Differential Proteomic Analysis of Extracellular Vesicles Produced by Granulicatella adiacens in Biofilm vs. Planktonic Lifestyle
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Culture Conditions
2.2. Biofilm and Planktonic Cultures
2.3. Isolation of Extracellular Vesicles (EVs)
2.4. Scanning Electron Microscopy (SEM)
2.5. Protein Quantification and SDS-PAGE
2.6. Sample Preparation for Mass Spectrometry
2.7. LC-MS/MS Analysis
2.8. Data Processing
2.9. Data Pre-Processing and Quality Control
2.10. Differential Protein Expression Analysis
2.11. Data Visualization and Functional Analysis
2.12. Cytokine Quantification
2.13. Statistical Analysis
2.14. Ethical Approval
3. Results
3.1. EV Preparation from G. adiacens Biofilm and Planktonic Cells
3.2. Differential Protein Expression in Vesicles Produced Under Biofilm and Planktonic Conditions
3.3. Differentially Expressed Proteins Exhibit Consistent and Robust Expression Patterns
3.4. Protein Overlaps and Distribution Between Vesicles from Biofilm and Planktonic Conditions
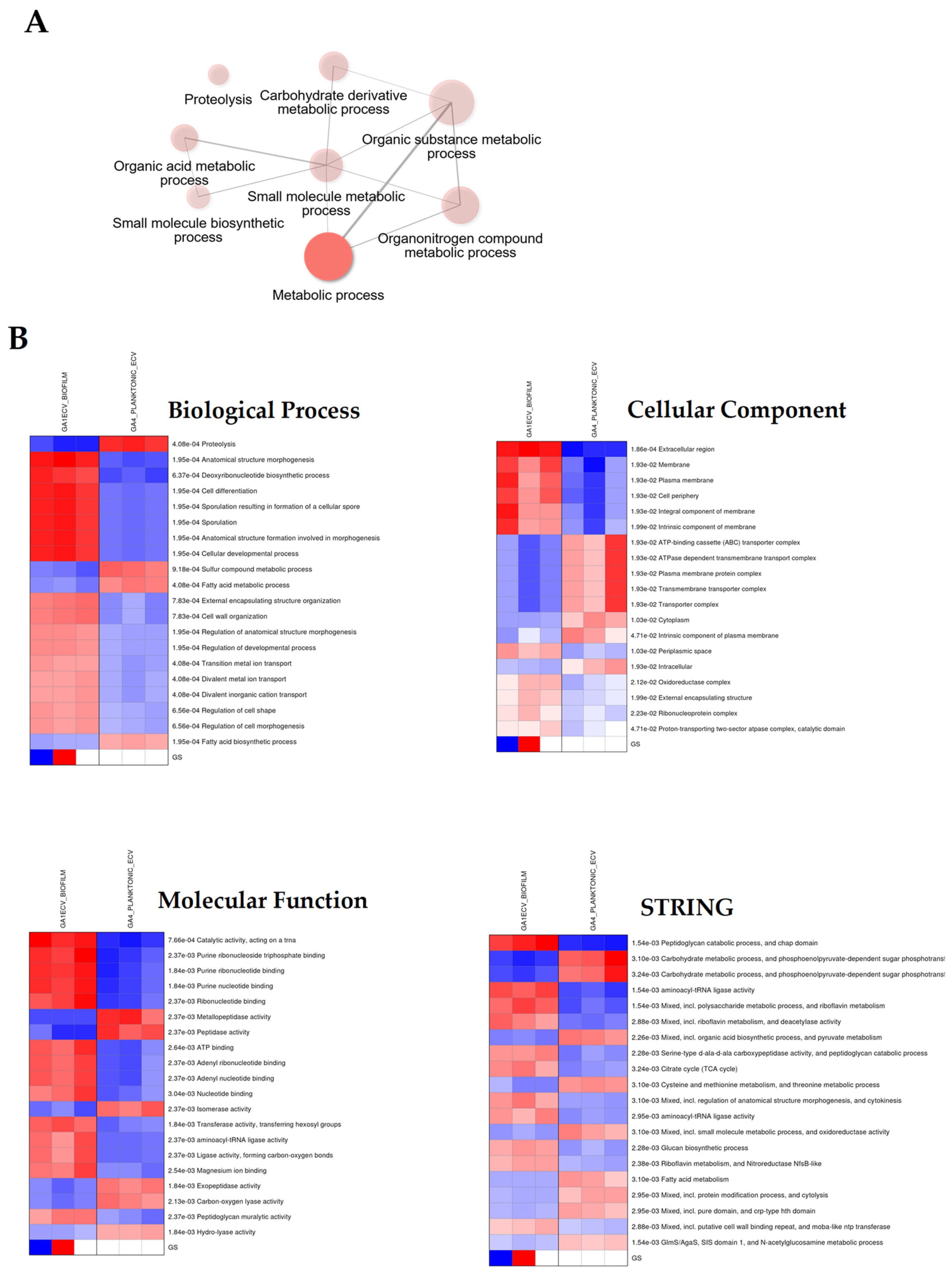
3.5. Gene Ontology and Functional Annotation of Differentially Expressed Proteins
3.6. Proteins with Predicted Virulence Potential
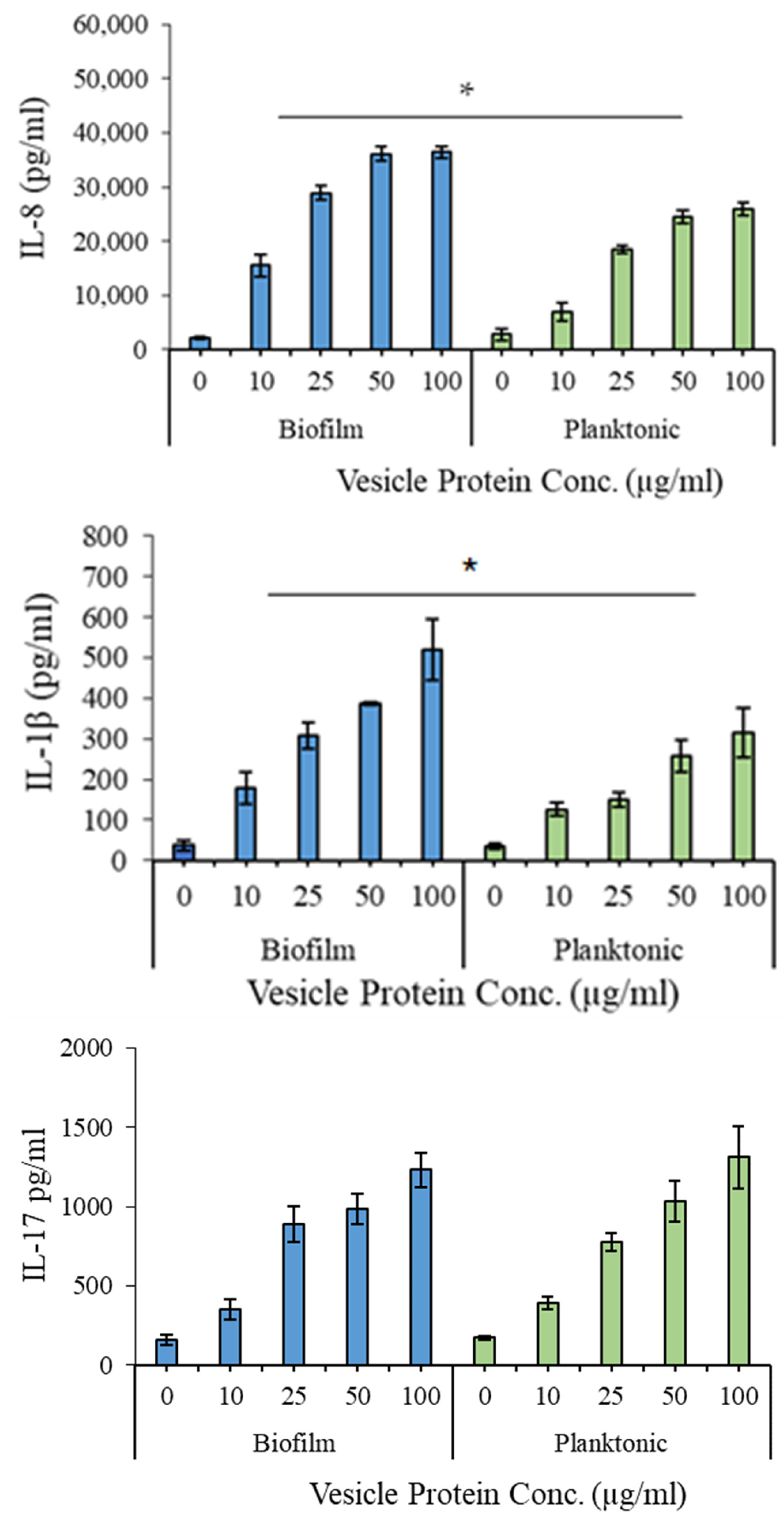
3.7. Proinflammatory Potential of EV Preparations
4. Discussion
5. Limitations and Future Directions
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lockhart, P.B.; Brennan, M.T.; Thornhill, M.; Michalowicz, B.S.; Noll, J.; Bahrani-Mougeot, F.K.; Sasser, H.C. Poor oral hygiene as a risk factor for infective endocarditis-related bacteremia. J. Am. Dent. Assoc. 2009, 140, 1238–1244. [Google Scholar] [CrossRef]
- Perez-Chaparro, P.J.; Meuric, V.; De Mello, G.; Bonnaure-Mallet, M. Bacteremia of oral origin. Rev. Stomatol. Chir. Maxillofac. 2011, 112, 300–303. [Google Scholar] [CrossRef]
- Adam, E.L.; Siciliano, R.F.; Gualandro, D.M.; Calderaro, D.; Issa, V.S.; Rossi, F.; Caramelli, B.; Mansur, A.J.; Strabelli, T.M.V. Case series of infective endocarditis caused by Granulicatella species. Int. J. Infect. Dis. 2015, 31, 56–58. [Google Scholar] [CrossRef]
- Kanasi, E.; Dewhirst, F.; Chalmers, N.; Kent, R., Jr.; Moore, A.; Hughes, C.; Pradhan, N.; Loo, C.; Tanner, A.C.R. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010, 44, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Shimoyama, Y.; Ishikawa, T.; Kodama, Y.; Tajika, S.; Kimura, S. Contribution of different adherent properties of Granulicatella adiacens and Abiotrophia defectiva to their associations with oral colonization and the risk of infective endocarditis. J. Oral Sci. 2020, 62, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef] [PubMed]
