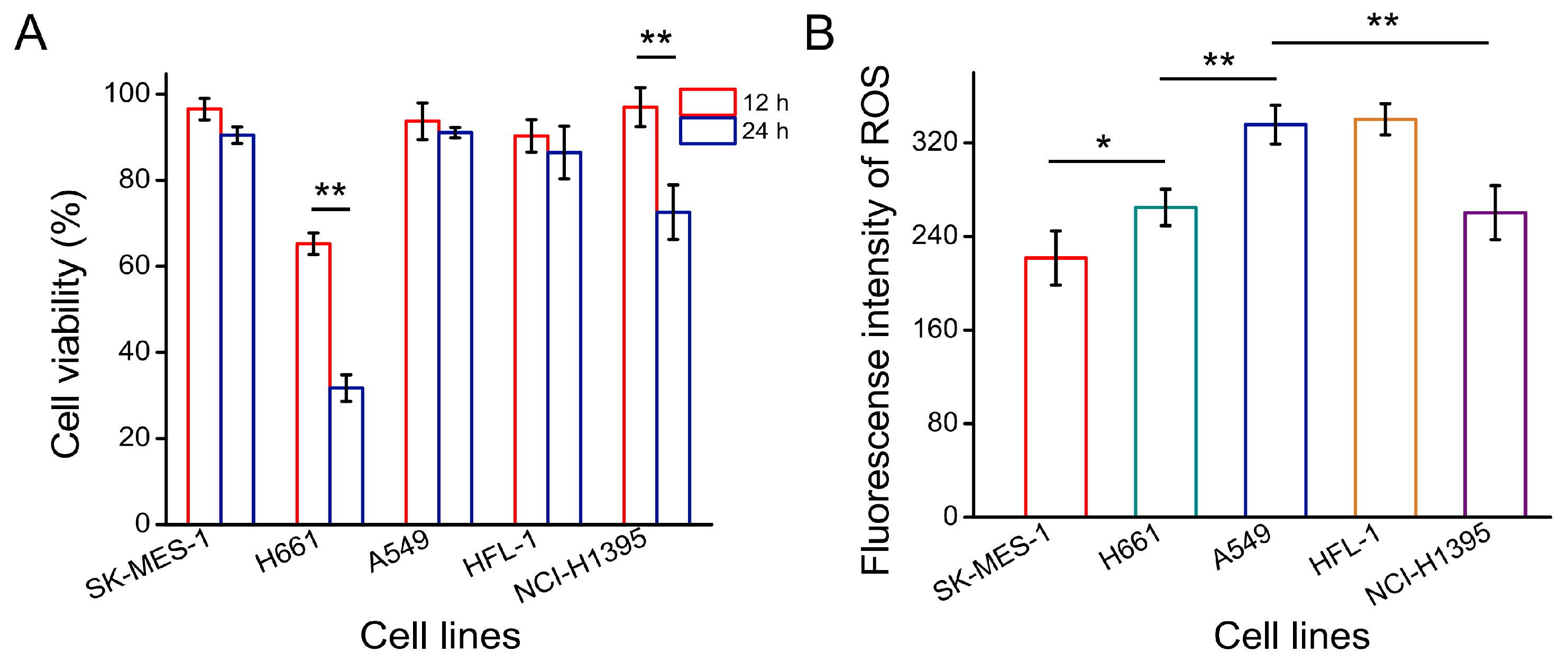
3.1. Optimal Cell Line for Oxidative Stress Damage Model
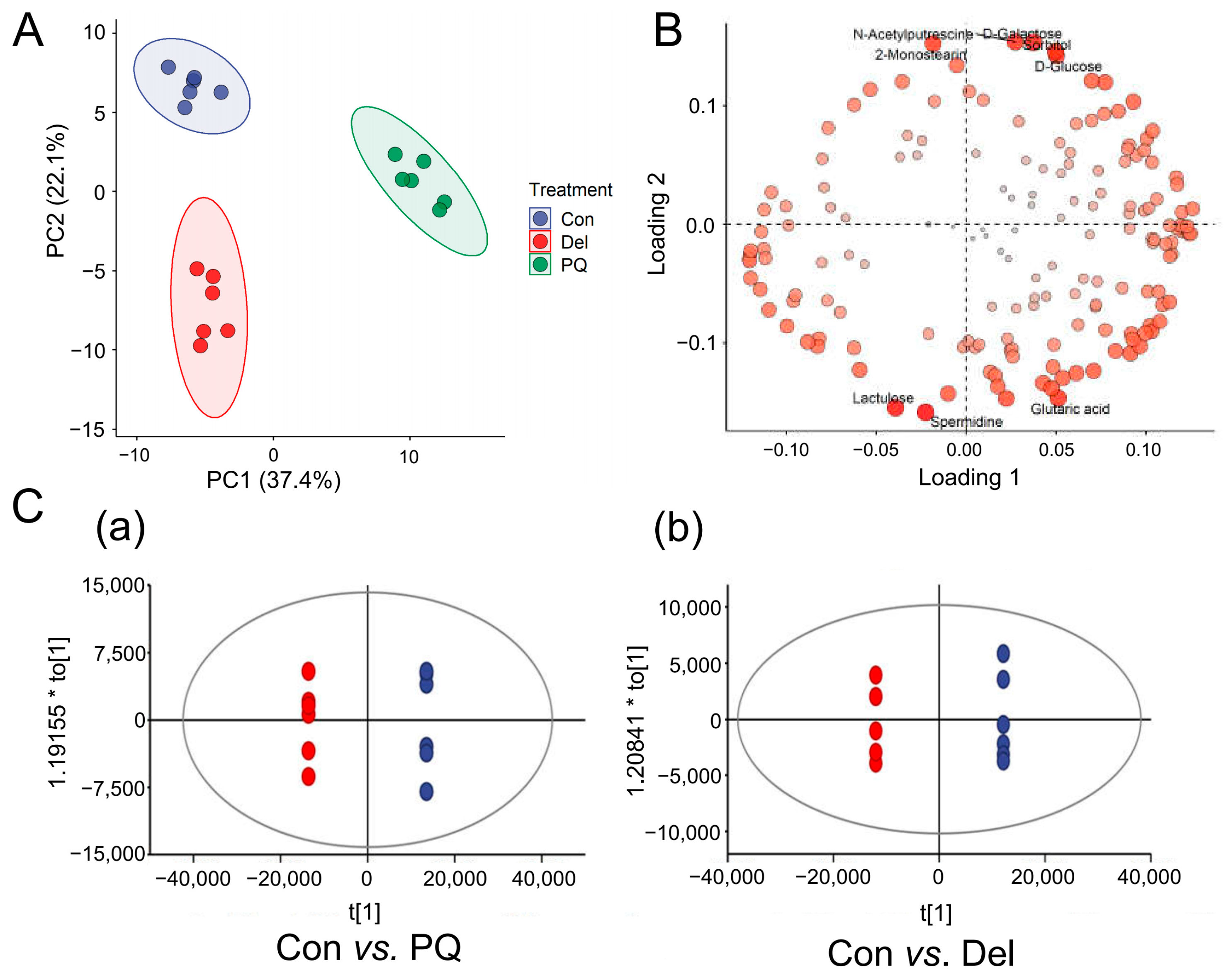
3.2. Multivariate Statistical Analysis to Visualize Metabolites
3.3. Identification of Different Metabolites and Prediction of Biomarkers
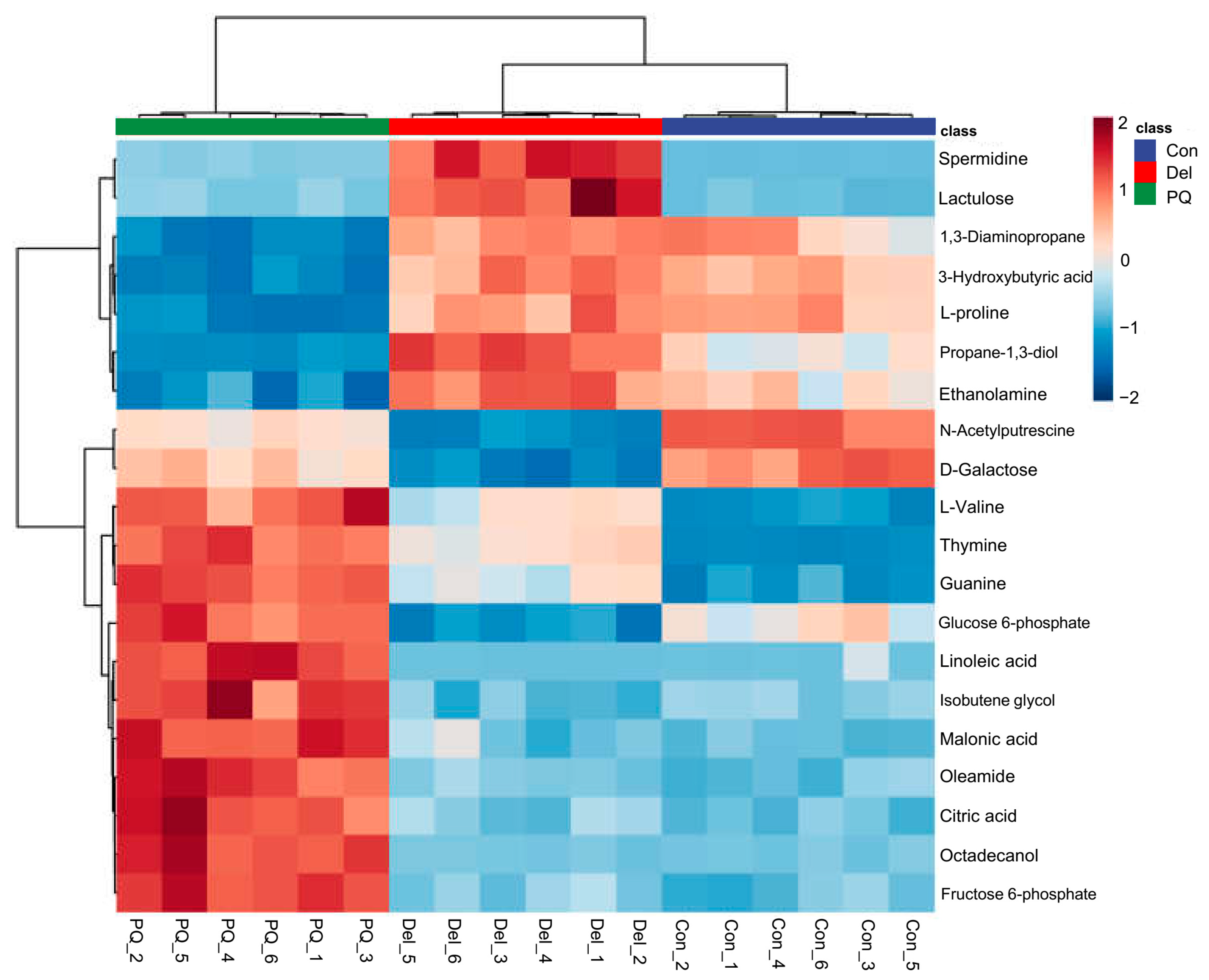
3.3.1. Differential Metabolite Screening
3.3.2. Prediction and Verification of Biomarkers
3.4. Metabolic Pathway Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUC | area under the ROC |
| Del | delphinidin |
| FC | fold change |
| GC-TOF/MS | gas chromatography-time of flight mass spectrometry |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| OPLS-DA | orthogonal projection to latent structure discriminant analysis |
| PCA | principal component analysis |
| PQ | paraquat |
| QC | quality control |
| ROC | receiver operating characteristic curve |
| ROS | reactive oxygen species |
| TCA | citric acid cycle |
| VIP | variable importance for the projection |
References
- Chen, J.; Li, H.Y.; Wang, D.; Guo, X.Z. Delphinidin protects β2m-/Thy1+ bone marrow-derived hepatocyte stem cells against TGF-beta1-induced oxidative stress and apoptosis through the PI3K/Akt pathway in vitro. Chem. Biol. Interact. 2019, 297, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Ni, T.; Yang, W.; Xing, Y. Protective effects of delphinidin against H2O2-induced oxidative injuries in human retinal pigment epithelial cells. Biosci. Rep. 2019, 39, BSR20190689. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Park, Y.J.; Song, M.G.; Kim, D.R.; Zada, S.; Kim, D.H. Cytoprotective Effects of Delphinidin for Human Chondrocytes against Oxidative Stress through Activation of Autophagy. Antioxidants 2020, 9, 83. [Google Scholar] [CrossRef] [PubMed]
- Sauer, R.S.; Krummenacher, I.; Bankoglu, E.E.; Yang, S.; Oehler, B.; Schoppler, F.; Mohammadi, M.; Guntzel, P.; Ben-Kraiem, A.; Holzgrabe, U.; et al. Stabilization of Delphinidin in Complex with Sulfobutylether-beta-Cyclodextrin Allows for Antinociception in Inflammatory Pain. Antioxid. Redox Signal. 2021, 34, 1260–1279. [Google Scholar] [CrossRef] [PubMed]
- Harada, G.; Onoue, S.; Inoue, C.; Hanada, S.; Katakura, Y. Delphinidin-3-glucoside suppresses lipid accumulation in HepG2 cells. Cytotechnology 2018, 70, 1707–1712. [Google Scholar] [CrossRef]
