Emergence and Genomic Characterization of the First Reported optrA-Carrying Linezolid-Resistant Enterococci Isolated from Retail Broiler Meat in the United Arab Emirates
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Setting and Chicken Samples
2.2. Isolates Identification and Screening for Antimicrobial Resistance
2.3. Whole-Genome Sequencing Analysis
2.4. Genome Sequence Data Availability
3. Results
3.1. Phenotypic and PCR-Based Confirmation of Linezolid Resistance
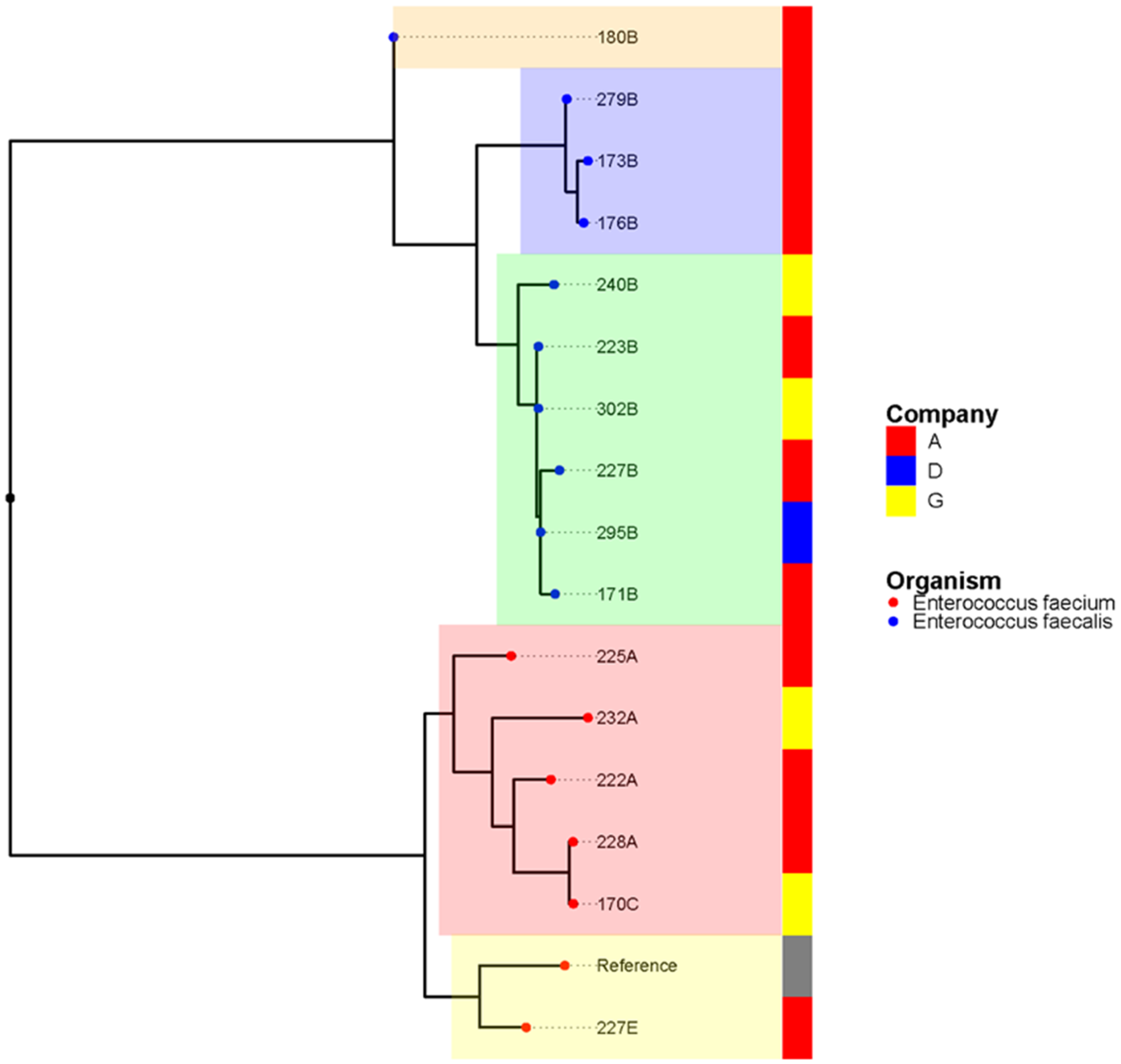
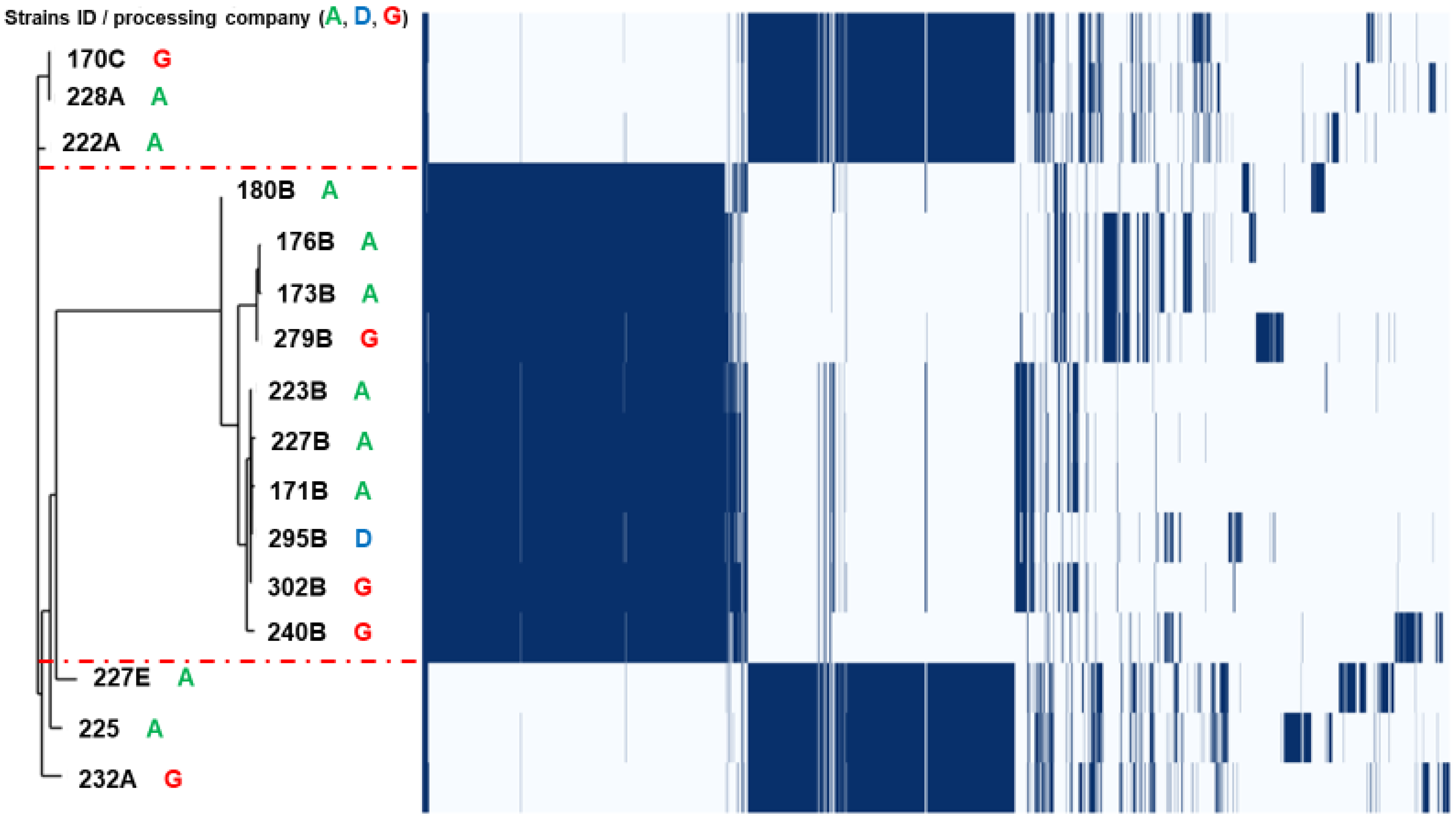
3.2. Population Structure and Clonal Relationship
3.3. Genomic Analysis of Antimicrobial Resistance
3.4. Genomic Insight into Virulence Traits
4. Discussion
4.1. A Concern for the Chicken Industry and Human Health
4.2. Clonal Relationship from a One Health Perspective
4.3. Genomic Insight into Multidrug Resistance and Virulence Traits
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cattoir, V. The multifaceted lifestyle of enterococci: Genetic diversity, ecology and risks for public health. Curr. Opin. Microbiol. 2022, 65, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Somily, A.M.; Al-Mohizea, M.M.; Absar, M.M.; Fatani, A.J.; Ridha, A.M.; Al-Ahdal, M.N.; Senok, A.C.; Al-Qahtani, A.A. Molecular epidemiology of vancomycin resistant enterococci in a tertiary care hospital in Saudi Arabia. Microb. Pathog. 2016, 97, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Graham, K.; Stack, H.; Rea, R. Safety, beneficial and technological properties of enterococci for use in functional food applications—A review. Crit. Rev. Food Sci. Nutr. 2020, 60, 3836–3861. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Lester, C.H.; Heuer, O.E. Antimicrobial-resistant enterococci in animals and meat: A human health hazard? Foodborne Pathog. Dis. 2010, 7, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.A.; Bodolea, C.; Vlase, L.; Hiriscau, E.I.; Popa, A. Linezolid Administration to Critically Ill Patients: Intermittent or Continuous Infusion? A Systematic Literature Search and Review. Antibiotics 2022, 11, 436. [Google Scholar] [CrossRef] [PubMed]
- Sadowy, E. Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid 2018, 99, 89–98. [Google Scholar] [CrossRef]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; Leon-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6, 185–227. [Google Scholar] [CrossRef]
