Point-of-Care Lateral Flow Detection of Viable Escherichia coli O157:H7 Using an Improved Propidium Monoazide-Recombinase Polymerase Amplification Method
Abstract
1. Introduction
2. Results
2.1. Confirmation of Viable and Non-Viable Cells
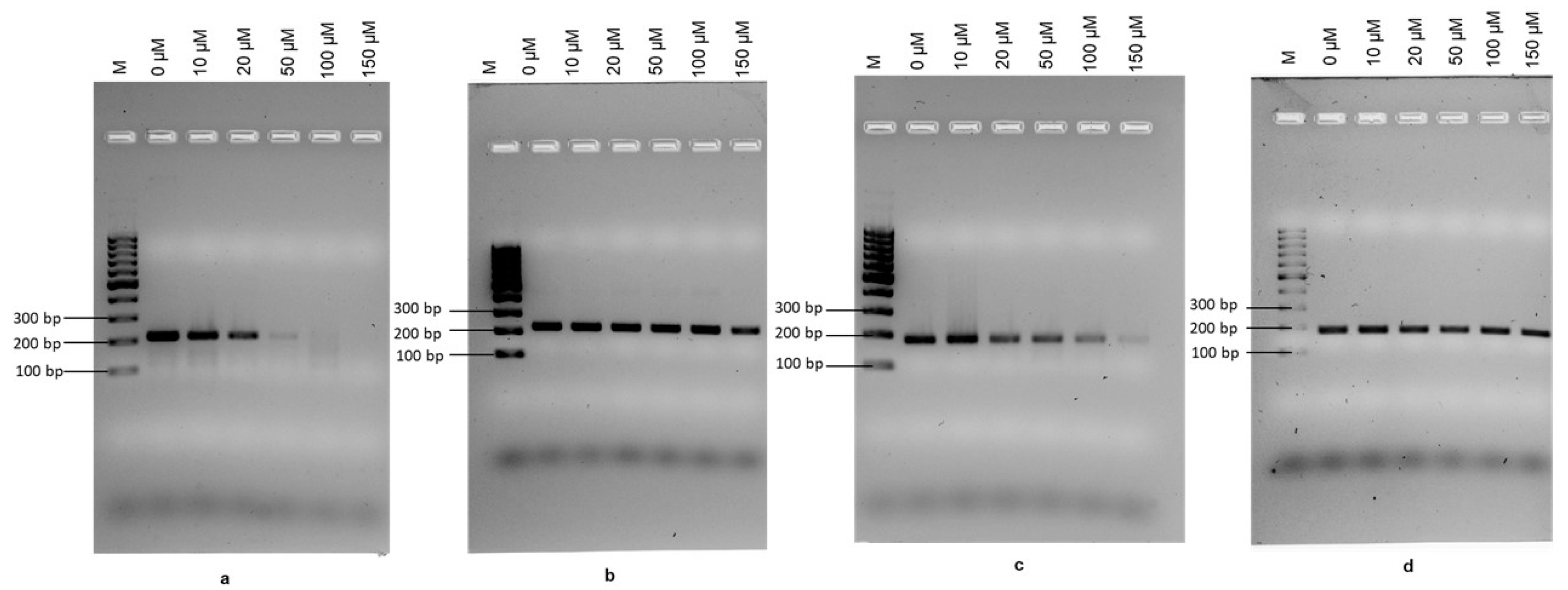
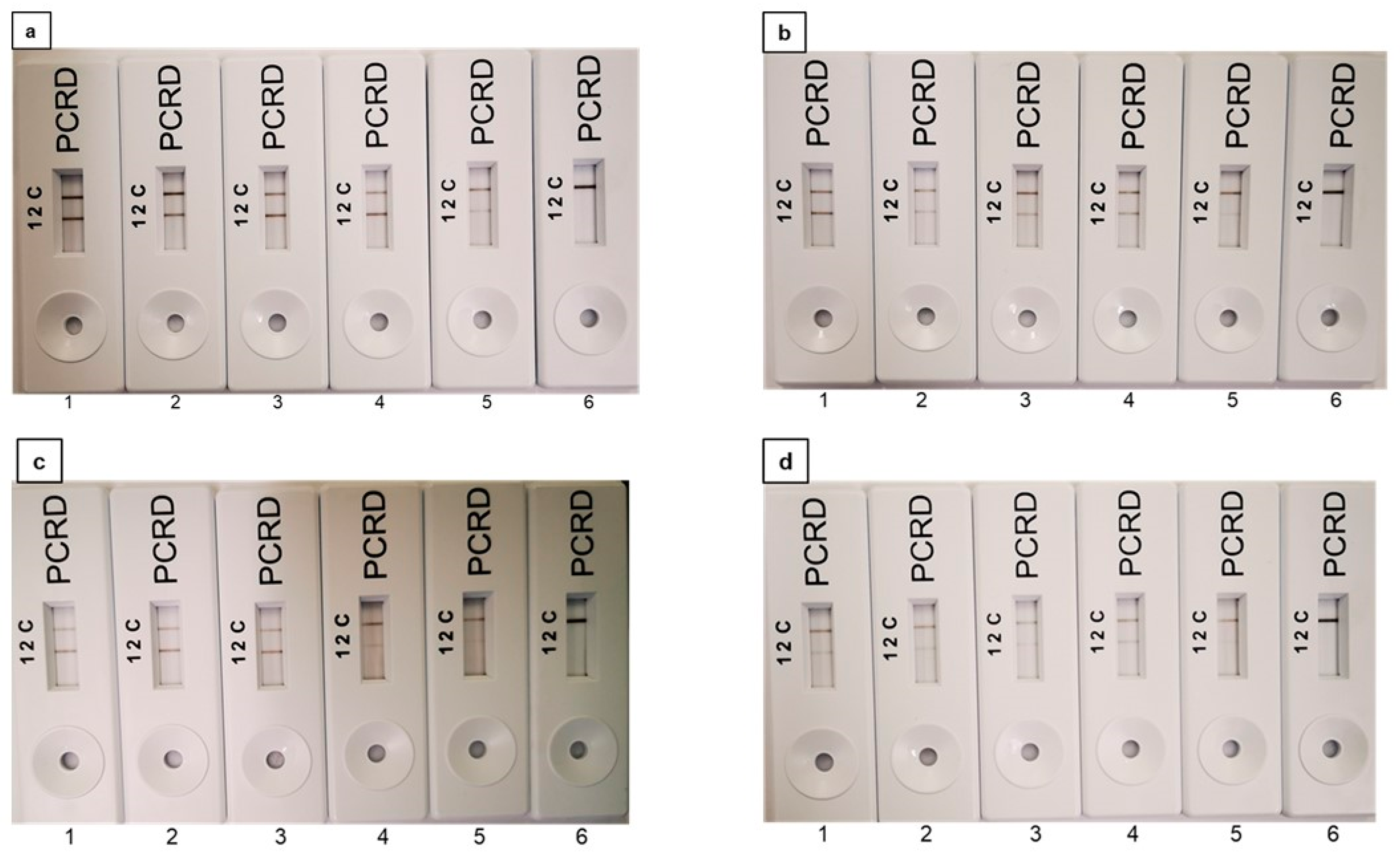
2.2. Optimisation of PMAxx Concentration for the Recombinase Polymerase Amplification Assay (RPA)
2.3. Specificity Assessment
2.4. Assessment of the Interference of PMAxx to the Recombinase Polymerase Amplification (RPA)
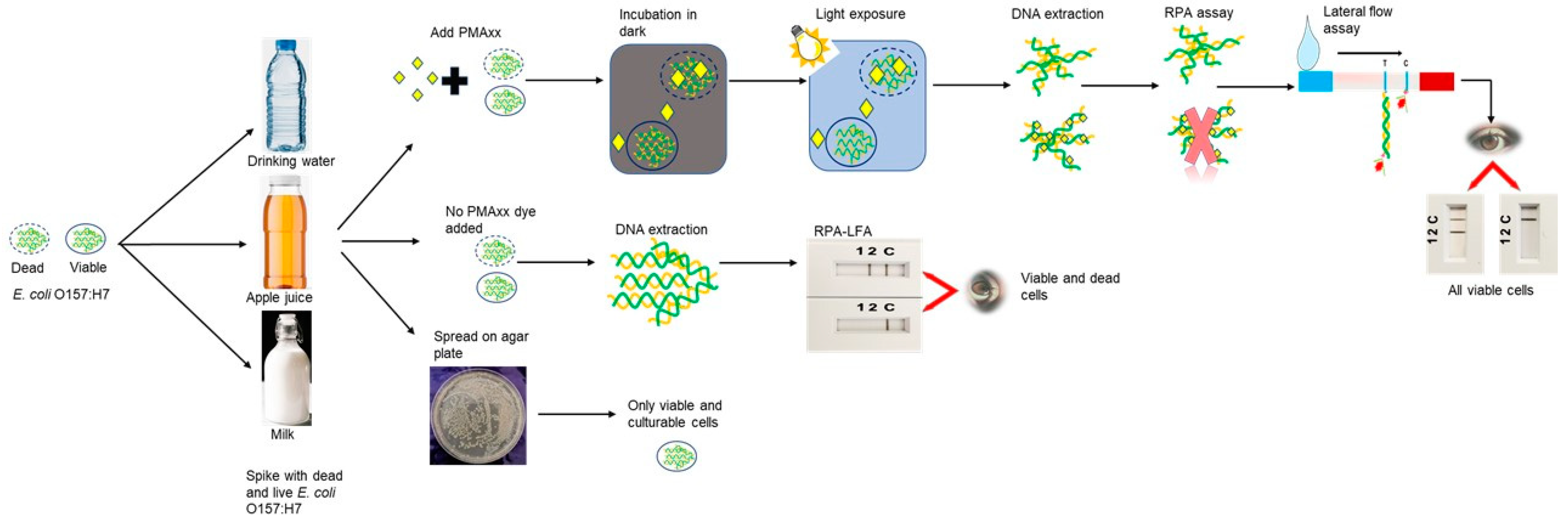
2.5. Validation of the PMAxx-Recombinase Polymerase Amplification (RPA) in Spiked Commercial Samples
2.6. Induction of Viable but Non Culturable (VBNC) E. coli O157:H7
2.7. Analysis of Viable but Non Culturable (VBNC) E. coli O157:H7 in Spiked Samples
2.8. Effect of pH on the Efficiency of Assay
3. Discussion
4. Material and Methods
4.1. Bacterial Isolates and Culture Conditions
4.2. Preparation of Non-Viable Cells
4.3. Propidium Monoazide Optimisation and DNA Extraction
4.4. Propidium Monoazide-Recombinase Polymerase Assay (PMAxx-RPA Assay)
4.5. Specificity of the PMAxx-Recombinase Polymerase Assay-Lateral Flow Assay (RPA-LFA)
4.6. Assessment of PMAxx-Recombinase Polymerase Assay for Pure E. coli O157:H7 Cultures and Spiked Commercial Samples
4.7. Induction of Viable but Non Culturable (VBNC) E. coli O157:H7
