Biochemical and Structural Properties of a High-Temperature-Active Laccase from Bacillus pumilus and Its Application in the Decolorization of Food Dyes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmid, and Chemicals
2.2. Strain Screening and Cultivation
2.3. Phylogenetic Analysis of the Laccase-Producing Strain
2.4. Heterologous Expression of Laccase
2.5. Molecular Docking and Molecular Dynamics Simulation Analysis of rLAC
2.6. Purification of rLAC
2.7. Enzyme Assay
2.8. Characterization of rLAC
2.9. Dye Decolorization
3. Results and Discussion
3.1. Identification of the Strain with Laccase Activity
3.2. Heterologous Expression of Laccase
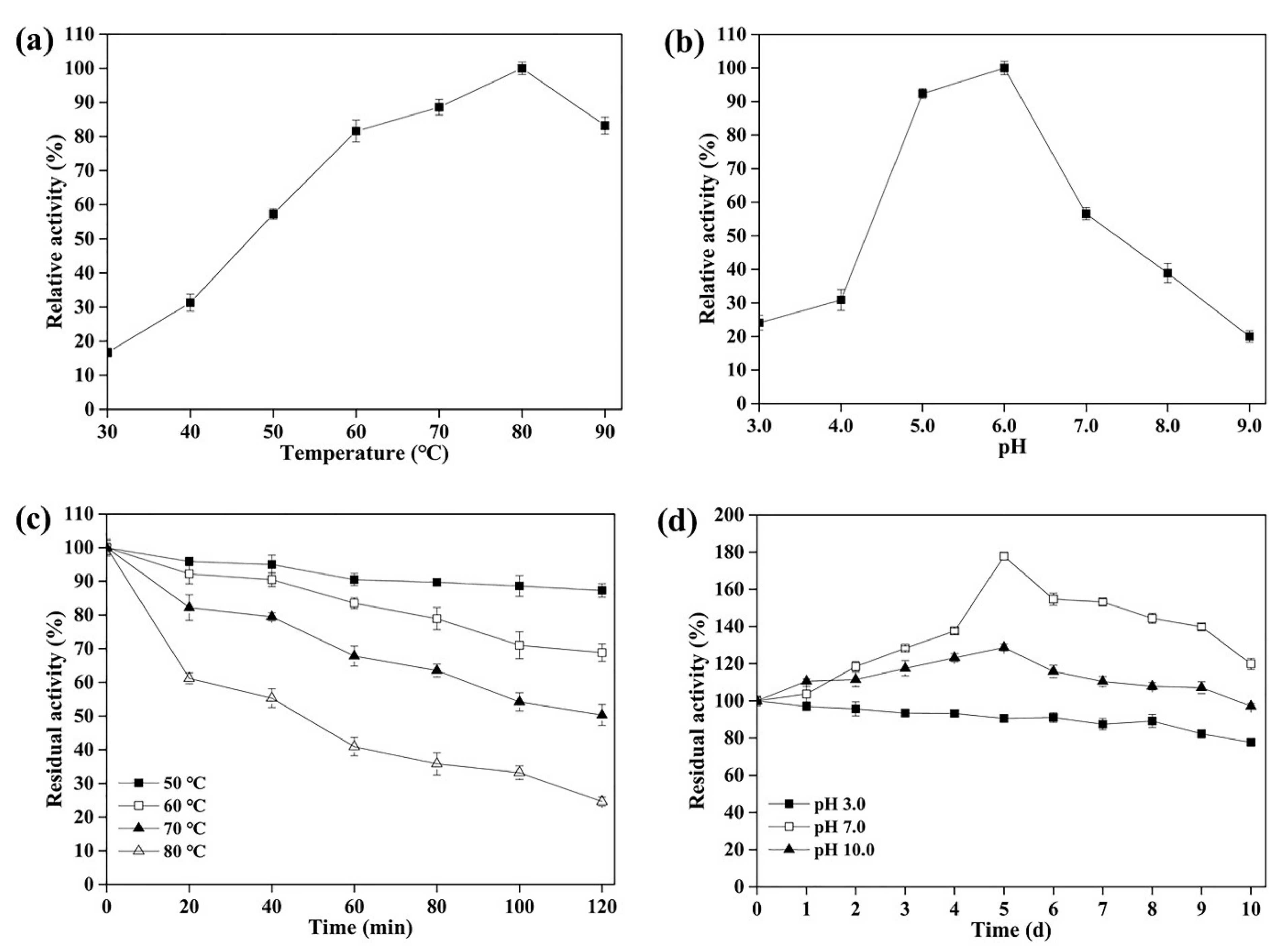
3.3. Effect of Temperature and pH on the Activity and Stability of rLAC
3.4. Influence of Metal Ions and Inhibitors on the Activity of rLAC
3.5. Investigation of the Thermophilic Features of rLAC via Molecular Docking and MD Simulations
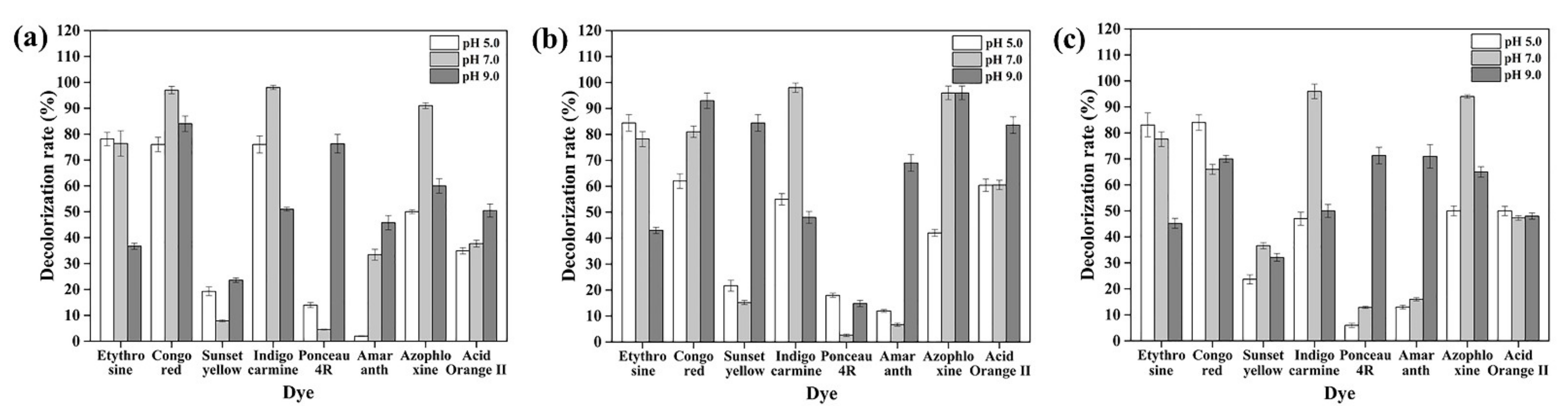
3.6. Dye Decolorization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ghodake, G.S.; Yang, J.; Shinde, S.S.; Mistry, B.M.; Kim, D.Y.; Sung, J.S.; Kadam, A.A. Paper waste extracted α-cellulose fibers super-magnetized and chitosan functionalized for covalent laccase immobilization. Bioresour. Technol. 2018, 261, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Couto, S. Laccases for denim bleaching: An eco-friendly alternative. Open Text. J. 2012, 5, 1–7. [Google Scholar] [CrossRef]
- Sharma, B.; Dangi, A.K.; Shukla, P. Contemporary enzyme based technologies for bioremediation: A review. J. Environ. Manage. 2018, 210, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Forootanfar, H.; Faramarzi, M.A. Insights into laccase producing organisms, fermentation states, purification strategies, and biotechnological applications. Biotechnol. Prog. 2015, 31, 1443–1463. [Google Scholar] [CrossRef] [PubMed]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Sondhi, S.; Sharma, P.; Saini, S.; Puri, N.; Gupta, N. Purification and characterization of an extracellular, thermo-alkali-stable, metal tolerant laccase from Bacillus tequilensis SN4. PLoS ONE 2014, 9, e96951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augustine, A.J.; Kragh, M.E.; Sarangi, R.; Fujii, S.; Liboiron, B.D.; Stoj, C.S.; Kosman, D.J.; Hodgson, K.O.; Hedman, B.; Solomon, E.I. Spectroscopic studies of perturbed T1 Cu sites in the multicopper oxidases Saccharmyces cerevisiae Fet3p and Rhus vernicifera laccase: Allosteric coupling between the T1 and trinuclear Cu sites. Biochemistry 2008, 47, 2036–2045. [Google Scholar] [CrossRef]