- Asikainen, S.; Dogan, B.; Turgut, Z.; Paster, B.J.; Bodur, A.; Oscarsson, J. Specified species in gingival crevicular fluid predict bacterial diversity. PLoS ONE 2010, 5, e13589. [Google Scholar] [CrossRef]
- Hsiao, W.W.L.; Li, K.L.; Liu, Z.; Jones, C.; Fraser-Liggett, C.M.; Fouad, A.F. Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Genom. 2012, 13, 345. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr.; Rocas, I.N. Catonella morbi and Granulicatella adiacens: New species in endodontic infections. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontology 2006, 102, 259–264. [Google Scholar] [CrossRef]
- Ellis, T.N.; Kuehn, M.J. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 2010, 74, 81–94. [Google Scholar] [CrossRef]
- Kuehn, M.J.; Kesty, N.C. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005, 19, 2645–2655. [Google Scholar] [CrossRef] [PubMed]
- Avila-Calderón, E.D.; Araiza-Villanueva, M.G.; Cancino-Diaz, J.C.; López-Villegas, E.O.; Sriranganathan, N.; Boyle, S.M.; Contreras-Rodríguez, A. Roles of bacterial membrane vesicles. Arch. Microbiol. 2015, 197, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-H.; Choi, C.W.; Park, E.C.; Lee, S.-Y.; Kim, S.I. Isolation and proteomic characterization of bacterial extracellular membrane vesicles. Curr. Protein Pept. Sci. 2014, 15, 719–731. [Google Scholar] [CrossRef]
- Liu, Y.; Defourny, K.A.Y.; Smid, E.J.; Abee, T. Gram-Positive Bacterial Extracellular Vesicles and Their Impact on Health and Disease. Front. Microbiol. 2018, 9, 1502. [Google Scholar] [CrossRef] [PubMed]
- Sangiorgio, G.; Nicitra, E.; Bivona, D.; Bonomo, C.; Bonacci, P.; Santagati, M.; Musso, N.; Bongiorno, D.; Stefani, S. Interactions of Gram-Positive Bacterial Membrane Vesicles and Hosts: Updates and Future Directions. Int. J. Mol. Sci. 2024, 25, 2904. [Google Scholar] [CrossRef]
- Xu, Y.; Xie, C.; Liu, Y.; Qin, X.; Liu, J. An update on our understanding of Gram-positive bacterial membrane vesicles: Discovery, functions, and applications. Front. Cell. Infect. Microbiol. 2023, 13, 1273813. [Google Scholar] [CrossRef]
- Toyofuku, M.; Cárcamo-Oyarce, G.; Yamamoto, T.; Eisenstein, F.; Hsiao, C.-C.; Kurosawa, M.; Gademann, K.; Pilhofer, M.; Nomura, N.; Eberl, L. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 2017, 8, 481. [Google Scholar] [CrossRef]