4.8. Detection of Viable but Not Culturable (VBNC) E. coli O157:H7 in Artificially Contaminated Samples
4.9. Effect of pH on the PMAxx-Recombinase Polymerase Assay-Lateral Flow Assay (RPA-LFA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, M.; Xu, L.; Jiang, H.; Zhou, Z.; Zhou, S.; Zhang, L. PMA-LAMP for rapid detection of Escherichia coli and shiga toxins from viable but non-culturable state. Microb. Pathog. 2017, 105, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Hara-Kudo, Y.; Takatori, K. Contamination level and ingestion dose of foodborne pathogens associated with infections. Epidemiol. Infect. 2011, 139, 1505–1510. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic Colitis Associated with a Rare Escherichia coli Serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, K.; Hong, W.; Bai, C.; Chen, L.; Fu, X.; Huang, T.; Liu, J. Development and Application of a Simple “Easy To Operate” Propidium Monoazide-Crossing Priming Amplification on Detection of Viable and Viable But Non-culturable Cells of O157 Escherichia coli. Front. Microbiol. 2020, 11, 569105. [Google Scholar] [CrossRef] [PubMed]
- Gill, A.; Huszczynski, G. Enumeration of Escherichia coli O157:H7 in Outbreak-Associated Beef Patties. J. Food Prot. 2016, 79, 1266–1268. [Google Scholar] [CrossRef]