- Habib, I.; Lakshmi, G.B.; Mohamed, M.I.; Ghazawi, A.; Khan, M.; Li, D. Enumeration, Antimicrobial Resistance, and Virulence Genes Screening of Enterococcus spp. Isolated from Retail Chicken Carcasses in the United Arab Emirates. Foodborne Pathog. Dis. 2022, 19, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Kathariou, S.; Winkler, A.; Skandamis, P.; Saint-Cyr, M.J.; Rouzeau-Szynalski, K.; Amezquita, A. Next generation microbiological risk assessment: Opportunities of whole genome sequencing (WGS) for foodborne pathogen surveillance, source tracking and risk assessment. Int. J. Food Microbiol. 2018, 287, 3–9. [Google Scholar] [CrossRef]
- Boss, R.; Overesch, G.; Baumgartner, A. Antimicrobial Resistance of Escherichia coli, Enterococci, Pseudomonas aeruginosa, and Staphylococcus aureus from Raw Fish and Seafood Imported into Switzerland. J. Food Prot. 2016, 79, 1240–1246. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Egan, S.A.; Shore, A.C.; O’Connell, B.; Brennan, G.I.; Coleman, D.C. Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: High prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J. Antimicrob. Chemother. 2020, 75, 1704–1711. [Google Scholar] [CrossRef]
- Morroni, G.; Brenciani, A.; Antonelli, A.; D’Andrea, M.M.; Di Pilato, V.; Fioriti, S.; Mingoia, M.; Vignaroli, C.; Cirioni, O.; Biavasco, F.; et al. Characterization of a Multiresistance Plasmid Carrying the optrA and cfr Resistance Genes from an Enterococcus faecium Clinical Isolate. Front. Microbiol. 2018, 9, 2189. [Google Scholar] [CrossRef]
- Chen, H.; Wang, X.; Yin, Y.; Li, S.; Zhang, Y.; Wang, Q.; Wang, H. Molecular characteristics of oxazolidinone resistance in enterococci from a multicenter study in China. BMC Microbiol. 2019, 19, 162. [Google Scholar] [CrossRef]
- Nuesch-Inderbinen, M.; Haussmann, A.; Treier, A.; Zurfluh, K.; Biggel, M.; Stephan, R. Fattening Pigs Are a Reservoir of Florfenicol-Resistant Enterococci Harboring Oxazolidinone Resistance Genes. J. Food Prot. 2022, 85, 740–746. [Google Scholar] [CrossRef]
- Freitas, A.R.; Tedim, A.P.; Novais, C.; Lanza, V.F.; Peixe, L. Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb. Genom. 2020, 6, e000350. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial resistance in the food chain: A review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef]
- Kim, Y.B.; Seo, K.W.; Jeon, H.Y.; Lim, S.K.; Sung, H.W.; Lee, Y.J. Molecular characterization of erythromycin and tetracycline-resistant Enterococcus faecalis isolated from retail chicken meats. Poult. Sci. 2019, 98, 977–983. [Google Scholar] [CrossRef]