- Wu, C.-H.; Huang, C.-C.; Hung, C.-H.; Yao, F.-Y.; Wang, C.-J.; Chang, Y.-C. Delphinidin-rich extracts of Hibiscus sabdariffa L. trigger mitochondria-derived autophagy and necrosis through reactive oxygen species in human breast cancer cells. J. Funct. Foods 2016, 25, 279–290. [Google Scholar] [CrossRef]
- Goszcz, K.; Deakin, S.J.; Duthie, G.G.; Stewart, D.; Megson, I.L. Bioavailable concentrations of delphinidin and its metabolite, gallic acid, induce antioxidant protection associated with increased intracellular glutathione in cultured endothelial cells. Oxidative Med. Cell. Longev. 2017, 2017, 9260701. [Google Scholar] [CrossRef]
- Czank, C.; Cassidy, A.; Zhang, Q.; Morrison, D.J.; Preston, T.; Kroon, P.A.; Botting, N.P.; Kay, C.D. Human metabolism and elimination of the anthocyanin, cyanidin-3-glucoside: A 13C-tracer study. Am. J. Clin. Nutr. 2013, 97, 995–1003. [Google Scholar] [CrossRef]
- Prior, R.L.; Wu, X. Anthocyanins: Structural characteristics that result in unique metabolic patterns and biological activities. Free. Radic. Res. 2006, 40, 1014–1028. [Google Scholar] [CrossRef]
- Xu, Y.; Ji, J.; Wu, H.; Pi, F.; Blazenovic, I.; Zhang, Y.; Sun, X. Untargeted GC-TOFMS-based cellular metabolism analysis to evaluate ozone degradation effect of deoxynivalenol. Toxicon 2019, 168, 49–57. [Google Scholar] [CrossRef]
- Ji, J.; Wang, Q.; Wu, H.; Xia, S.; Guo, H.; Blazenovic, I.; Zhang, Y.; Sun, X. Insights into cellular metabolic pathways of the combined toxicity responses of Caco-2 cells exposed to deoxynivalenol, zearalenone and Aflatoxin B1. Food Chem. Toxicol. 2019, 126, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Nunez-Garcia, M.; Fernandez-Tussy, P.; Barbier-Torres, L.; Fernandez-Ramos, D.; Gomez-Santos, B.; Buque, X.; Lopitz-Otsoa, F.; Goikoetxea-Usandizaga, N.; Serrano-Macia, M.; et al. Targeting hepatic glutaminase 1 ameliorates non-alcoholic steatohepatitis by restoring very-low-density lipoprotein triglyceride assembly. Cell Metab. 2020, 31, 605–622.e610. [Google Scholar] [CrossRef]
- Jia, S.; Guan, T.; Zhang, X.; Liu, Y.; Liu, Y.; Zhao, X. Serum metabonomics analysis of quercetin against the toxicity induced by cadmium in rats. J. Biochem. Mol. Toxicol. 2020, 34, e22448. [Google Scholar] [CrossRef]
- Dong, X.W.; Yao, S.Q.; Wu, H.Y.; Zhang, Y.B.; Wang, C.; Na, X.L.; Wu, W.D. Urine Metabonomic Analysis of Interventions Effect of Soy Isoflavones on Rats Exposed to Di-(2-ethylhexyl) Phthalate. Biomed. Environ. Sci. 2020, 33, 77–88. [Google Scholar] [PubMed]