- Furukawa, I.; Suzuki, M.; Masaoka, T.; Nakajima, N.; Mitani, E.; Tasaka, M.; Teranishi, H.; Matsumoto, Y.; Koizumi, M.; Ogawa, A.; et al. Outbreak of Enterohemorrhagic Escherichia coli O157:H7 Infection Associated with Minced Meat Cutlets Consumption in Kanagawa, Japan. Jpn. J. Infect. Dis. 2018, 71, 436–441. [Google Scholar] [CrossRef]
- Gill, A.; Oudit, D.; Alexander, P. Enumeration of Escherichia coli O157 in Outbreak-Associated Gouda Cheese Made with Raw Milk. J. Food Prot. 2015, 78, 1733–1737. [Google Scholar] [CrossRef]
- Heijnen, L.; Medema, G. Quantitative detection of E. coli, E. coli O157 and other shiga toxin producing E. coli in water samples using a culture method combined with real-time PCR. J. Water Health 2006, 4, 487–498. [Google Scholar] [CrossRef]
- CDC. Multistate Outbreak of E. coli O157:H7 Infections Linked to Romaine Lettuce (Final Update); Centers for Disease Control and Prevention: Atlanta, GA, USA, 2018.
- Zhou, B.; Liang, T.; Zhan, Z.; Liu, R.; Li, F.; Xu, H. Rapid and simultaneous quantification of viable Escherichia coli O157: H7 and Salmonella spp. in milk through multiplex real-time PCR. J. Dairy Sci. 2017, 100, 8804–8813. [Google Scholar] [CrossRef]
- Zhao, X.; Zhong, J.; Wei, C.; Lin, C.-W.; Ding, T. Current Perspectives on Viable but Non-culturable State in Foodborne Pathogens. Front. Microbiol. 2017, 8, 580. [Google Scholar] [CrossRef]