- Cauwerts, K.; Decostere, A.; De Graef, E.M.; Haesebrouck, F.; Pasmans, F. High prevalence of tetracycline resistance in Enterococcus isolates from broilers carrying the erm(B) gene. Avian Pathol. 2007, 36, 395–399. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef]
- Sanderson, H.; Ortega-Polo, R.; Zaheer, R.; Goji, N.; Amoako, K.K.; Brown, R.S.; Majury, A.; Liss, S.N.; McAllister, T.A. Comparative genomics of multidrug-resistant Enterococcus spp. isolated from wastewater treatment plants. BMC Microbiol. 2020, 20, 20. [Google Scholar] [CrossRef]
- Ben Said, L.; Klibi, N.; Dziri, R.; Borgo, F.; Boudabous, A.; Ben Slama, K.; Torres, C. Prevalence, antimicrobial resistance and genetic lineages of Enterococcus spp. from vegetable food, soil and irrigation water in farm environments in Tunisia. J. Sci. Food Agric. 2016, 96, 1627–1633. [Google Scholar] [CrossRef]
- Sparo, M.; Urbizu, L.; Solana, M.V.; Pourcel, G.; Delpech, G.; Confalonieri, A.; Ceci, M.; Sanchez Bruni, S.F. High-level resistance to gentamicin: Genetic transfer between Enterococcus faecalis isolated from food of animal origin and human microbiota. Lett. Appl. Microbiol. 2012, 54, 119–125. [Google Scholar] [CrossRef]
- Choi, J.M.; Woo, G.J. Transfer of tetracycline resistance genes with aggregation substance in foodborne Enterococcus faecalis. Curr. Microbiol. 2015, 70, 476–484. [Google Scholar] [CrossRef]
- O’Dea, M.; Sahibzada, S.; Jordan, D.; Laird, T.; Lee, T.; Hewson, K.; Pang, S.; Abraham, R.; Coombs, G.W.; Harris, T.; et al. Genomic, Antimicrobial Resistance, and Public Health Insights into Enterococcus spp. from Australian Chickens. J. Clin. Microbiol. 2019, 57, e00319-19. [Google Scholar] [CrossRef]
- Aslam, M.; Diarra, M.S.; Checkley, S.; Bohaychuk, V.; Masson, L. Characterization of antimicrobial resistance and virulence genes in Enterococcus spp. isolated from retail meats in Alberta, Canada. Int. J. Food Microbiol. 2012, 156, 222–230. [Google Scholar] [CrossRef]
- Hancock, L.E.; Perego, M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 2004, 186, 5629–5639. [Google Scholar] [CrossRef]
- Thurlow, L.R.; Thomas, V.C.; Narayanan, S.; Olson, S.; Fleming, S.D.; Hancock, L.E. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect. Immun. 2010, 78, 4936–4943. [Google Scholar] [CrossRef]