- Brookes, Z.L.; Belfield, L.A.; Ashworth, A.; Casas-Agustench, P.; Raja, M.; Pollard, A.J.; Bescos, R. Effects of chlorhexidine mouthwash on the oral microbiome. J. Dent. 2021, 113, 103768. [Google Scholar] [CrossRef]
- Baroudi, K.; Dagli, R.; Dagli, N.; Darwish, S. Oral Microbial Shift: Factors affecting the Microbiome and Prevention of Oral Disease. J. Contemp. Dent. Pract. 2016, 17, 90–96. [Google Scholar] [CrossRef]
- Kleine Bardenhorst, S.; Hagenfeld, D.; Matern, J.; Prior, K.; Harks, I.; Eickholz, P.; Lorenz, K.; Kim, T.-S.; Kocher, T.; Meyle, J.; et al. The role of the oral microbiota in the causal effect of adjunctive antibiotics on clinical outcomes in stage III-IV periodontitis patients. Microbiome 2024, 12, 220. [Google Scholar] [CrossRef]
- Alhazmi, Y.A.; Aljabri, M.Y.; Raafat, S.N.; Gomaa, S.M.; Shamel, M. Exploring the Effects of Low-Level Laser Therapy on the Cytocompatibility and Osteo/Odontogenic Potential of Gingival-Derived Mesenchymal Stem Cells: Preliminary Report. Appl. Sci. 2023, 13, 8490. [Google Scholar] [CrossRef]
- De Angelis, M.; Siragusa, S.; Campanella, D.; Di Cagno, R.; Gobbetti, M. Comparative proteomic analysis of biofilm and planktonic cells of Lactobacillus plantarum DB200. Proteomics 2015, 15, 2244–2257. [Google Scholar] [CrossRef] [PubMed]
- Dumitrache, A.; Klingeman, D.M.; Natzke, J.; Rodriguez, M., Jr.; Giannone, R.J.; Hettich, R.L.; Davison, B.H.; Brown, S.D. Specialized activities and expression differences for Clostridium thermocellum biofilm and planktonic cells. Sci Rep. 2017, 7, 43583. [Google Scholar] [CrossRef] [PubMed]
- Marinacci, B.; D’aMbrosio, C.; Vitale, I.; Di Sotto, A.; Cairone, F.; Spano, M.; Carradori, S.; Scaloni, A.; Gullì, M.; Puca, V.; et al. Biochemical and functional properties of vesicles from planktonic and biofilm phenotypes of Limosilactobacillus reuteri DSM 17938. Sci. Rep. 2025, 15, 18889. [Google Scholar] [CrossRef]
- Alkandari, S.A.; Bhardwaj, R.G.; Ellepola, A.; Karched, M. Proteomics of extracellular vesicles produced by Granulicatella adiacens, which causes infective endocarditis. PLoS ONE 2020, 15, e0227657. [Google Scholar] [CrossRef]
- Karched, M.; Bhardwaj, R.G.; Tiss, A.; Asikainen, S. Proteomic Analysis and Virulence Assessment of Granulicatella adiacens Secretome. Front. Cell. Infect. Microbiol. 2019, 9, 104. [Google Scholar] [CrossRef]