- Baldrian, P. Fungal laccases-occurrence and properties. FEMS Microbiol. Rev. 2006, 30, 215–242. [Google Scholar] [CrossRef] [Green Version]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Hilde’n, K.; Hakala, T.; Lundell, T. Thermotolerant and thermostable laccases. Biotechnol. Lett. 2009, 31, 1117–1128. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Goradia, B.; Saxena, A. Bacterial laccase: Recent update on production and industrial applications. 3 Biotech. 2017, 7, 323. [Google Scholar] [CrossRef]
- Martins, L.O.; Soares, C.M.; Pereira, M.M.; Teixeira, M.; Costa, T.; Jones, G.H.; Henriques, A.O. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J. Biol. Chem. 2002, 277, 18849–18859. [Google Scholar] [CrossRef] [Green Version]
- Brander, S.; Mikkelsen, J.D.; Kepp, K.P. Characterization of an alkali- and halide-resistant laccase expressed in E. coli: CotA from Bacillus clausii. PLoS ONE 2014, 9, e99402. [Google Scholar]
- Christensen, N.J.; Kepp, K.P. Stability mechanisms of a thermophilic laccase probed by molecular dynamics. PLoS ONE 2013, 8, e61985. [Google Scholar] [CrossRef] [Green Version]
- Farnet, A.M.; Criquet, S.; Cigna, M.; Gil, G.; Ferré, E. Purification of a laccase from Marasmius quercophilus induced with ferulic acid: Reactivity towards natural and xenobiotic aromatic compounds. Enzym. Microb. Technol. 2004, 34, 549–554. [Google Scholar] [CrossRef]
- Papinutti, L.; Dimitriu, P.; Forchiassin, F. Stabilization studies of Fomes sclerodermeus laccases. Bioresource Technol. 2008, 99, 419–424. [Google Scholar] [CrossRef]
- Liu, Y.H.; Huang, L.; Guo, W.; Jia, L.B.; Fu, Y.; Gui, S.; Lu, F.P. Cloning, expression, and characterization of a thermostable and pH-stable laccase from Klebsiella pneumoniae and its application to dye decolorization. Process. Biochem. 2017, 53, 125–134. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.H.; Huang, L.; Shan, M.Y.; Sang, J.C.; Li, Y.Z.; Jia, L.G.; Wang, N.; Wang, S.; Shao, S.L.; Liu, F.F.; et al. Enhancing the activity and thermostability of Streptomyces mobaraensis transglutaminase by directed evolution and molecular dynamics simulation. Biochem. Eng. J. 2019, 151, 107333. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Kittl, R.; Mueangtoom, K.; Gonaus, C.; Khazaneh, S.T.; Sygmund, C.; Haltrich, D.; Ludwig, R. A chloride tolerant laccase from the plant pathogen ascomycete Botrytis aclada expressed at high levels in Pichia pastoris. J. Biotechnol. 2012, 157, 304–314. [Google Scholar] [CrossRef]