- Semedo, T.; Santos, M.A.; Lopes, M.F.; Figueiredo Marques, J.J.; Barreto Crespo, M.T.; Tenreiro, R. Virulence factors in food, clinical and reference Enterococci: A common trait in the genus? Syst. Appl. Microbiol. 2003, 26, 13–22. [Google Scholar] [CrossRef]
- Seputiene, V.; Bogdaite, A.; Ruzauskas, M.; Suziedeliene, E. Antibiotic resistance genes and virulence factors in Enterococcus faecium and Enterococcus faecalis from diseased farm animals: Pigs, cattle and poultry. Pol. J. Vet. Sci. 2012, 15, 431–438. [Google Scholar]
- Eaton, T.J.; Gasson, M.J. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 2001, 67, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
| Species | Strains | Company | Antimicrobial Resistance Phenotype * |
|---|---|---|---|
| Enterococcus faecalis | 171B | A | CIP-ERYTH-LIN-TET |
| 223B | A | CIP-ERYTH-LIN | |
| 227B | A | CIP-ERYTH-LIN-TET | |
| 173B | A | CIP-ERYTH-LIN-TET | |
| 176B | A | CIP-ERYTH-LIN-TET | |
| 180B | A | HLS-CIP-ERYTH-LIN-TET | |
| 295B | D | HLG-CIP-ERYTH-LIN-TET | |
| 279B | G | HLG-CIP-ERYTH-LIN-TET | |
| 302B | G | HLG-HLS-CIP-ERYTH-LIN-TET | |
| 240B | G | HLG-HLS-CIP-ERYTH-LIN-TET | |
| 288B ** | G | HLG-CIP-ERYTH-LIN-TET | |
| Enterococcus faecium | 227E | A | HLG-CIP-ERYTH-LIN-TET |
| 222A | A | HLG-HLS-CIP-ERYTH-LIN-TET | |
| 225A | A | ERYTH-LIN-TET | |
| 228A | A | HLS-CIP-ERYTH-LIN-TET | |
| 170C | G | HLG-HLS-CIP-ERYTH-LIN-TET | |
| 232A | G | HLG-HLS-CIP-ERYTH-LIN-TET |
| Species | Strains | ST | Quinolone Point Mutation | Antimicrobial Resistance Genes * | Plasmid Replicon Type | MGEs Associated with ARGs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Linezolid | Lincosamide | Aminoglycoside | Macrolide | Tetracycline | Trimethoprim | Phenicol | ||||||
| Enterococcus faecalis | 171B | 476 | gyrA_S83I and parC_S80I | optrA | - | ant(9)-la and aph(3’)-lll | erm(A), lsa(A), and erm(B) | tet(L) | - | cat and fexA | - | - |
| 223B | 476 | optrA | - | ant(9)-la | erm(A), Isa(A), and erm(B) | - | - | fexA | - | - | ||
| 227B | 476 | gyrA_S83I and parC_S80I | optrA | - | ant(9)-la and aph(3’)-lll | erm(A), lsa(A), and erm(B) | tet(L) | dfrG | cat and fexA | rep9a | - | |
| 173B | 1184 | gyrA_S83I and parC_S80I | optrA | lnu(G) | aph(3’)-lll | Isa(A), erm(A), and erm(B) | tet(L) | dfrG | cat and fexA | rep9a and repUS43 | tet(L), erm(B), aph(3’)-lll, and cat on repUS43 | |
