Do Concurrent Peri-Implantitis and Periodontitis Share Their Microbiotas? A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Type of Study
2.2. Study Participants
2.3. Sampling and Microbial DNA Extraction
2.4. Microbial DNA Extraction
2.5. Library Preparation
2.6. Bioinformatics and Microbial Diversity Analysis
3. Results
3.1. Study Participants
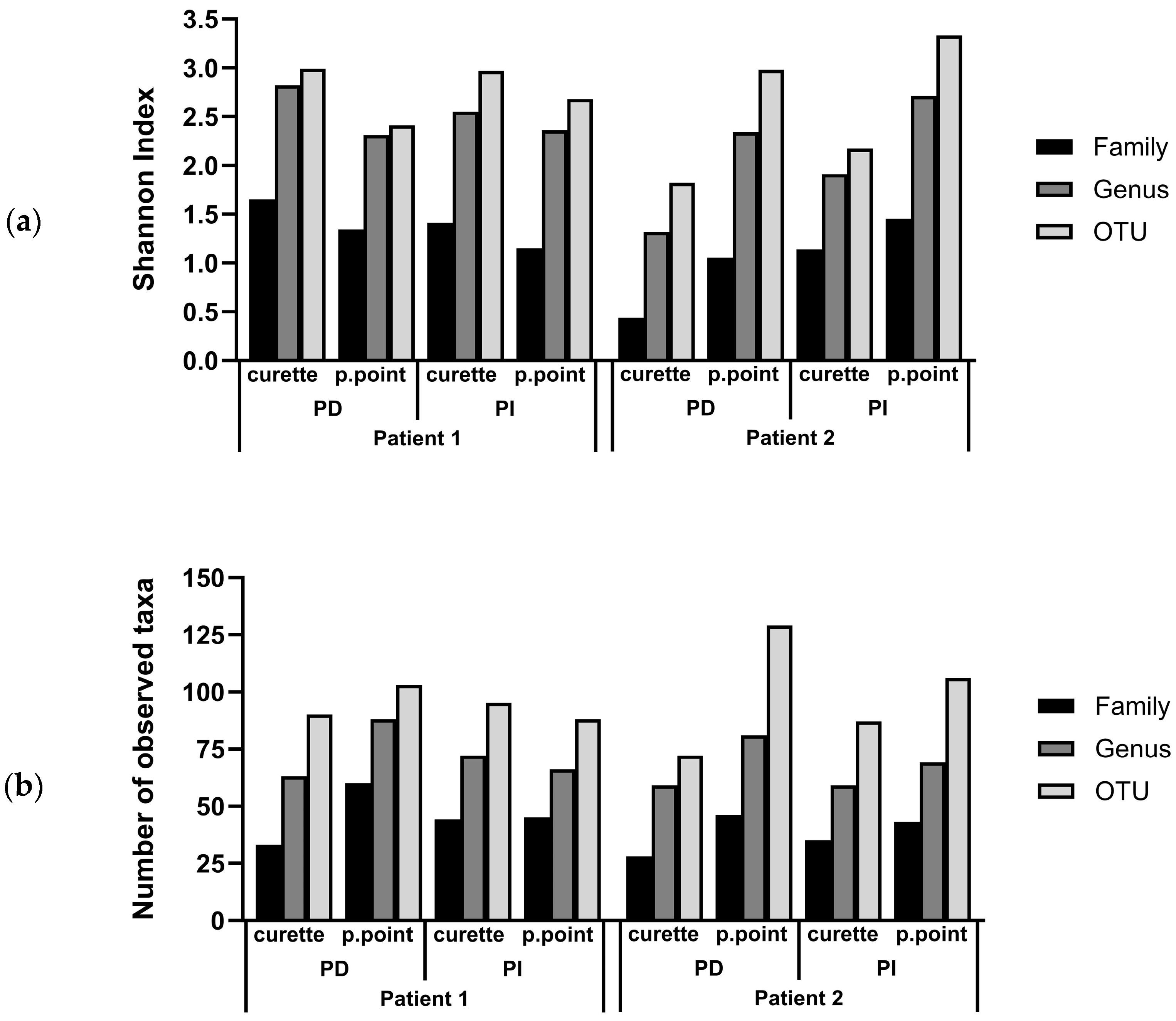
3.2. Comparison of the Subgingival Microbial Diversity in Biofilms Recovered Using Paper Points or Curette Collection
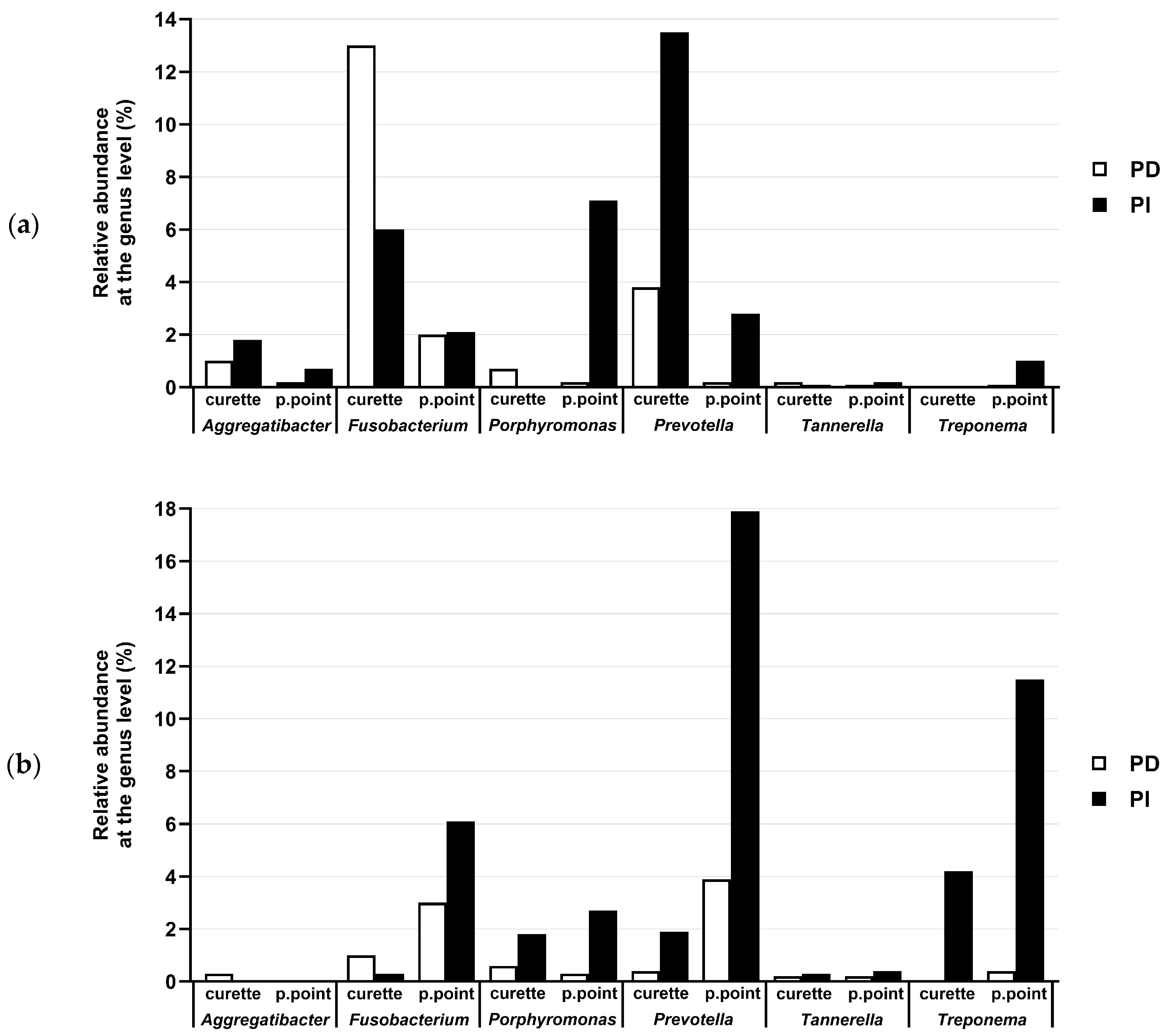
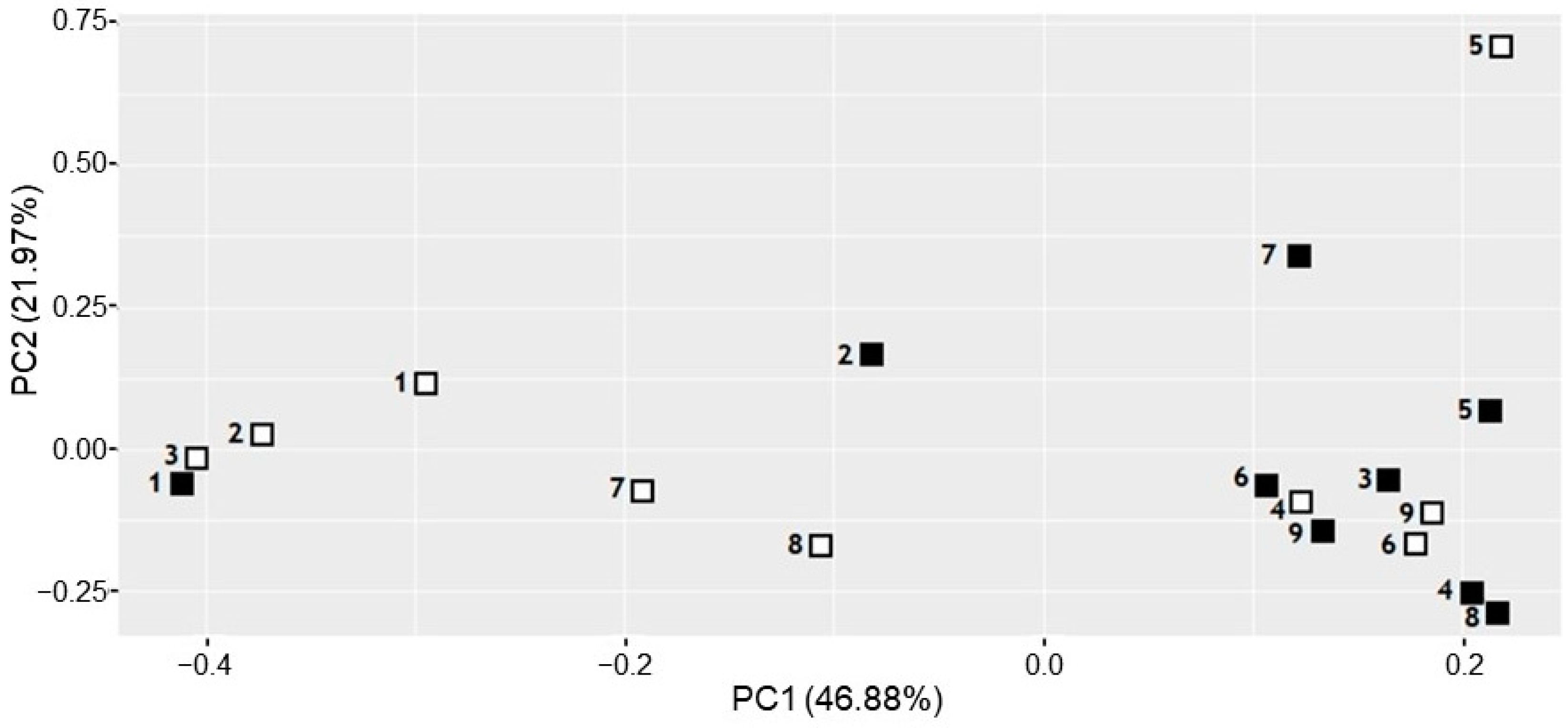
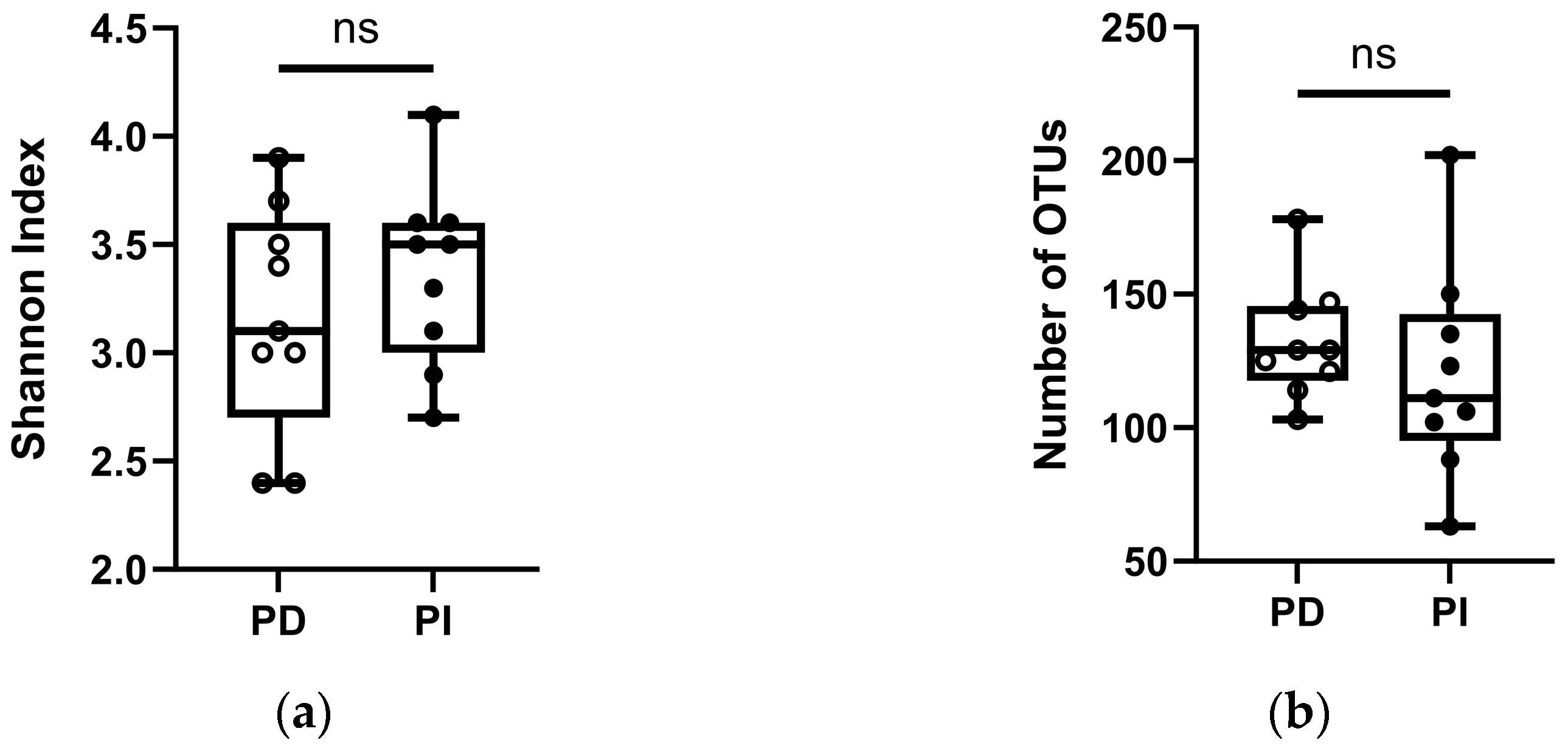
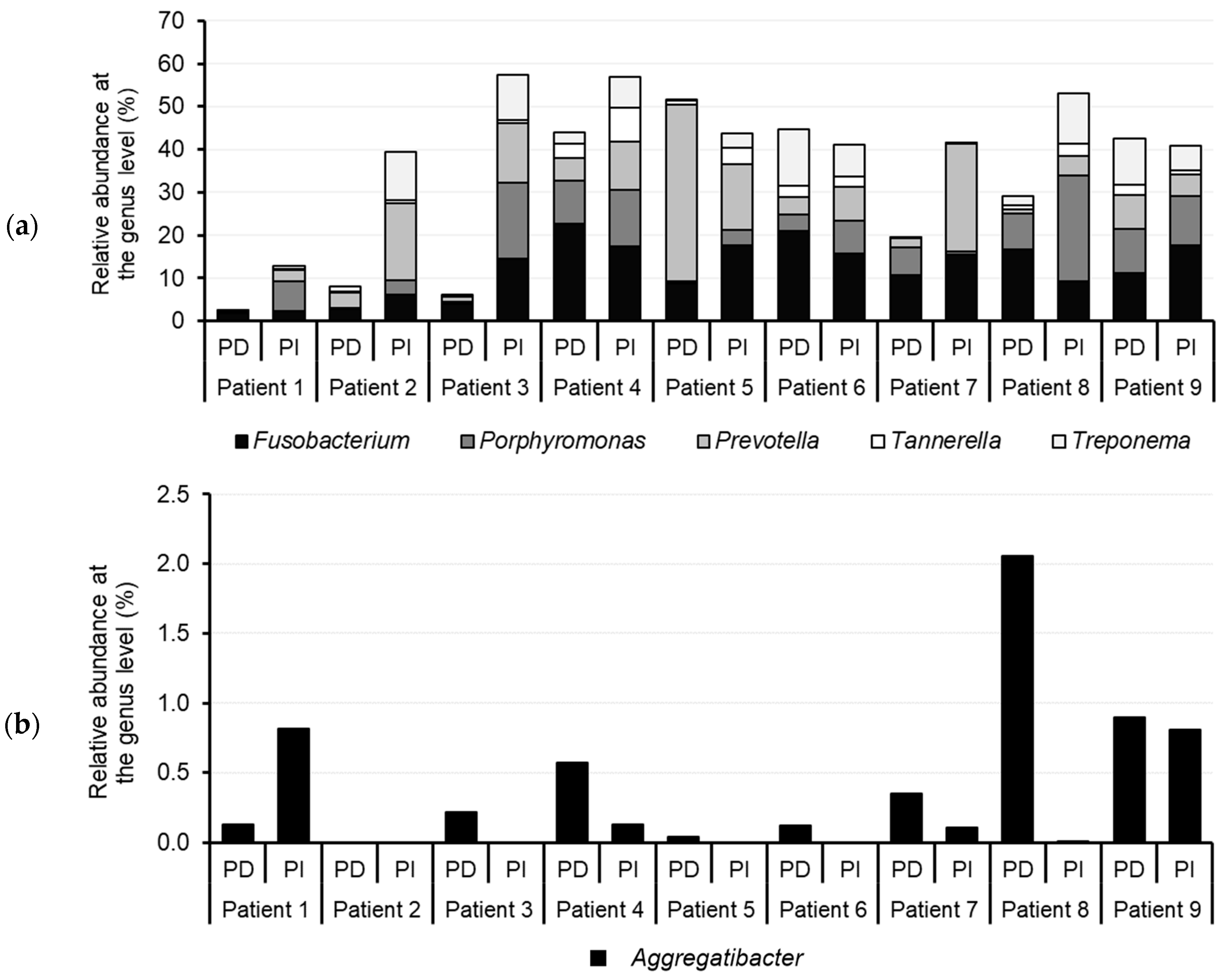