- Dai, Y.; Liu, J.; Leng, J.; Ma, Z.; Wang, H. 1H-NMR-Based Metabonomics Study on the Restorative Effect of Soybean Polypeptide in Rats of Oxidative Damaged Induced by d-Galactose. Int. J. Pept. Res. Ther. 2016, 23, 37–47. [Google Scholar] [CrossRef]
- Chagas-Paula, D.A.; Oliveira, T.B.; Zhang, T.; Edrada-Ebel, R.; Da Costa, F.B. Prediction of Anti-inflammatory Plants and Discovery of Their Biomarkers by Machine Learning Algorithms and Metabolomic Studies. Planta Med. 2015, 81, 450–458. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, B.; Cheng, X.; Ding, Y.; Tian, X.; Shi, Y.; Le, G. Metabolomic studies on the systemic responses of mice with oxidative stress induced by short-term oxidized tyrosine administration. RSC Adv. 2017, 7, 28591–28605. [Google Scholar] [CrossRef]
- Alberghina, L.; Gaglio, D. Redox control of glutamine utilization in cancer. Cell Death Dis. 2014, 5, e1561. [Google Scholar] [CrossRef]
- Datta, R.; Heylman, C.; George, S.C.; Gratton, E. Label-free imaging of metabolism and oxidative stress in human induced pluripotent stem cell-derived cardiomyocytes. Biomed. Opt. Express 2016, 7, 1690–1701. [Google Scholar] [CrossRef]
- Ye, Y.; Ji, J.; Pi, F.; Yang, H.; Liu, J.; Zhang, Y.; Xia, S.; Wang, J.; Xu, D.; Sun, X. A novel electrochemical biosensor for antioxidant evaluation of phloretin based on cell-alginate/L-cysteine/gold nanoparticle-modified glassy carbon electrode. Biosens. Bioelectron. 2018, 119, 119–125. [Google Scholar] [CrossRef]
- Liu, K.; Pi, F.; Zhang, H.; Ji, J.; Xia, S.; Cui, F.; Sun, J.; Sun, X. Metabolomics Analysis To Evaluate the Anti-Inflammatory Effects of Polyphenols: Glabridin Reversed Metabolism Change Caused by LPS in RAW 264.7 Cells. J. Agric. Food Chem. 2017, 65, 6070–6079. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Geng, S.; Ji, J.; Ye, Y.; Xu, D.; Zhang, Y.; Sun, X. Separation and identification of antioxidant chemical components in Diaphragma juglandis Fructus and functional evaluation in Caenorhabditis elegans. J. Funct. Foods 2021, 80, 104422. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, C.; Xu, S.; Gao, L.; Blazenovic, I.; Ji, J.; Wang, J.; Sun, X. Untargeted metabolomics analysis by gas chromatography/time-of-flight mass spectrometry of human serum from methamphetamine abusers. Addict. Biol. 2021, 26, e13062. [Google Scholar] [CrossRef] [PubMed]