- Zhang, S.; Ye, C.; Lin, H.; Lv, L.; Yu, X. UV Disinfection Induces a Vbnc State in Escherichia coli and Pseudomonas aeruginosa. Environ. Sci. Technol. 2015, 49, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 2010, 34, 415–425. [Google Scholar] [CrossRef]
- Han, L.; Wang, K.; Ma, L.; Delaquis, P.; Bach, S.; Feng, J.; Lu, X. Viable but Nonculturable Escherichia coli O157:H7 and Salmonella enterica in Fresh Produce: Rapid Determination by Loop-Mediated Isothermal Amplification Coupled with a Propidium Monoazide Treatment. Appl. Environ. Microbiol. 2020, 86, e02566-19. [Google Scholar] [CrossRef] [PubMed]
- Ramamurthy, T.; Ghosh, A.; Pazhani, G.P.; Shinoda, S. Current perspectives on viable but non-culturable (VBNC) pathogenic bacteria. Front. Public Health 2014, 2, 103. [Google Scholar] [CrossRef] [PubMed]
- Highmore, C.J.; Warner, J.C.; Rothwell, S.D.; Wilks, S.A.; Keevil, C.W. Viable-but-Nonculturable Listeria monocytogenes and Salmonella enterica Serovar Thompson Induced by Chlorine Stress Remain Infectious. mBio 2018, 9, e00540-18. [Google Scholar] [CrossRef]
- Makino, S.I.; Kii, T.; Asakura, H.; Shirahata, T.; Ikeda, T.; Takeshi, K.; Itoh, K. Does enterohemorrhagic Escherichia coli O157: H7 enter the viable but nonculturable state in salted salmon roe? Appl. Environ. Microbiol. 2000, 66, 5536–5539. [Google Scholar] [CrossRef][Green Version]
- Health Canada Compendium of Analytical Methods; Health Canada: Ottawa, ON, Canada, 2015.