- Lu, L.; Zhao, M.; Li, G.F.; Li, J.; Wang, T.N.; Li, D.B.; Xu, T.F. Decolorization of synthetic dyes by immobilized spore from Bacillus amyloliquefaciens. Catal. Commun. 2012, 26, 58–62. [Google Scholar] [CrossRef]
- Reiss, R.; Ihssen, J.; Thöny-Meyer, L. Bacillus pumilus laccase: A heat stable enzyme with a wide substrate spectrum. BMC Biotechnol. 2011, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Guan, Z.B.; Chen, Y.; Zhang, F.; Cai, Y.J.; Xu, C.W.; Chen, X.S.; Liao, X.R. Production of spore laccase from Bacillus pumilus W3 and its application in dye decolorization after immobilization. Water Sci. Technol. 2017, 76, 147–154. [Google Scholar] [CrossRef]
- Zhang, C.; Diao, H.W.; Lu, F.X.; Bie, X.M.; Wang, Y.F.; Lu, Z.X. Degradation of triphenylmethane dyes using a temperature and pH stable spore laccase from a novel strain of Bacillus vallismortis. J. Bioresour. Technol. 2012, 126, 80–86. [Google Scholar] [CrossRef]
- Wang, H.B.; Huang, L.; Li, Y.Z.; Ma, J.Y.; Wang, S.; Zhang, Y.F.; Ge, X.Q.; Wang, N.; Lu, F.P.; Liu, Y.H. Characterization and application of a novel laccase derived from Bacillus amyloliquefaciens. Int. J. Biol. Macromol. 2020, 150, 982–990. [Google Scholar] [CrossRef]
- Guan, Z.B.; Zhang, N.; Song, C.M.; Zhou, W.; Zhou, L.X.; Zhao, H.; Xu, C.W.; Cai, Y.J.; Liao, X.R. Molecular cloning, characterization, and dye-decolorizing ability of a temperature-and pH-stable laccase from Bacillus subtilis X1. Appl. Biochem. Biotech. 2014, 172, 1147–1157. [Google Scholar] [CrossRef]
- Li, T.; Huang, L.; Li, Y.Z.; Xu, Z.H.; Ge, X.Q.; Zhang, Y.F.; Wang, N.; Wang, S.; Yang, W.; Lu, F.P.; et al. The heterologous expression, characterization, and application of a novel laccase from Bacillus velezensis. Sci. Total Environ. 2020, 713, 136713. [Google Scholar] [CrossRef]
- Koschorreck, K.; Richter, S.M.; Ene, A.B.; Roduner, E.; Schmid, R.D.; Urlacher, V.B. Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic acids. Appl. Microbiol. Biotechnol. 2008, 79, 217–224. [Google Scholar] [CrossRef]
- Solomon, E.I.; Sundaram, U.M.; Machonkin, T.E. Multicopper oxidases and oxygenases. Chem. Rev. 1996, 96, 2563–2606. [Google Scholar] [CrossRef] [PubMed]