- da Motta, L.L.; De Bastiani, M.A.; Stapenhorst, F.; Klamt, F. Oxidative stress associates with aggressiveness in lung large-cell carcinoma. Tumor Biol. 2015, 36, 4681–4688. [Google Scholar] [CrossRef]
- Podder, B.; Kim, Y.S.; Zerin, T.; Song, H.Y. Antioxidant effect of silymarin on paraquat-induced human lung adenocarcinoma A549 cell line. Food Chem. Toxicol. 2012, 50, 3206–3214. [Google Scholar] [CrossRef]
- Mercer, P.F.; Johns, R.H.; Scotton, C.J.; Krupiczojc, M.A.; Konigshoff, M.; Howell, D.C.; McAnulty, R.J.; Das, A.; Thorley, A.J.; Tetley, T.D.; et al. Pulmonary epithelium is a prominent source of proteinase-activated receptor-1-inducible CCL2 in pulmonary fibrosis. J. Respir. Cell Mol. Biol. 2009, 179, 414–425. [Google Scholar] [CrossRef]
- Cui, S.; Nian, Q.; Chen, G.; Wang, X.; Zhang, J.; Qiu, J.; Zhang, Z. Ghrelin ameliorates A549 cell apoptosis caused by paraquat via p38-MAPK regulated mitochondrial apoptotic pathway. Toxicology 2019, 426, 152267. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H.; Li, S.; Zhong, X.; Wang, H.; Liu, X. Comparative metabolic profiling of Ophiocordyceps sinensis and its cultured mycelia using GC-MS. Food Res. Int. 2020, 134, 109241. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline mechanisms of stress survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef]
- Zhang, L.; Alfano, J.R.; Becker, D.F. Proline metabolism increases katG expression and oxidative stress resistance in Escherichia coli. J. Bacteriol. 2015, 197, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Bauduin, S.; Latini, M.; Belleggia, I.; Migliore, M.; Biancucci, M.; Mattioli, R.; Francioso, A.; Mosca, L.; Funck, D.; Trovato, M. Interplay between proline metabolism and ROS in the fine tuning of root-meristem size in arabidopsis. Plants 2022, 11, 1512. [Google Scholar] [CrossRef]
- Ferreira, A.G.K.; Biasibetti-Brendler, H.; Sidegum, D.S.V.; Loureiro, S.O.; Figueiró, F.; Wyse, A.T.S. Effect of proline on cell death, cell cycle, and oxidative stress in c6 glioma cell line. Neurotox. Res. 2021, 39, 327–334. [Google Scholar] [CrossRef]
- Christgen, S.L.; Becker, D.F. Role of proline in pathogen and host interactions. Antioxid. Redox Signal. 2019, 30, 683–709. [Google Scholar] [CrossRef] [PubMed]
- Mierziak, J.; Burgberger, M.; Wojtasik, W. 3-Hydroxybutyrate as a metabolite and a signal molecule regulating processes of living organisms. Biomolecules 2021, 11, 402. [Google Scholar] [CrossRef]