- Krzyściak, W.; Jurczak, A.; Kościelniak, D.; Bystrowska, B.; Skalniak, A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 499–515. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Wu, Z.; Lu, C. Reduced virulence is an important characteristic of biofilm infection of Streptococcus suis. FEMS Microbiol. Lett. 2011, 316, 36–43. [Google Scholar] [CrossRef]
- Warrier, A.; Satyamoorthy, K.; Murali, T.S. Quorum-sensing regulation of virulence factors in bacterial biofilm. Futur. Microbiol. 2021, 16, 1003–1021. [Google Scholar] [CrossRef]
- Karched, M.; Bhardwaj, R.G.; Inbamani, A.; Asikainen, S. Quantitation of biofilm and planktonic life forms of coexisting periodontal species. Anaerobe 2015, 35 Pt A, 13–20. [Google Scholar] [CrossRef]
- Effah, C.Y.; Ding, X.; Drokow, E.K.; Li, X.; Tong, R.; Sun, T. Bacteria-derived extracellular vesicles: Endogenous roles, therapeutic potentials and their biomimetics for the treatment and prevention of sepsis. Front. Immunol. 2024, 15, 1296061. [Google Scholar] [CrossRef]
- Peregrino, E.S.; Castañeda-Casimiro, J.; Vázquez-Flores, L.; Estrada-Parra, S.; Wong-Baeza, C.; Serafín-López, J.; Wong-Baeza, I. The Role of Bacterial Extracellular Vesicles in the Immune Response to Pathogens, and Therapeutic Opportunities. Int. J. Mol. Sci. 2024, 25, 6210. [Google Scholar] [CrossRef]
- Chakravarty, S.; Melton, C.N.; Bailin, A.; Yahr, T.L.; Anderson, G.G. Pseudomonas aeruginosa Magnesium Transporter MgtE Inhibits Type III Secretion System Gene Expression by Stimulating rsmYZ Transcription. J. Bacteriol. 2017, 199, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.d.l.P.; Wong, T.Y.; Damron, F.H.; Fernández, J.; Sisti, F. Cyclic di-GMP Regulates the Type III Secretion System and Virulence in Bordetella bronchiseptica. Infect. Immun. 2022, 90, e0010722. [Google Scholar] [CrossRef] [PubMed]
- Płaczkiewicz, J.; Adamczyk-Popławska, M.; Lasek, R.; Bącal, P.; Kwiatek, A. Inactivation of Genes Encoding MutL and MutS Proteins Influences Adhesion and Biofilm Formation by Neisseria gonorrhoeae. Microorganisms 2019, 7, 647. [Google Scholar] [CrossRef]
- Diene, S.M.; Pontarotti, P.; Azza, S.; Armstrong, N.; Pinault, L.; Chabrière, E.; Colson, P.; Rolain, J.-M.; Raoult, D. Origin, Diversity, and Multiple Roles of Enzymes with Metallo-β-Lactamase Fold from Different Organisms. Cells 2023, 12, 1752. [Google Scholar] [CrossRef]
- DeJesus, M.A.; Gerrick, E.R.; Xu, W.; Park, S.W.; Long, J.E.; Boutte, C.C.; Rubin, E.J.; Schnappinger, D.; Ehrt, S.; Fortune, S.M.; et al. Comprehensive Essentiality Analysis of the Mycobacterium tuberculosis Genome via Saturating Transposon Mutagenesis. mBio 2017, 8, e02133-16. [Google Scholar] [CrossRef]