- Ruijssenaars, H.J.; Hartmans, S. A cloned Bacillus halodurans multicopper oxidase exhibiting alkaline laccase activity. Appl. Microbiol. Biotechnol. 2004, 65, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Xu, W.Q.; Pan, X.R.; Lu, L. A novel non-blue laccase from Bacillus amyloliquefaciens: Secretory expression and characterization. Int. J. Biol. Macromol. 2015, 76, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, H.B.; Li, J.W.; Jiang, L.Y.; Kang, H.W.; Guo, Z.H.; Wang, C.; Yang, W.; Liu, F.F.; Lu, F.P.; et al. Enzymatic characterization, molecular dynamics simulation, and application of a novel Bacillus licheniformis laccase. Int. J. Biol. Macromol. 2021, 167, 1393–1405. [Google Scholar] [CrossRef]
- Aung, T.; Jiang, H.; Chen, C.C.; Liu, G.L.; Hu, Z.; Chi, Z.M.; Chi, Z. Production, gene cloning, and overexpression of a laccase in the marine-derived yeast Aureobasidium melanogenum strain 11-1 and characterization of the recombinant laccase. Mar. Biotechnol. 2019, 21, 76–87. [Google Scholar] [CrossRef]
- Wang, S.S.; Ning, Y.J.; Wang, S.N.; Zhang, J.; Zhang, G.Q.; Chen, Q.J. Purification, characterization, and cloning of an extracellular laccase with potent dye decolorizing ability from white rot fungus Cerrena unicolor GSM-01. Int. J. Biol. Macromol. 2017, 95, 920–927. [Google Scholar] [CrossRef]
- Wang, S.N.; Chen, Q.J.; Zhu, M.J.; Xue, F.Y.; Li, W.C.; Zhao, T.J.; Li, G.D.; Zhang, G.Q. An extracellular yellow laccase from white rot fungus Trametes sp. F1635 and its mediator systems for dye decolorization. Biochimie 2018, 148, 46–54. [Google Scholar] [CrossRef]
- Shraddha; Shekher, R.; Sehgal, S.; Kamthania, M.; Kumar, A. Laccase: Microbial sources, production, purification, and potential biotechnological applications. Enzym. Res. 2011, 2011, 217861. [Google Scholar]
- Yang, Q.H.; Zhang, M.L.; Zhang, M.M.; Wang, C.Q.; Liu, Y.Y.; Fan, X.J.; Li, H. Characterization of a novel, cold-adapted, and thermostable laccase-like enzyme with high tolerance for organic solvents and salt and potent dye decolorization ability, derived from a marine metagenomic library. Front. Microbiol. 2018, 9, 2998. [Google Scholar] [CrossRef] [Green Version]
- Ossiadacz, J.; Al-Adhami, A.J.H.; Bajraszewska, D.; Fischer, P.; Peczyñska-Czoch, W. On the use of trametes versicolor laccase for the conversion of 4-methyl-3-hydroxyanthranilic acid to actinocin chromophore. J. Biotechnol. 1999, 72, 141–149. [Google Scholar] [CrossRef]