- Majrashi, M.; Altukri, M.; Ramesh, S.; Govindarajulu, M.; Schwartz, J.; Almaghrabi, M.; Smith, F.; Thomas, T.; Suppiramaniam, V.; Moore, T.; et al. β-hydroxybutyric acid attenuates oxidative stress and improves markers of mitochondrial function in the HT-22 hippocampal cell line. J. Integr. Neurosci. 2021, 20, 321–329. [Google Scholar] [CrossRef]
- Hu, W.; Yang, P.; Fu, Z.; Wang, Y.; Zhou, Y.; Ye, Z.; Gong, Y.; Huang, A.; Sun, L.; Zhao, Y.; et al. High L-valine concentrations associate with increased oxidative stress and newly-diagnosed type 2 diabetes mellitus: A cross-sectional study. Diabetes Metab. Syndr. Obes. 2022, 15, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Turpeinen, A.M.; Basu, S.; Mutanen, M. A high linoleic acid diet increases oxidative stress in vivo and affects nitric oxide metabolism in humans. Prostaglandins Leukot. Essent. Fat. Acids 1998, 59, 229–233. [Google Scholar] [CrossRef]
- Senizza, A.; Rocchetti, G.; Callegari, M.L.; Lucini, L.; Morelli, L. Linoleic acid induces metabolic stress in the intestinal microorganism Bifidobacterium breve DSM 20213. Sci. Rep. 2020, 10, 5997. [Google Scholar] [CrossRef]
- Kalonia, H.; Kumar, P.; Kumar, A. Targeting oxidative stress attenuates malonic acid induced Huntington like behavioral and mitochondrial alterations in rats. Eur. J. Pharmacol. 2010, 634, 46–52. [Google Scholar] [CrossRef]
- Efferth, T.; Schwarzl, S.; Smith, J.; Osieka, R. Role of glucose-6-phosphate dehydrogenase for oxidative stress and apoptosis. Cell Death Differ. 2006, 13, 527–528. [Google Scholar] [CrossRef] [PubMed]
- Akram, M. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem. Biophys. 2014, 68, 475–478. [Google Scholar] [PubMed]
- Estacion, M.; Weinberg, J.S.; Sinkins, W.G.; Schilling, W.P. Blockade of maitotoxin-induced endothelial cell lysis by glycine and L-alanine. Am. J. Physiol. Cell Physiol. 2003, 284, C1006–C1020. [Google Scholar] [CrossRef]
- Grosser, N.; Oberle, S.; Berndt, G.; Erdmann, K.; Hemmerle, A.; Schroder, H. Antioxidant action of L-alanine: Heme oxygenase-1 and ferritin as possible mediators. Biochem. Biophys. Res. Commun. 2004, 314, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ye, Y.; Ji, J.; Yang, X.; Xu, J.; Wang, J.; Han, X.; Zhang, T.; Sun, X. Diet composition affects long-term zearalenone exposure on the gut–blood–liver axis metabolic dysfunction in mice. Ecotoxicol. Environ. Saf. 2022, 236, 113466. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gui, X.; Wu, L.; Tian, S.; Wang, H.; Xie, L.; Wu, W. Amino acid metabolism, lipid metabolism, and oxidative stress are associated with post-stroke depression: A metabonomics study. BMC Neurol. 2020, 20, 250. [Google Scholar] [CrossRef]