- Figueiredo, T.A.; Ludovice, A.M.; Sobral, R.G. Contribution of peptidoglycan amidation to beta-lactam and lysozyme resistance in different genetic lineages of Staphylococcus aureus. Microb. Drug Resist. 2014, 20, 238–249. [Google Scholar] [CrossRef]
- Thanassi, J.A.; Hartman-Neumann, S.L.; Dougherty, T.J.; Dougherty, B.A.; Pucci, M.J. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 2002, 30, 3152–3162. [Google Scholar] [CrossRef]
- Schwab, A.; Meyering, S.S.; Lepene, B.; Iordanskiy, S.; van Hoek, M.L.; Hakami, R.M.; Kashanchi, F. Extracellular vesicles from infected cells: Potential for direct pathogenesis. Front. Microbiol. 2015, 6, 1132. [Google Scholar] [CrossRef]
- Turner, S.; Raisley, B.; Roach, K.; Bajaña, S.; Munroe, M.E.; James, J.A.; Coggeshall, K.M.; Kovats, S. Gram-Positive Bacteria Cell Wall Peptidoglycan Polymers Activate Human Dendritic Cells to Produce IL-23 and IL-1β and Promote TH17 Cell Differentiation. Microorganisms 2023, 11, 173. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Zimmermann, C.; Kieseier, B.C.; Hartung, H.-P.; Hofstetter, H.H. Bacteria and their cell wall components uniformly co-activate interleukin-17-producing thymocytes. Clin. Exp. Immunol. 2014, 178, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Araujo, I.R.; Ferrari, T.C.; Teixeira-Carvalho, A.; Campi-Azevedo, A.C.; Rodrigues, L.V.; Guimaraes Junior, M.H.; Barros, T.L.S.; Gelape, C.L.; Sousa, G.R.; Nunes, M.C.P.N.; et al. Cytokine Signature in Infective Endocarditis. PLoS ONE 2015, 10, e0133631. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, Y.; Kubota, T.; Sasaguri, K.; Sato, S.; Suzuki, Y.; Kumada, H.; Umemoto, T. Inflammatory cytokine gene expression in human periodontal ligament fibroblasts stimulated with bacterial lipopolysaccharides. Infect. Immun. 1995, 63, 3576–3581. [Google Scholar] [CrossRef]
- Briaud, P.; Carroll, R.K. Extracellular Vesicle Biogenesis and Functions in Gram-Positive Bacteria. Infect. Immun. 2020, 88, 10–1128. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Thay, B.; Damm, A.; Kufer, T.A.; Wai, S.N.; Oscarsson, J. Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-kappaB activation. Infect. Immun. 2014, 82, 4034–4046. [Google Scholar] [CrossRef]
- Maldonado, R.; Wei, R.; Kachlany, S.; Kazi, M.; Balashova, N. Cytotoxic effects of Kingella kingae outer membrane vesicles on human cells. Microb. Pathog. 2011, 51, 22–30. [Google Scholar] [CrossRef]