- Siroosi, M.; Amoozegar, M.A.; Khajeh, K.; Dabirmanesh, B. Decolorization of dyes by a novel sodium azide-resistant spore laccase from a halotolerant bacterium, Bacillus safensis sp. strain S31. Water Sci. Technol. 2018, 77, 2867–2875. [Google Scholar] [CrossRef]
- Jimenez-Juarez, N.; Roman-Miranda, R.; Baeza, A.; Sánchez-Amat, A.; VazquezDuhalt, R.; Valderrama, B. Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea. J. Biotechnol. 2005, 117, 73–82. [Google Scholar] [CrossRef]
- Enguita, F.J.; Martins, L.O.; Henriques, A.O.; Carrondo, M.A. Crystal structure of a bacterial endospore coat component: A laccase with enhanced thermostability properties. J. Biol. Chem. 2003, 278, 19416–19425. [Google Scholar] [CrossRef] [Green Version]
- Guan, Z.B.; Song, C.M.; Zhang, N.; Zhou, W.; Xu, C.W.; Zhou, L.X.; Zhao, H.; Cai, Y.J.; Liao, X.R. Overexpression, characterization, and dye-decolorizing ability of a thermostable, pH-stable, and organic solvent-tolerant laccase from Bacillus pumilus W3. J. Mol. Catal. B-Enzym. 2014, 101, 1–6. [Google Scholar] [CrossRef]
- Rodrigues, C.S.D.; Madeira, L.M.; Boaventura, R.A.R. Treatment of textile effluent by chemical (Fenton’s Reagent) and biological (sequencing batch reactor) oxidation. J. Hazard. Mater. 2009, 172, 1551–1559. [Google Scholar] [CrossRef]
- Telke, A.A.; Kadam, A.A.; Jagtap, S.S.; Jadhav, J.P.; Govindwar, S.P. Biochemical characterization and potential for textile dye degradation of blue laccase from Aspergillus ochraceus NCIM-1146. Biotechnol. Bioprocess. Eng. 2010, 15, 696–703. [Google Scholar] [CrossRef]
- Miyazaki, K. A hyperthermophilic laccase from Thermus thermophiles HB27. Extremophiles 2005, 9, 415–425. [Google Scholar] [CrossRef]
- Forootanfar, H.; Faramarzi, M.A.; Shahverdi, A.R.; Yazdi, M.T. Purification and biochemical characterization of extracellular laccase from the ascomycete Paraconiothyrium variabile. Bioresour. Technol. 2011, 102, 1808–1814. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, Y.; Wang, X.J.; Shang, J.Z.; Li, Y.; Zhang, H.T.; Lu, F.P.; Liu, F.F. Biochemical characterization and molecular mechanism of acid denaturation of a novel α-amylase from Aspergillus niger. Biochem. Eng. J. 2018, 137, 222–231. [Google Scholar] [CrossRef]
- Mollania, N.; Heidari, M.; Khajeh, K. Catalytic activation of Bacillus laccase after temperature treatment: Structural & biochemical characterization. Int. J. Biol. Macromol. 2018, 109, 49–56. [Google Scholar]