| 176B | 1184 | gyrA_S83I and parC_S80I | optrA | lnu(G) | aph(3’)-lll | Isa(A), erm(A), and erm(B) | tet(L) | dfrG | cat and fexA | rep9a and repUS43 | tet(L), erm(B), aph(3’)-lll, and cat on repUS43 | |
| 180B | 1291 * | gyrA_S83Y and parC_S80I | optrA | lnu(G) | aph(3’)-lll and ant(6)-la | Isa(A) and erm(B) | tet(L) | - | fexA | rep6, rep1, rep9b, and repUS43 | tet(L) on repUS43--- aph(3’)-lll, and ant(6)-la on rep1 | |
| 295B | 476 | gyrA_S83I and parC_S80I | optrA | - | aph(3’)-lll, ant(9)-la, and aac(6’)-aph(2’’) | erm(A), erm(B), and lsa(A) | tet(L) | - | fexA | repUS11 and rep9b | - | |
| 279B | 314 | eatA_T450I | optrA | lnu(G) | aph(3’)-lll and aac(6’)-aph(2’’) | erm(A), erm(B), and lsa(A) | tet(L) | - | cat and fexA | repUS11, repUS43, and rep9a | tet(L), erm(B), and cat on repUS43 | |
| 302B | 476 | gyrA_S83Y and parC_S80I | optrA | lnu(B) | aph(3’)-lll, ant(9)-la, and aac(6’)-aph(2’’) | erm(A), erm(B), Isa(A), and Isa(E) | tet(L) | dfrG | cat and fexA | repUS43 | tet(L) on repUS43 | |
| 240B | 1290 * | eatA_T450I | optrA | lnu(B) | aph(3’)-lll, ant(9)-la, and aac(6’)-aph(2’’) | erm(A), erm(B), lsa(A), and lsa(E) | tet(L) | dfrG | fexA | repUS11, rep9a, and rep6 | - | |
| Enterococcus faecium | 227E | 195 | optrA, poxtA | lnu(G) | aac(6’)-li and aadD | erm(A), msr(C), and erm(B) | tet(L) | dfrG | fexA and fexB | rep2, rep14a, repUS15, and rep29 | - | |
| 222A | 2236 * | gyrA_S83Y and parC_S80I | optrA | - | aph(3’)-lll, aac(6’)-li, and aac(6’)-aph(2’’) | erm(A), msr(C), and erm(B) | tet(M) | dfrE | fexA | repUS1 and repUS15 | optrA, ermA, and fexA on repUS1 | |
| 225A | 2239 * | gyrA_S83I | optrA | - | ant(9)-la and aac(6’)-li | erm(A) and msr(C) | tet(L) | - | fexA | repUS43, repUS15, rep22, and repUS1 | tet(L) on repUS43 | |
| 228A | 2236 * | eatA_T450I | optrA | - | ant(6)-la, aph(3’’)-lll, and aac(6’)-li | msr(C) and ermB | tet(L) | - | fexA | rep2 and repUS15 | - | |
| 170C | 2236 * | eatA_T450I | optrA | - | ant(6)-la, aph(3’’)-lll, aac(6’)-li, and aac(6’)-aph(2’’) | erm(A), msr(C), and erm(B) | tet(L) | dfrE | fexA | rep2 and repUS15 | - | |
| 232A | 2238 * | optrA | lnu(B) | ant(6)-la, aph(3’)-lll, aac(6’)-li, and aac(6’)-aph(2’’) | erm(A), erm(B), and lsa(E) | tet(L) and tet(S) | dfrG | fexA | rep22, repUS43, and repUS15 | tet(L) on rep22 | ||
| Strains * | Virulence Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sex Pheromones | Adhesion | Invasion | Aggregation | Cytolytic Toxin | Biofilm Formation | Antiphagocytic | Immunity | Protease | |