- Guo, Y.; Wan, S.; Han, M.; Zhao, Y.; Li, C.; Cai, G.; Zhang, S.; Sun, Z.; Hu, X.; Cao, H.; et al. Plasma metabolomics analysis identifies abnormal energy, lipid, and amino acid metabolism in abdominal aortic aneurysms. Med. Sci. Monit. 2020, 26, e926766. [Google Scholar] [CrossRef]
- Yudkoff, M.; Daikhin, Y.; Melø, T.M.; Nissim, I.; Sonnewald, U.; Nissim, I. The ketogenic diet and brain metabolism of amino acids: Relationship to the anticonvulsant effect. Annu. Rev. Nutr. 2007, 27, 415–430. [Google Scholar] [CrossRef]
- Gamberino, W.C.; Berkich, D.A.; Lynch, C.J.; Xu, B.; LaNoue, K.F. Role of pyruvate carboxylase in facilitation of synthesis of glutamate and glutamine in cultured astrocytes. J. Neurochem. 1997, 69, 2312–2325. [Google Scholar] [CrossRef]
- Scoville, E.A.; Allaman, M.M.; Brown, C.T.; Motley, A.K.; Horst, S.N.; Williams, C.S.; Koyama, T.; Zhao, Z.; Adams, D.W.; Beaulieu, D.B.; et al. Alterations in Lipid, Amino Acid, and Energy Metabolism Distinguish Crohn’s Disease from Ulcerative Colitis and Control Subjects by Serum Metabolomic Profiling. Metabolomics 2018, 14, 17. [Google Scholar] [CrossRef]
- Selamnia, M.; Robert, V.; Mayeur, C.; Duée, P.H.; Blachier, F. Effects of L-valine on growth and polyamine metabolism in human colon carcinoma cells. Biochim. Biophys. Acta 1998, 1379, 151–160. [Google Scholar] [CrossRef]
- Chen, Q.; Tang, L.; Xin, G.; Li, S.; Ma, L.; Xu, Y.; Zhuang, M.; Xiong, Q.; Wei, Z.; Xing, Z.; et al. Oxidative stress mediated by lipid metabolism contributes to high glucose-induced senescence in retinal pigment epithelium. Free Radic. Biol. Med. 2019, 130, 48–58. [Google Scholar] [CrossRef]
- Bartolacci, C.; Andreani, C.; El-Gammal, Y.; Scaglioni, P.P. Lipid metabolism regulates oxidative stress and ferroptosis in ras-driven cancers: A perspective on cancer progression and therapy. Front. Mol. Biosci. 2021, 8, 706650. [Google Scholar] [CrossRef]
- Kim, H.M.; Lee, D.K.; Long, N.P.; Kwon, S.W.; Park, J.H. Uptake of nanopolystyrene particles induces distinct metabolic profiles and toxic effects in Caenorhabditis elegans. Environ. Pollut. 2019, 246, 578–586. [Google Scholar] [CrossRef]