- Abe, F.R.; Machado, A.L.; Soares, A.M.V.M.; Oliveira, D.P.; Pestana, J.L.T. Life history and behavior effects of synthetic and natural dyes on Daphnia magna. Chemosphere 2019, 236, 124390. [Google Scholar] [CrossRef]
- Gao, J.F.; Zhang, Q.; Wang, J.H.; Wu, X.L.; Wang, S.Y.; Peng, Y.Z. Contributions of functional groups and extracellular polymeric substances on the biosorption of dyes by aerobic granules. Bioresour. Technol. 2011, 102, 805–813. [Google Scholar] [CrossRef]
- Abe, F.R.; Soares, A.M.V.M.; Oliveira, D.P.; Gravato, C. Toxicity of dyes to zebrafish at the biochemical level: Cellular energy allocation and neurotoxicity. Environ. Pollut. 2018, 235, 255–262. [Google Scholar] [CrossRef]
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S.P. Bacterial decolorization and degradation of azo dyes: A review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157. [Google Scholar] [CrossRef]
- Imran, M.; Ashraf, M.; Hussain, S.; Mustafa, A. Microbial biotechnology for detoxification of azo-dye loaded textile effluents: A critical review. Int. J. Agric. Bio. 2019, 22, 1138–1154. [Google Scholar]
- Wang, F.H.; Xu, Z.H.; Wang, C.; Guo, Z.H.; Yuan, Z.T.; Kang, H.W.; Li, J.W.; Lu, F.P.; Liu, Y.H. Biochemical characterization of a tyrosinase from Bacillus aryabhattai and its application. Int. J. Biol. Macromol. 2021, 176, 37–46. [Google Scholar] [CrossRef]
- Couto, S.R.; Herrera, J.L.T. Industrial and biotechnological applications of laccases: A review. Biotechnol. Adv. 2006, 24, 500–513. [Google Scholar] [CrossRef]
- Santhanam, N.; Vivanco, M.; Decker, S.R.; Reardon, K.F. Expression of industrially relevant laccases: Prokaryotic style. Trends Biotechnol. 2011, 29, 480–489. [Google Scholar] [CrossRef]
- Enguita, F.J.; Matias, P.M.; Martins, L.O.; Placido, D.; Henriques, A.O.; Carrondo, M.A. Spore-coat laccase CotA from Bacillus subtilis: Crystallization and preliminary X-ray characterization by the MAD method. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 1490–1493. [Google Scholar] [CrossRef]





| Metal Ions/Inhibitors | Concentration (mM) | Relative Activity (%) a |
|---|---|---|
| None | - | 100.0 ± 1.5 |
| KCl | 0.5 | 102.6 ± 2.7 |
| 5 | 93.2 ± 1.9 | |
| CaCl2 | 0.5 | 96.2 ± 3.2 |
| 5 | 90.8 ± 1.6 | |
| CuCl2 | 0.5 | 95.4 ± 2.1 |
| 5 | 92.7 ± 2.0 | |
| MgCl2 | 0.5 | 98.0 ± 2.9 |