3.3. Comparison of the Microbiota of PD and PI Sites Using NGS
4. Discussion
Limitations of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, C.T.; Huang, Y.W.; Zhu, L.; Weltman, R. Prevalences of peri-implantitis and peri-implant mucositis: Systematic review and meta-analysis. J. Dent. 2017, 62, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Derks, J.; Ichioka, Y.; Dionigi, C.; Trullenque-Eriksson, A.; Berglundh, J.; Tomasi, C.; Graziani, F. Prevention and management of peri-implant mucositis and peri-implantitis: A systematic review of outcome measures used in clinical studies in the last 10 years. J. Clin. Periodontol. 2023, 50 (Suppl. S25), 55–66. [Google Scholar] [CrossRef] [PubMed]
- Carra, M.C.; Blanc-Sylvestre, N.; Courtet, A.; Bouchard, P. Primordial and primary prevention of peri-implant diseases: A systematic review and meta-analysis. J. Clin. Periodontol. 2023, 50 (Suppl. S26), 77–112. [Google Scholar] [CrossRef] [PubMed]
- Kordbacheh Changi, K.; Finkelstein, J.; Papapanou, P.N. Peri-implantitis prevalence, incidence rate, and risk factors: A study of electronic health records at a U.S. dental school. Clin. Oral Implant. Res. 2019, 30, 306–314. [Google Scholar] [CrossRef]
- Belibasakis, G.N.; Charalampakis, G.; Bostanci, N.; Stadlinger, B. Peri-implant infections of oral biofilm etiology. Adv. Exp. Med. Biol. 2015, 830, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Robitaille, N.; Reed, D.N.; Walters, J.D.; Kumar, P.S. Periodontal