- Iwabuchi, Y.; Yoshida, H.; Kamei, S.; Uematsu, T.; Saito, M.; Senpuku, H. Formation of Mono-Organismal and Mixed Staphylococcus aureus and Streptococcus mutans Biofilms in the Presence of NaCl. Microorganisms 2025, 13, 1118. [Google Scholar] [CrossRef]
- Yumoto, H.; Hirota, K.; Hirao, K.; Ninomiya, M.; Murakami, K.; Fujii, H.; Miyake, Y. The Pathogenic Factors from Oral Streptococci for Systemic Diseases. Int. J. Mol. Sci. 2019, 20, 4571. [Google Scholar] [CrossRef]
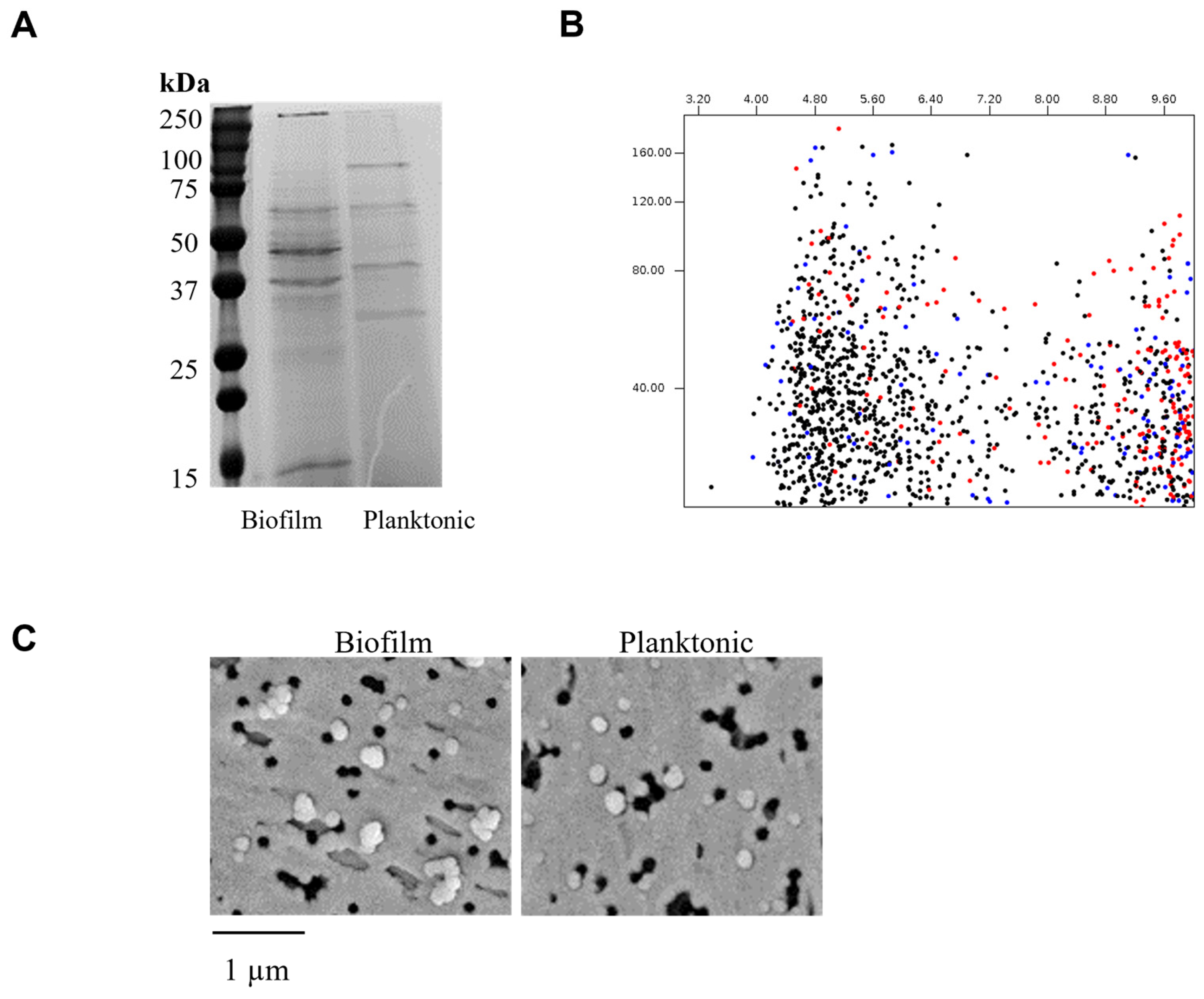
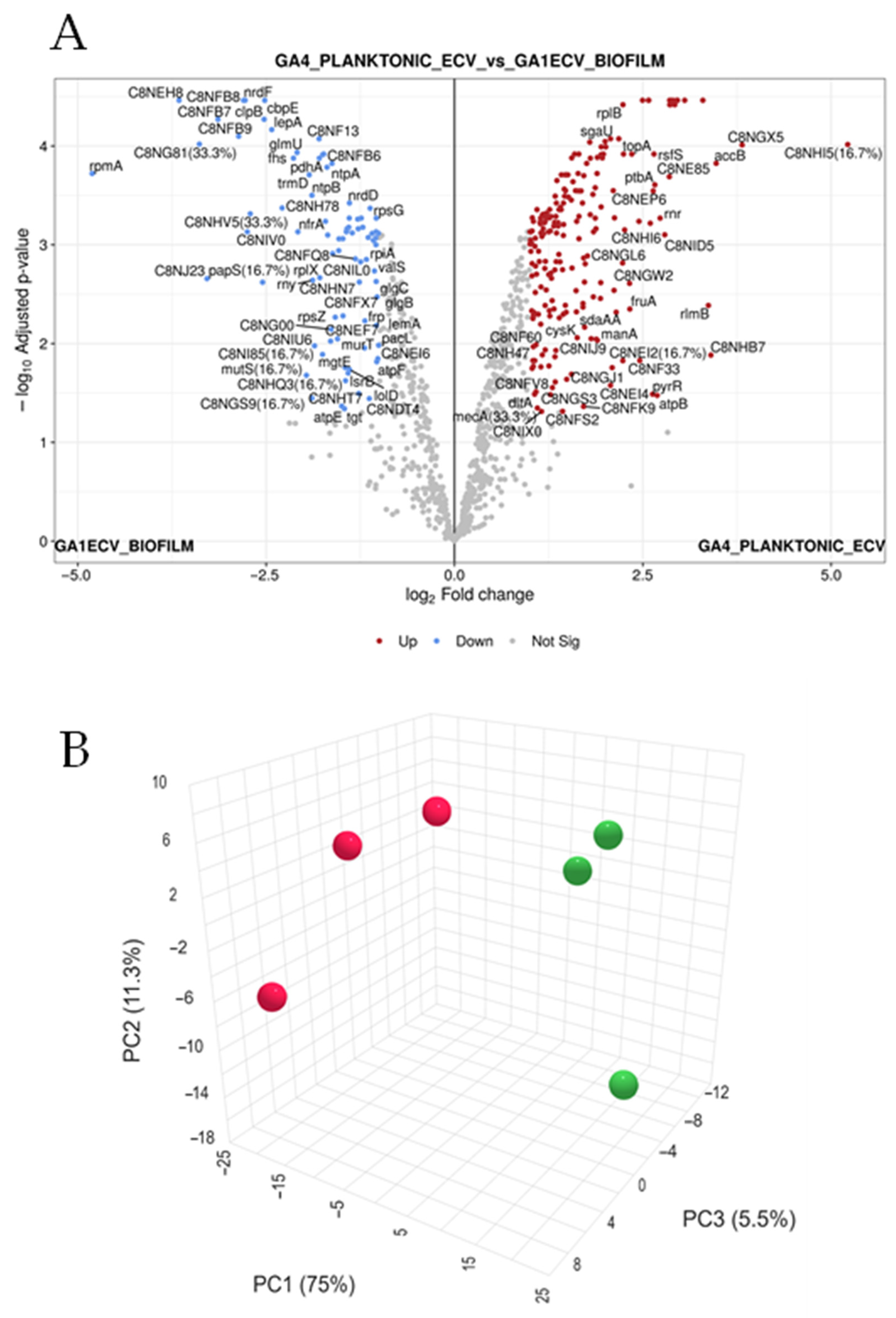
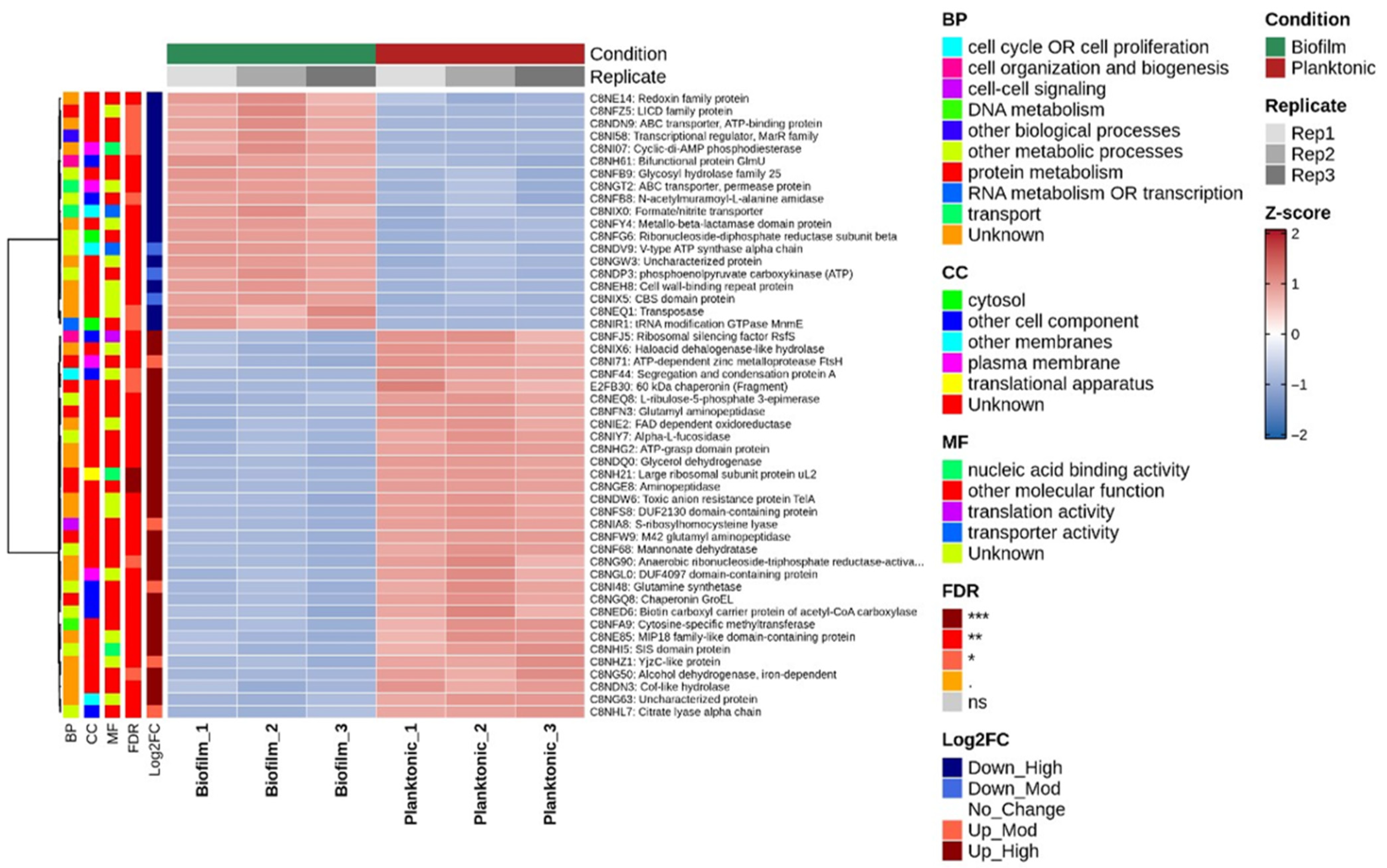
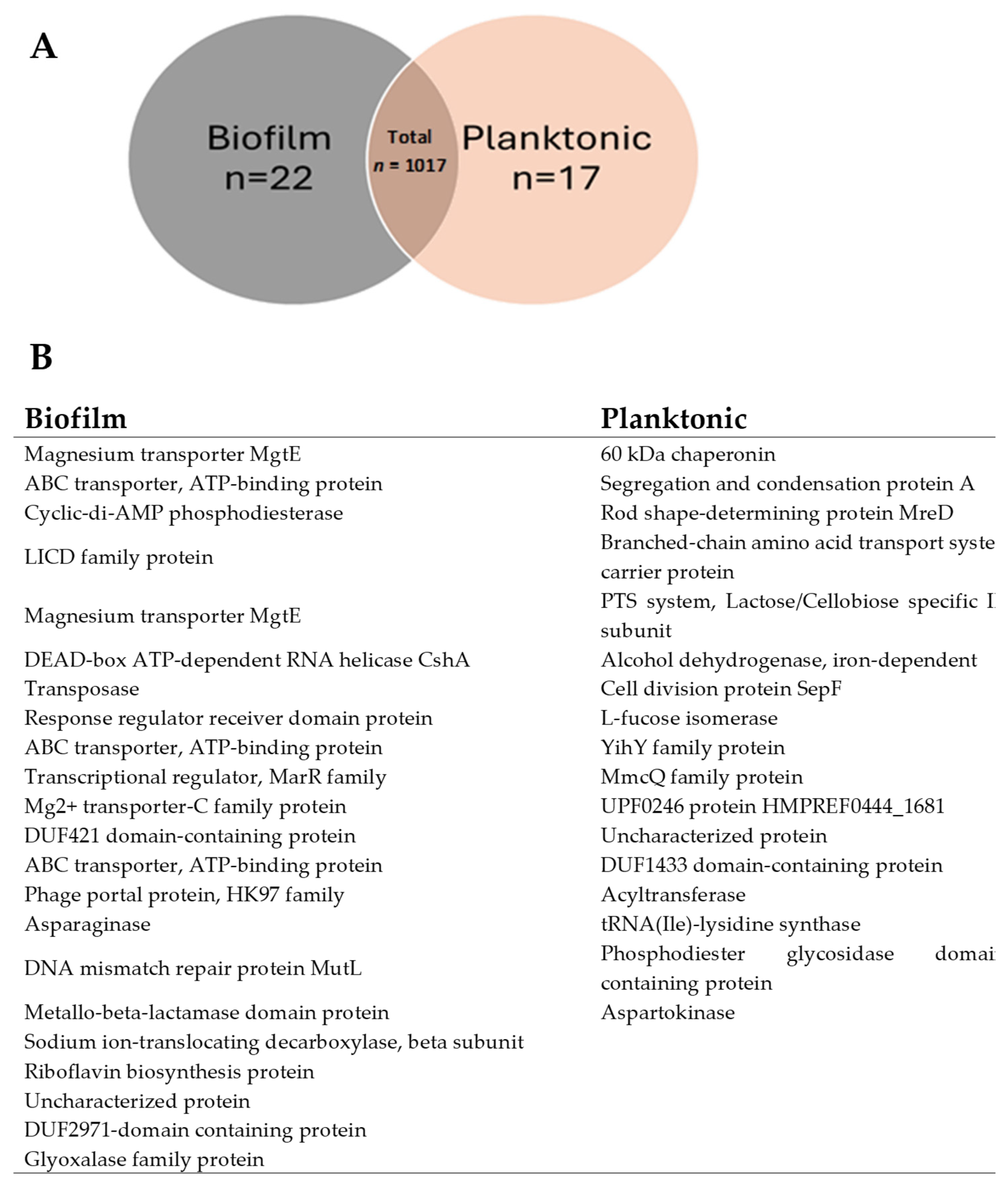
| S. No. | Protein Name | Predicted Scores |
|---|---|---|
| 1 | DUF2130 domain-containing protein | 0.9537 |
| 2 | Glycosyl hydrolase family 25 | 1.0436 |
| 3 | Aminopeptidase | 0.632 |
| 4 | Toxic anion resistance protein TelA | 0.9069 |
| 5 | SIS domain protein | 1.0157 |
| 6 | Large ribosomal subunit protein uL2 | 0.5151 |
| 7 | Endonuclease GajA/Old nuclease/RecF-like AAA domain-containing protein | 1.0891 |
| 8 | Peptidoglycan hydrolase | 0.8508 |
| 9 | Ribonucleoside-diphosphate reductase subunit beta | 0.824 |
| 10 | Metallo-beta-lactamase domain protein | 0.9978 |
| 11 | Type VII secretion protein, YukD family | 1.11 |
| 12 | Uncharacterized protein | 1.0537 |
| 13 | Cell wall-binding repeat protein | 1.0257 |
| 14 | Cell wall-binding repeat protein | 1.0221 |
| 15 | Antitoxin | 0.5697 |
| 16 | ABC-2 type transporter transmembrane domain-containing protein | 0.1668 |
| 17 | Ferritin-like protein | 1.1171 |
| 18 | CBS domain protein | 0.9741 |
| 19 | Uncharacterized protein | 0.8529 |
| 20 | Large ribosomal subunit protein uL24 | 0.6124 |
| 21 | ACT domain protein | 1.0264 |
| 22 | YjzC-like protein | 1.1051 |
| 23 | NYN domain-containing protein | 0.8158 |
| 24 | CHAP domain protein | 0.8252 |
| 25 | Periplasmic binding protein | 0.9848 |
| 26 | Oxidoreductase, NAD-binding domain protein | 1.0851 |
| 27 | Dipeptidase | 0.9934 |
| 28 | Replication initiation and membrane attachment protein | 0.3368 |
| 29 | Alpha amylase, catalytic domain protein | 0.312 |
| 30 | Polysaccharide biosynthesis protein | 0.3913 |
| 31 | Lipoprotein | 0.9981 |
| 32 | LPXTG-motif cell wall anchor domain protein | 1.0466 |
| 33 | Putative dGTPase | 0.9897 |
| 34 | 3D domain protein | 1.0095 |
| 35 | Lipoprotein | 0.8274 |
| 36 | BadF/BadG/BcrA/BcrD ATPase family protein | 1.002 |
| 37 | Ribitol-5-phosphate cytidylyltransferase | 0.4136 |
| 38 | Oxidoreductase, short chain dehydrogenase/reductase family protein | 0.3596 |