| 5 | 91.1 ± 3.1 | |
| ZnSO4 | 0.5 | 97.4 ± 2.9 |
| 5 | 105.5 ± 1.5 | |
| BaCl2 | 0.5 | 101.7 ± 1.2 |
| 5 | 87.1 ± 0.5 | |
| NiSO4 | 0.5 | 90.9 ± 0.6 |
| 5 | 105.1 ± 3.2 | |
| CoCl2 | 0.5 | 77.5 ± 0.5 |
| 5 | 24.0 ± 0.7 | |
| FeSO4 | 0.5 | 75.8 ± 1.0 |
| 5 | 42.1 ± 0.7 | |
| FeCl3 | 0.5 | 94.0 ± 3.5 |
| 5 | 79.1 ± 1.8 | |
| MnCl2 | 0.5 | 17.5 ± 2.3 |
| 5 | 9.4 ± 0.8 | |
| NaCl | 0.5 | 105.5 ± 3.0 |
| 5 | 89.5 ± 2.3 | |
| 10 | 79.9 ± 2.6 | |
| 100 | 68.7 ± 1.7 | |
| 500 | 50.7 ± 0.9 | |
| 1000 | 0 | |
| Dithiothreitol | 0.5 | 6.2 ± 1.0 |
| 5 | 0 | |
| L-Cysteine | 0.5 | 35.2 ± 3.5 |
| 5 | 27.6 ± 1.6 | |
| β-Mercaptoethanol | 0.5 | 6.5 ± 2.0 |
| 5 | 0 | |
| SDS | 0.5 | 108.7 ± 0.7 |
| 5 | 103.9 ± 2.1 | |
| EDTA | 0.5 | 90.8 ± 1.1 |
| 5 | 62.1 ± 2.8 |
| Temperature (K) | Binding Energy a (kJ/mol) | van der Waals Energy (kJ/mol) | Electrostatic Energy (kJ/mol) | Polar Solvation Energy (kJ/mol) | SASA Energy (kJ/mol) |
|---|---|---|---|---|---|
| 325 | −106.77 ± 21.59 | −189.47 ± 20.04 | −51.29 ± 20.60 | 155.13 ± 27.40 | −21.14 ± 1.61 |
| 355 | −154.14 ± 27.74 | −251.44 ± 30.52 | −40.50 ± 13.12 | 160.48 ± 17.43 | −22.67 ± 1.42 |
| 365 | −135.00 ± 22.68 | −234.27 ± 27.52 | −28.83 ± 16.34 | 150.44 ± 27.52 | −22.34 ± 1.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Chu, X.; Yuan, Z.; Yao, Z.; Li, J.; Lu, F.; Liu, Y. Biochemical and Structural Properties of a High-Temperature-Active Laccase from Bacillus pumilus and Its Application in the Decolorization of Food Dyes. Foods 2022, 11, 1387. https://doi.org/10.3390/foods11101387
Li T, Chu X, Yuan Z, Yao Z, Li J, Lu F, Liu Y. Biochemical and Structural Properties of a High-Temperature-Active Laccase from Bacillus pumilus and Its Application in the Decolorization of Food Dyes. Foods. 2022; 11(10):1387. https://doi.org/10.3390/foods11101387
Chicago/Turabian StyleLi, Tao, Xiuxiu Chu, Zhaoting Yuan, Zhiming Yao, Jingwen Li, Fuping Lu, and Yihan Liu. 2022. "Biochemical and Structural Properties of a High-Temperature-Active Laccase from Bacillus pumilus and Its Application in the Decolorization of Food Dyes" Foods 11, no. 10: 1387. https://doi.org/10.3390/foods11101387
APA StyleLi, T., Chu, X., Yuan, Z., Yao, Z., Li, J., Lu, F., & Liu, Y. (2022). Biochemical and Structural Properties of a High-Temperature-Active Laccase from Bacillus pumilus and Its Application in the Decolorization of Food Dyes. Foods, 11(10), 1387. https://doi.org/10.3390/foods11101387