- Kumar, S.S.; Ghosh, A.R. Assessment of bacterial viability: A comprehensive review on recent advances and challenges. Microbiology 2019, 165, 593–610. [Google Scholar] [CrossRef]
- Waswa, J.; Irudayaraj, J.; DebRoy, C. Direct detection of E. coli O157:H7 in selected food systems by a surface plasmon resonance biosensor. LWT Food Sci. Technol. 2007, 40, 187–192. [Google Scholar] [CrossRef]
- Rani, A.; Ravindran, V.B.; Surapaneni, A.; Mantri, N.; Ball, A.S. Review: Trends in point-of-care diagnosis for Escherichia coli O157:H7 in food and water. Int. J. Food Microbiol. 2021, 349, 109233. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.; Chousalkar, K.K. Development of PMAxxTM-Based qPCR for the Quantification of Viable and Non-viable Load of Salmonella from Poultry Environment. Front. Microbiol. 2020, 11, 581201. [Google Scholar] [CrossRef]
- Rani, A.; Donovan, N.; Mantri, N. The future of plant pathogen diagnostics in a nursery production system. Biosens. Bioelectron. 2019, 145, 111631. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J.; von Stetten, F. A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2018, 144, 31–67. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Zhang, B.; Wang, J.; Li, B.; He, K.; Zhang, X. Rapid Detection of Escherichia coli O157:H7 by Loop-Mediated Isothermal Amplification Coupled with a Lateral Flow Assay Targeting the z3276 Genetic Marker. Food Anal. Methods 2022, 15, 908–916. [Google Scholar] [CrossRef]
- Xie, R.; Gao, J.; Li, H.; Yu, W.; Zhang, J.; Wang, N.; Chen, A. Real-time Fluorescence and Visual Colorimetric Loop–Mediated Isothermal Amplification Assays for the Rapid and Visual Identification of the Genus Diodon. Food Anal. Methods 2022, 15, 2487–2495. [Google Scholar] [CrossRef]
- Baymiev, A.K.; Kuluev, B.R.; Shvets, K.Y.; Yamidanov, R.S.; Matniyazov, R.T.; Chemeris, D.A.; Zubov, V.V.; Alekseev, Y.I.; Mavzyutov, A.R.; Ivanenkov, Y.A.; et al. Modern Approaches to Differentiation of Live and Dead Bacteria Using Selective Amplification of Nucleic Acids. Microbiology 2020, 89, 13–27. [Google Scholar] [CrossRef]
- Nocker, A.; Cheung, C.-Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.; Zhou, D.; Xie, G.; Liu, J.; Wang, Z.; Xiong, Q.; Xu, H. Real-time recombinase-aided amplification with improved propidium monoazide for the rapid detection of viable Escherichia coli O157:H7 in milk. J. Dairy Sci. 2022, 105, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Truchado, P.; Gil, M.I.; Kostic, T.; Allende, A. Optimization and validation of a PMA qPCR method for Escherichia coli quantification in primary production. Food Control 2016, 62, 150–156. [Google Scholar] [CrossRef]
- Gao, S.; Sun, C.; Hong, H.; Gooneratne, R.; Mutukumira, A.; Wu, X. Rapid detection of viable Cronobacter sakazakii in powdered infant formula using improved propidium monoazide (PMAxx) and quantitative recombinase polymerase amplification (qRPA) assay. Food Control 2021, 124, 107899. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Ravindran, V.B.; Surapaneni, A.; Shahsavari, E.; Haleyur, N.; Mantri, N.; Ball, A.S. Evaluation and comparison of recombinase polymerase amplification coupled with lateral-flow bioassay for Escherichia coli O157:H7 detection using different genes. Sci. Rep. 2021, 11, 1881. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, Y.; Su, H.; Ding, H.; Sun, X.; Gao, H.; Geng, Y.; Wang, Z. Rapid analysis of Escherichia coli O157:H7 using isothermal recombinase polymerase amplification combined with triple-labeled nucleotide probes. Mol. Cell. Probes 2020, 50, 101501. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.; Jung, J.H.; Park, B.H.; Oh, S.J.; Seo, J.H.; Choi, J.S.; Kim, D.H.; Seo, T.S. A centrifugal direct recombinase polymerase amplification (direct-RPA) microdevice for multiplex and real-time identification of food poisoning bacteria. Lab A Chip 2016, 16, 2309–2316. [Google Scholar] [CrossRef] [PubMed]
- March, S.B.; Ratnam, S. Latex agglutination test for detection of Escherichia coli serotype O157. J. Clin. Microbiol. 1989, 27, 1675–1677. [Google Scholar] [CrossRef] [PubMed]
- Van Alfen, N.K. Food Microbiology. In Encyclopedia of Agriculture and Food Systems; Van Alfen, N.K., Ed.; Academic Press: Oxford, UK, 2014; pp. 213–231. [Google Scholar]
- Foddai, A.C.G.; Grant, I.R. Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef] [PubMed]
- Ravan, H.; Amandadi, M.; Sanadgol, N. A highly specific and sensitive loop-mediated isothermal amplification method for the detection of Escherichia coli O157:H7. Microb. Pathog. 2016, 91, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Erfanmanesh, M.; Afshar, D.; Mohammadi, M.; Ghaderi, O.; Haghnazari, A. Visual Detection of Enterohemorrhagic Escherichia coli O157:H7 Using Loop-Mediated Isothermal Amplification. Electron. Physician 2016, 8, 2576–2585. [Google Scholar] [CrossRef] [PubMed]
- Girones, R.; Ferrús, M.A.; Alonso, J.L.; Rodriguez-Manzano, J.; Calgua, B.; Corrêa, A.D.A.; Hundesa, A.; Carratalà, A.; Bofill-Mas, S. Molecular detection of pathogens in water—The pros and cons of molecular techniques. Water Res. 2010, 44, 4325–4339. [Google Scholar] [CrossRef] [PubMed]
- Lampel, K.A.; Wilson, G. Chapter 1—Food industry current status. In Molecular Microbial Diagnostic Methods; Cook, N., D’Agostino, M., Thompson, K.C., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 1–17. [Google Scholar]
- Zhang, Z.; Liu, W.; Xu, H.; Aguilar, Z.P.; Shah, N.P.; Wei, H. Propidium monoazide combined with real-time PCR for selective detection of viable Staphylococcus aureus in milk powder and meat products. J. Dairy Sci. 2015, 98, 1625–1633. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yan, L.; Wu, X.; Li, F.; Wang, D.; Xu, H. Multiplex PCR coupled with propidium monoazide for the detection of viable Cronobacter sakazakii, Bacillus cereus, and Salmonella spp. in milk and milk products. J. Dairy Sci. 2017, 100, 7874–7882. [Google Scholar] [CrossRef] [PubMed]
- El-Aziz, N.K.; Tartor, Y.H.; El-Aziz Gharib, A.A.; Ammar, A.M. Propidium Monoazide Quantitative Real-Time Polymerase Chain Reaction for Enumeration of Some Viable but Nonculturable Foodborne Bacteria in Meat and Meat Products. Foodborne Pathog. Dis. 2018, 15, 226–234. [Google Scholar] [CrossRef]
- Li, B.; Chen, J.-Q. Real-Time PCR Methodology for Selective Detection of Viable Escherichia coli O157:H7 Cells by Targeting Z3276 as a Genetic Marker. Appl. Environ. Microbiol. 2012, 78, 5297–5304. [Google Scholar] [CrossRef]