- Zhou, X.; He, L.; Wu, C.; Zhang, Y.; Wu, X.; Yin, Y. Serine alleviates oxidative stress via supporting glutathione synthesis and methionine cycle in mice. Mol. Nutr. Food Res. 2017, 61, 1700262. [Google Scholar] [CrossRef]
- Galati, G.; Lin, A.; Sultan, A.M.; O’Brien, P.J. Cellular and in vivo hepatotoxicity caused by green tea phenolic acids and catechins. Free Radic. Biol. Med. 2006, 40, 570–580. [Google Scholar] [CrossRef]
- van der Reest, J.; Lilla, S.; Zheng, L.; Zanivan, S.; Gottlieb, E. Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nat. Commun. 2018, 9, 1581. [Google Scholar] [CrossRef]
- Martinez, Y.; Li, X.; Liu, G.; Bin, P.; Yan, W.; Mas, D.; Valdivie, M.; Hu, C.A.; Ren, W.; Yin, Y. The role of methionine on metabolism, oxidative stress, and diseases. Amino Acids 2017, 49, 2091–2098. [Google Scholar] [CrossRef]
- Gusarov, I.; Pani, B.; Gautier, L.; Smolentseva, O.; Eremina, S.; Shamovsky, I.; Katkova-Zhukotskaya, O.; Mironov, A.; Nudler, E. Glycogen controls Caenorhabditis elegans lifespan and resistance to oxidative stress. Nat. Commun. 2017, 8, 15868. [Google Scholar] [CrossRef]
- Hou, C. Comparing the new and existing hypotheses on energy metabolism and longevity. Bioessays 2018, 40, e1800110. [Google Scholar] [CrossRef]
- Ratnasekhar, C.; Sonane, M.; Satish, A.; Mudiam, M.K. Metabolomics reveals the perturbations in the metabolome of Caenorhabditis elegans exposed to titanium dioxide nanoparticles. Nanotoxicology 2015, 9, 994–1004. [Google Scholar] [CrossRef]
- Saulite, L.; Jekabsons, K.; Klavins, M.; Muceniece, R.; Riekstina, U. Effects of malvidin, cyanidin and delphinidin on human adipose mesenchymal stem cell differentiation into adipocytes, chondrocytes and osteocytes. Phytomedicine 2019, 53, 86–95. [Google Scholar] [CrossRef]
- Bhagavathi, S.; Periyanaina, K.; Chaiyavat, C. The influence of supplementation of anthocyanins on obesity-associated comorbidities: A concise review. Foods 2020, 9, 687. [Google Scholar]
- Crockett, E.L. The cold but not hard fats in ectotherms: Consequences of lipid restructuring on susceptibility of biological membranes to peroxidation, a review. J. Comp. Physiol. B 2008, 178, 795–809. [Google Scholar] [CrossRef] [PubMed]
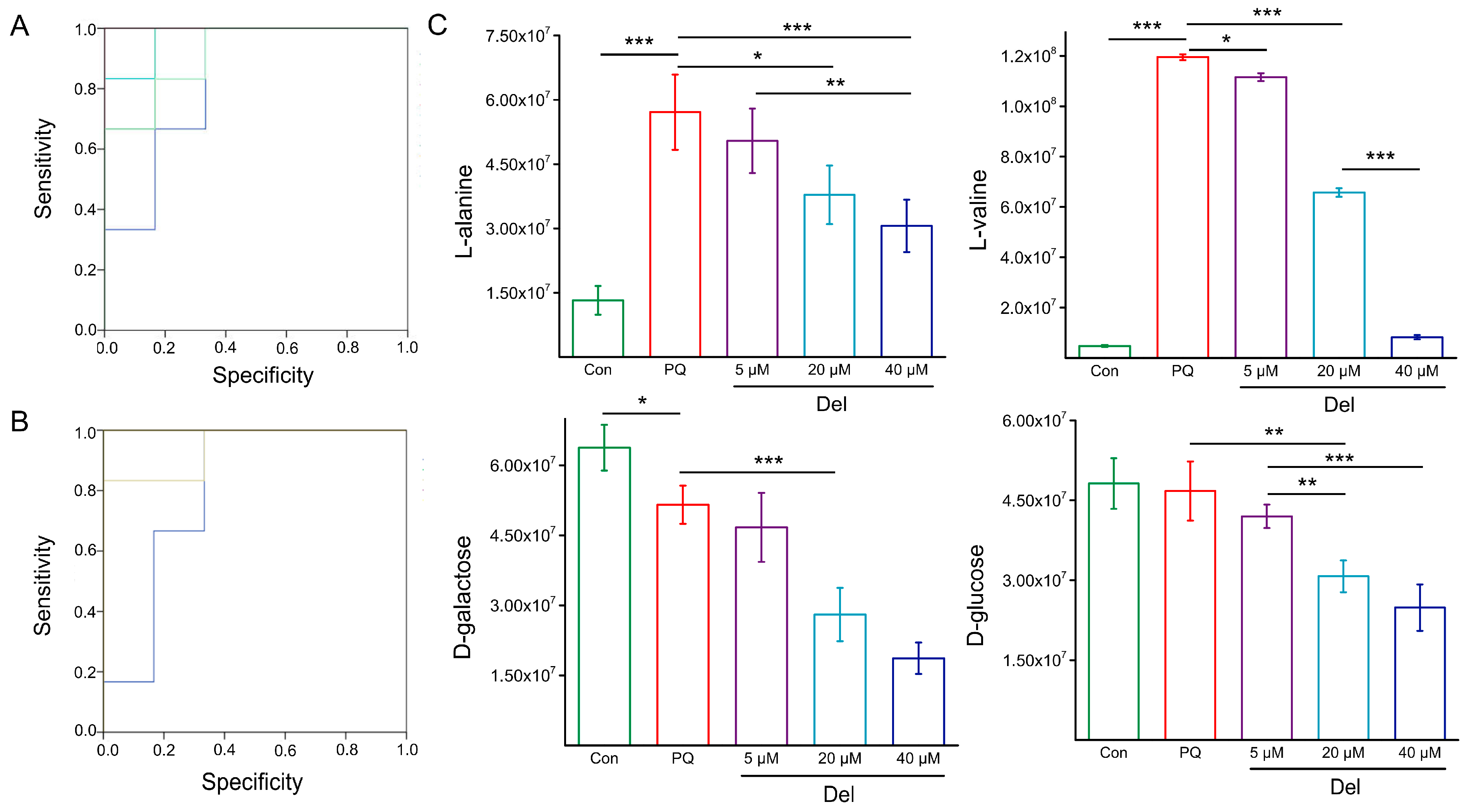
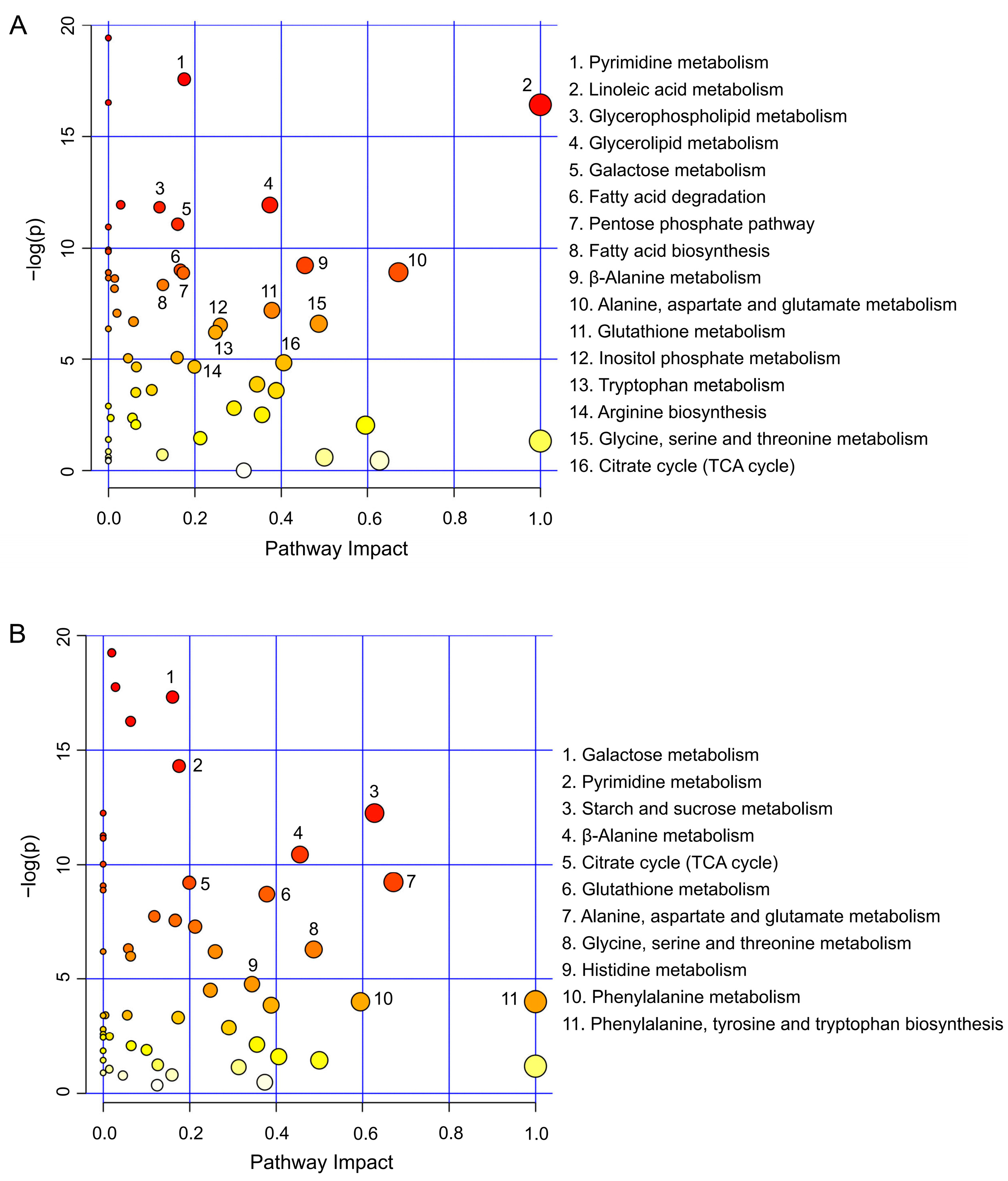
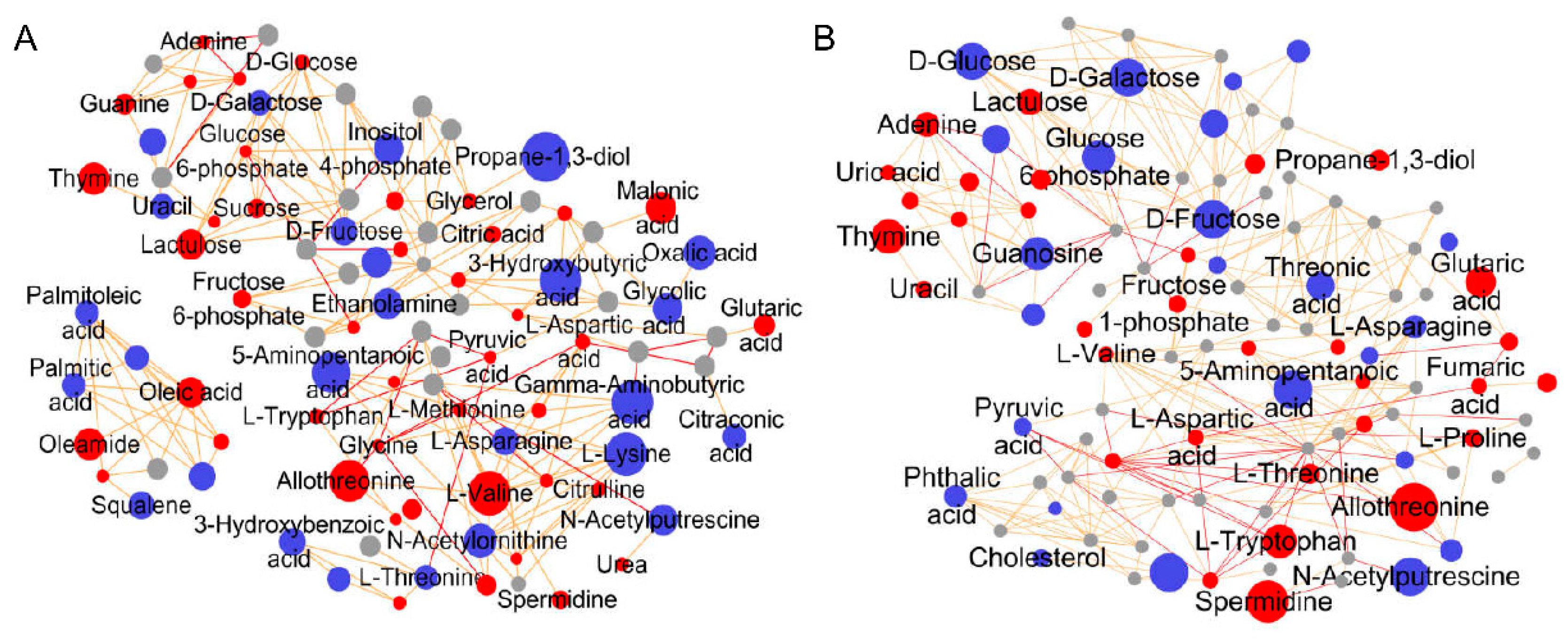