| 39 | Zinc-ribbon domain-containing protein | 0.721 |
| 40 | DltD domain protein | 0.3036 |
| 41 | DNA-binding helix-turn-helix protein | 0.9929 |
| 42 | Lipoprotein | 0.5198 |
| 43 | Type VII secretion system accessory factor EsaA | 1.0691 |
| 44 | Uncharacterized protein | 1.0868 |
| 45 | ABC transporter, ATP-binding protein | 0.9232 |
| 46 | Small ribosomal subunit protein uS14 | 1.0552 |
| 47 | Large ribosomal subunit protein bL27 | 1.1145 |
| 48 | Helix-turn-helix domain-containing protein | 0.1066 |
| 49 | Large ribosomal subunit protein bL20 | 0.987 |
| 50 | Flagellar FliJ protein | 0.9929 |
| 51 | Cell wall-binding repeat protein | 1.0173 |
| 52 | Uncharacterized protein | 0.0284 |
| 53 | Response regulator receiver domain protein | 1.0023 |
| 54 | LPXTG-motif cell wall anchor domain protein | 1.0137 |
| 55 | CAAX amino terminal protease family protein | 0.5992 |
| 56 | Small ribosomal subunit protein uS10 | 0.7333 |
| 57 | Excalibur domain protein | 1.0081 |
| 58 | Small ribosomal subunit protein bS20 | 1.0387 |
| 59 | DivIVA domain protein | 1.0225 |
| 60 | Arylsulfotransferase N-terminal domain-containing protein | 0.998 |
| 61 | Glycosyltransferase, group 2 family protein | 1.0253 |
| 62 | LPXTG-motif cell wall anchor domain protein | 1.007 |
| 63 | Acyl carrier protein | 0.0809 |
| 64 | Uncharacterized protein | 0.9996 |
| 65 | Nucleoside triphosphate/diphosphate phosphatase | 0.5432 |
| 66 | N-acetylmuramoyl-L-alanine amidase | 1.0077 |
| 67 | DNA-directed RNA polymerase subunit epsilon | 0.5563 |
| 68 | YbbR-like protein | 1.0947 |
| 69 | Signal recognition particle protein | 0.3172 |
| 70 | CBS domain protein | 0.4193 |
| 71 | LXG domain-containing protein | 0.5803 |
| 72 | ABC transporter, ATP-binding protein | 0.4808 |
| 73 | Phage major tail protein, phi13 family | 0.993 |
| 74 | RelA/SpoT domain protein | 1.0544 |
| 75 | Transcriptional regulator, GntR family | 0.1018 |
| 76 | Ribosomal protein L7Ae | 1.1258 |
| 77 | 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase | 0.9949 |
| 78 | DUF2207 domain-containing protein | 0.5563 |
| 79 | LemA family protein | 1.0085 |
| 80 | DUF4176 domain-containing protein | 1.1326 |
| 81 | ESAT-6-like protein | 1.0188 |
| 82 | YqeY-like protein | 0.1766 |
| 83 | Putative hyalurononglucosaminidase | 1.0868 |
| 84 | Small ribosomal subunit protein uS17 | 0.8802 |
| 85 | Outer surface protein | 0.9101 |
| 86 | Pilin isopeptide linkage domain protein | 1.0169 |
| 87 | Bleomycin resistance protein | 1.011 |