| 171B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | hylA | - | - | - | tpx | ElrA | - |
| 223B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | hylA | - | - | - | tpx | ElrA | - |
| 227B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | - | - | - | - | tpx | ElrA | - |
| 173B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | hylB | - | - | fsrB | tpx | ElrA | gelE |
| 176B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | hylB | - | - | fsrB | tpx | ElrA | gelE |
| 180B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | hylA and hylB | - | - | fsrB | tpx | ElrA | gelE |
| 295B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | hylA | - | cylB, cylL, and cylM | - | tpx | ElrA | - |
| 279B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | hylB | - | - | fsrB | tpx | ElrA | gelE |
| 302B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | hylA | - | - | - | tpx | ElrA | - |
| 240B | cCF10, cOB1, cad, and camE | SrtA, ebpA, ebpC, and efaAfs | hylA | - | - | - | tpx | ElrA | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habib, I.; Ghazawi, A.; Lakshmi, G.B.; Mohamed, M.-Y.I.; Li, D.; Khan, M.; Sahibzada, S. Emergence and Genomic Characterization of the First Reported optrA-Carrying Linezolid-Resistant Enterococci Isolated from Retail Broiler Meat in the United Arab Emirates. Foods 2022, 11, 3190. https://doi.org/10.3390/foods11203190
Habib I, Ghazawi A, Lakshmi GB, Mohamed M-YI, Li D, Khan M, Sahibzada S. Emergence and Genomic Characterization of the First Reported optrA-Carrying Linezolid-Resistant Enterococci Isolated from Retail Broiler Meat in the United Arab Emirates. Foods. 2022; 11(20):3190. https://doi.org/10.3390/foods11203190
Chicago/Turabian StyleHabib, Ihab, Akela Ghazawi, Glindya Bhagya Lakshmi, Mohamed-Yousif Ibrahim Mohamed, Dan Li, Mushtaq Khan, and Shafi Sahibzada. 2022. "Emergence and Genomic Characterization of the First Reported optrA-Carrying Linezolid-Resistant Enterococci Isolated from Retail Broiler Meat in the United Arab Emirates" Foods 11, no. 20: 3190. https://doi.org/10.3390/foods11203190
APA StyleHabib, I., Ghazawi, A., Lakshmi, G. B., Mohamed, M.-Y. I., Li, D., Khan, M., & Sahibzada, S. (2022). Emergence and Genomic Characterization of the First Reported optrA-Carrying Linezolid-Resistant Enterococci Isolated from Retail Broiler Meat in the United Arab Emirates. Foods, 11(20), 3190. https://doi.org/10.3390/foods11203190