| Number | Metabolites | PubChem ID/ * | KEGG ID | Fold Change Value | |
|---|---|---|---|---|---|
| PQ | Del | ||||
| 1 | Propane-1,3-diol | 10,442 | C02457 | 3.93 **Δ | 1.87 **Δ |
| 2 | L-Alanine | 5950 | C00041 | 3.93 **Δ | 2.49 **Δ |
| 3 | Oxalic acid | 971 | C00209 | 0.59 **Δ | 0.94 |
| 4 | D-Galactose | 6036 | C00984 | 0.79 **Δ | 0.36 **Δ |
| 5 | Maleimide | 10,935 | C07272 | 2.87 **Δ | 1.10 |
| 6 | L-Valine | 6287 | C00183 | 25.95 **Δ | 1.20 Δ |
| 7 | Urea | 1176 | C00086 | 1.54 *Δ | 1.04 |
| 8 | Malonic acid | 867 | C00383 | 5.13 **Δ | 1.21 |
| 9 | Isobutene Glycol | 68,410 | C21290 | 0.65 **Δ | 1.07 |
| 10 | Thymine | 5610 | C00483 | 5.30 **Δ | 3.65 **Δ |
| 11 | Oleic acid | 445,639 | C00712 | 3.91 **Δ | 1.21 |
| 12 | 3-Hydroxybutyric acid | 441 | C01089 | 0.26 **Δ | 1.14 |
| 13 | L-Threonine | 6288 | C00188 | 0.40 **Δ | 1.83 **Δ |
| 14 | D-Glucose | 5793 | C00031 | 1.06 | 0.53 **Δ |
| Combination | PQ Group vs. Con Group | Del Group vs. Con Group | ||
|---|---|---|---|---|
| Metabolites | AUC Value | Metabolites | AUC Value | |
| 1 | L-alanine/L-valine | 0.944 | L-alanine/L-Threonine | 0.889 |
| 2 | L-valine/urea | 0.972 | D-galactose/D-glucose | 0.944 |
| 3 | L-alanine/L-valine/urea | 0.972 | L-alanine/D-galactose/D-glucose | 0.998 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Y.; Ji, J.; Huang, Y.; Zhang, Y.; Sun, X. Metabolic Regulation Effect and Potential Metabolic Biomarkers of Pre-Treated Delphinidin on Oxidative Damage Induced by Paraquat in A549 Cells. Foods 2022, 11, 3575. https://doi.org/10.3390/foods11223575
Ye Y, Ji J, Huang Y, Zhang Y, Sun X. Metabolic Regulation Effect and Potential Metabolic Biomarkers of Pre-Treated Delphinidin on Oxidative Damage Induced by Paraquat in A549 Cells. Foods. 2022; 11(22):3575. https://doi.org/10.3390/foods11223575
Chicago/Turabian StyleYe, Yongli, Jian Ji, Yaoguang Huang, Yinzhi Zhang, and Xiulan Sun. 2022. "Metabolic Regulation Effect and Potential Metabolic Biomarkers of Pre-Treated Delphinidin on Oxidative Damage Induced by Paraquat in A549 Cells" Foods 11, no. 22: 3575. https://doi.org/10.3390/foods11223575
APA StyleYe, Y., Ji, J., Huang, Y., Zhang, Y., & Sun, X. (2022). Metabolic Regulation Effect and Potential Metabolic Biomarkers of Pre-Treated Delphinidin on Oxidative Damage Induced by Paraquat in A549 Cells. Foods, 11(22), 3575. https://doi.org/10.3390/foods11223575