- Dinu, L.-D.; Bach, S. Detection of viable but non-culturable Escherichia coli O157:H7 from vegetable samples using quantitative PCR with propidium monoazide and immunological assays. Food Control 2013, 31, 268–273. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, F.; Xu, H.; Aguilar, Z.P.; Niu, R.; Yuan, Y.; Sun, J.; You, X.; Lai, W.; Xiong, Y.; et al. Magnetic nano-beads based separation combined with propidium monoazide treatment and multiplex PCR assay for simultaneous detection of viable Salmonella Typhimurium, Escherichia coli O157:H7 and Listeria monocytogenes in food products. Food Microbiol. 2013, 34, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Powell, M.L.; Bowler, F.R.; Martinez, A.J.; Greenwood, C.J.; Armes, N.; Piepenburg, O. New Fpg probe chemistry for direct detection of recombinase polymerase amplification on lateral flow strips. Anal. Biochem. 2018, 543, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Murinda, S.E.; Ibekwe, A.M.; Zulkaffly, S.; Cruz, A.; Park, S.; Razak, N.; Paudzai, F.M.; Ab Samad, L.; Baquir, K.; Muthaiyah, K.; et al. Real-Time Isothermal Detection of Shiga Toxin–Producing Escherichia coli Using Recombinase Polymerase Amplification. Foodborne Pathog. Dis. 2014, 11, 529–536. [Google Scholar] [CrossRef]
- Ibekwe, M.A.; Murinda, S.E.; Park, S.; Obayiuwana, A.; Murry, M.A.; Schwartz, G.; Lundquist, T. Comparative Use of Quantitative PCR (qPCR), Droplet Digital PCR (ddPCR), and Recombinase Polymerase Amplification (RPA) in the Detection of Shiga Toxin-Producing E. coli (STEC) in Environmental Samples. Water 2020, 12, 3507. [Google Scholar] [CrossRef]
- Kaushik, R.; Balasubramanian, R. Discrimination of viable from non-viable Gram-negative bacterial pathogens in airborne particles using propidium monoazide-assisted qPCR. Sci. Total Environ. 2013, 449, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Zhao, X. Detection of viable but non-culturable Escherichia coli O157:H7 by PCR in combination with propidium monoazide. 3 Biotech 2018, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Salas-Massó, N.; Linh, Q.T.; Chin, W.H.; Wolff, A.; Andree, K.B.; Furones, M.D.; Figueras, M.J.; Bang, D.D. The Use of a DNA-Intercalating Dye for Quantitative Detection of Viable Arcobacter spp. Cells (v-qPCR) in Shellfish. Front. Microbiol. 2019, 10, 368. [Google Scholar] [CrossRef]
- Contreras, P.J.; Urrutia, H.; Sossa, K.; Nocker, A. Effect of PCR amplicon length on suppressing signals from membrane-compromised cells by propidium monoazide treatment. J. Microbiol. Methods 2011, 87, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Van Holm, W.; Ghesquière, J.; Boon, N.; Verspecht, T.; Bernaerts, K.; Zayed, N.; Chatzigiannidou, I.; Teughels, W. A Viability Quantitative PCR Dilemma: Are Longer Amplicons Better? Appl. Environ. Microbiol. 2021, 87, e02653-20. [Google Scholar] [CrossRef]
- Elizaquível, P.; Sánchez, G.; Aznar, R. Quantitative detection of viable foodborne E. coli O157:H7, Listeria monocytogenes and Salmonella in fresh-cut vegetables combining propidium monoazide and real-time PCR. Food Control 2012, 25, 704–708. [Google Scholar] [CrossRef]