| 88 | Sortase family protein | 1.0526 |
| 89 | Transposase | 1.0859 |
| 90 | Cell envelope-like function transcriptional attenuator common domain protein | 1.0305 |
| 91 | GtrA-like protein domain-containing protein | 0.9072 |
| 92 | Lipoprotein | 0.9882 |
| 93 | Large ribosomal subunit protein bL33 | 0.9998 |
| 94 | DNA-directed RNA polymerase subunit omega | 1.1517 |
| 95 | Ribonuclease P protein component | 0.3267 |
| 96 | Acyl carrier protein | 0.3664 |
| 97 | DNA-binding helix-turn-helix protein | 1.0418 |
| 98 | Phosphopantetheine adenylyltransferase | 1.0486 |
| 99 | Trypsin | 0.9085 |
| 100 | DnaD domain protein | 0.9444 |
| 101 | Putative ACR, COG1399 | 0.4689 |
| 102 | Pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickel insertion protein | 0.8938 |
| 103 | Response regulator receiver domain protein | 0.6493 |
| 104 | PTS system mannose/fructose/sorbose family IID component | 0.7343 |
| 105 | DUF1310 family protein | 1.0365 |
| 106 | Oxidoreductase, short chain dehydrogenase/reductase family protein | 0.5302 |
| 107 | Citrate lyase acyl carrier protein | 0.826 |
| 108 | Small ribosomal subunit protein bS18 | 1.0776 |
| 109 | Small ribosomal subunit protein uS15 | 0.6017 |
| 110 | LPXTG-motif cell wall anchor domain protein | 1.0351 |
| 111 | Putative cross-wall-targeting lipoprotein signal | 1.0018 |
| 112 | Uncharacterized protein | 0.9772 |
| 113 | Putative DNA/RNA non-specific endonuclease | 0.9956 |
| 114 | Transcriptional regulator, Crp/Fnr family | 0.4498 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karched, M.; Alkandari, S. Differential Proteomic Analysis of Extracellular Vesicles Produced by Granulicatella adiacens in Biofilm vs. Planktonic Lifestyle. Dent. J. 2025, 13, 557. https://doi.org/10.3390/dj13120557
Karched M, Alkandari S. Differential Proteomic Analysis of Extracellular Vesicles Produced by Granulicatella adiacens in Biofilm vs. Planktonic Lifestyle. Dentistry Journal. 2025; 13(12):557. https://doi.org/10.3390/dj13120557
Chicago/Turabian StyleKarched, Maribasappa, and Sarah Alkandari. 2025. "Differential Proteomic Analysis of Extracellular Vesicles Produced by Granulicatella adiacens in Biofilm vs. Planktonic Lifestyle" Dentistry Journal 13, no. 12: 557. https://doi.org/10.3390/dj13120557
APA StyleKarched, M., & Alkandari, S. (2025). Differential Proteomic Analysis of Extracellular Vesicles Produced by Granulicatella adiacens in Biofilm vs. Planktonic Lifestyle. Dentistry Journal, 13(12), 557. https://doi.org/10.3390/dj13120557