- Magajna, B.; Schraft, H. Evaluation of Propidium Monoazide and Quantitative PCR To Quantify Viable Campylobacter jejuni Biofilm and Planktonic Cells in Log Phase and in a Viable but Nonculturable State. J. Food Prot. 2015, 78, 1303–1311. [Google Scholar] [CrossRef]
- Lee, J.W.; Nguyen, V.D.; Seo, T.S. Paper-based Molecular Diagnostics for the Amplification and Detection of Pathogenic Bacteria from Human Whole Blood and Milk without a Sample Preparation Step. BioChip J. 2019, 13, 243–250. [Google Scholar] [CrossRef]
- Zou, Y.; Mason, M.G.; Wang, Y.; Wee, E.; Turni, C.; Blackall, P.J.; Trau, M.; Botella, J.R. Nucleic acid purification from plants, animals and microbes in under 30 seconds. PLoS Biol. 2017, 15, e2003916. [Google Scholar] [CrossRef] [PubMed]



| S. No. | Bacteria | Strain | Serotype | rfbE Gene | stx Gene | ||
|---|---|---|---|---|---|---|---|
| PMAxx-RPA-LFA | PMAxx-RPA-EF | PMAxx-RPA-LFA | PMAxx-RPA-EF | ||||
| 1. | E. coli | ATCC 43888 | O157:H7 | − | − | + | + |
| 2. | E. coli | ATCC 43895 | O157:H7 | − | − | + | + |
| 3. | E. coli | ATCC 8740 | O157:H7 | − | − | + | + |
| 4. | E. coli | NCTC 8620 | O26:H- | − | − | − | − |
| 5. | E. coli | ATCC 25922 | O6 biotype 1 | − | − | − | − |
| 6. | E. coli | NCTC 9001 | O1:K1:H7 | − | − | − | − |
| 7. | E. coli | ATCC 23542 | O129:H11 | − | − | − | − |
| 8. | E. coli | NCTC 11560 | − | − | − | − | |
| 9. | E. coli | ATCC 35218 | − | − | − | − | |
| 10. | E. coli | NCTC 8196 | O103 | − | − | − | − |
| 11. | E. coli | ATCC 27325 | W3110 | − | − | − | − |
| 12. | E. coli | ATCC 23716 | K-12 | − | − | − | − |
| 13. | E. coli | Unknown | BL21 | − | − | − | − |
| 14. | Staphylococcus aureus | ATCC 6538 | Unknown | − | − | − | − |
| 15. | Bacillus cereus | ATCC 11778 | Unknown | − | − | − | − |
| 16. | Klebsiella pneumonia | ATCC 13883 | Unknown | − | − | − | − |
| 17. | Pseudomonas aerouginosa | ATCC 27853 | Unknown | − | − | − | − |
| 18. | Shigella dysenteriae | NCTC 4837 | Unknown | − | − | − | − |
| 19. | Shigella flexneri | Unknown | Unknown | − | − | − | − |
| 20. | Salmonella enteritidis | ATCC 13076 | Unknown | − | − | − | − |
| 21. | Listeria monocytogenes | ATCC 19115 | Unknown | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rani, A.; Dike, C.C.; Mantri, N.; Ball, A. Point-of-Care Lateral Flow Detection of Viable Escherichia coli O157:H7 Using an Improved Propidium Monoazide-Recombinase Polymerase Amplification Method. Foods 2022, 11, 3207. https://doi.org/10.3390/foods11203207
Rani A, Dike CC, Mantri N, Ball A. Point-of-Care Lateral Flow Detection of Viable Escherichia coli O157:H7 Using an Improved Propidium Monoazide-Recombinase Polymerase Amplification Method. Foods. 2022; 11(20):3207. https://doi.org/10.3390/foods11203207
Chicago/Turabian StyleRani, Alka, Charles Chinyere Dike, Nitin Mantri, and Andrew Ball. 2022. "Point-of-Care Lateral Flow Detection of Viable Escherichia coli O157:H7 Using an Improved Propidium Monoazide-Recombinase Polymerase Amplification Method" Foods 11, no. 20: 3207. https://doi.org/10.3390/foods11203207
APA StyleRani, A., Dike, C. C., Mantri, N., & Ball, A. (2022). Point-of-Care Lateral Flow Detection of Viable Escherichia coli O157:H7 Using an Improved Propidium Monoazide-Recombinase Polymerase Amplification Method. Foods, 11(20), 3207. https://doi.org/10.3390/foods11203207