and peri-implant diseases: Identical or fraternal infections? Mol. Oral Microbiol. 2016, 31, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Chrcanovic, B.R.; Albrektsson, T.; Wennerberg, A. Periodontally compromised vs. periodontally healthy patients and dental implants: A systematic review and meta-analysis. J. Dent. 2014, 42, 1509–1527. [Google Scholar] [CrossRef] [PubMed]
- Salvi, G.E.; Stahli, A.; Imber, J.C.; Sculean, A.; Roccuzzo, A. Physiopathology of peri-implant diseases. Clin. Implant Dent. Relat. Res. 2023, 25, 629–639. [Google Scholar] [CrossRef]
- Becker, S.T.; Beck-Broichsitter, B.E.; Graetz, C.; Dörfer, C.E.; Wiltfang, J.; Häsler, R. Peri-implantitis versus periodontitis: Functional differences indicated by transcriptome profiling. Clin. Implant Dent. Relat. Res. 2014, 16, 401–411. [Google Scholar] [CrossRef]
- Gerber, J.; Wenaweser, D.; Heitz-Mayfield, L.; Lang, N.P.; Rutger Persson, G. Comparison of bacterial plaque samples from titanium implant and tooth surfaces by different methods. Clin. Oral Implant. Res. 2006, 17, 1–7. [Google Scholar] [CrossRef]
- Kensara, A.; Saito, H.; Mongodin, E.F.; Masri, R. Microbiological profile of peri-implantitis: Analyses of peri-implant microbiome. J. Prosthodont. 2024, 33, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, T.; Sakamoto, M.; Takeuchi, Y.; Ohkuma, M.; Izumi, Y. Analysis of microbiota associated with peri-implantitis using 16S rRNA gene clone library. J. Oral Microbiol. 2010, 2, 5104. [Google Scholar] [CrossRef] [PubMed]
- Dierens, M.; Vandeweghe, S.; Kisch, J.; Persson, G.R.; Cosyn, J.; De Bruyn, H. Long-term follow-up of turned single implants placed in periodontally healthy patients after 16 to 22 years: Microbiologic outcome. J. Periodontol. 2013, 84, 880–894. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.-F.; Watt, R.M.; Mattheos, N.; Si, M.-S.; Lai, H.-C.; Lang, N.P. Periodontal and peri-implant microbiota in patients with healthy and inflamed periodontal and peri-implant tissues. Clin. Oral Implant. Res. 2016, 27, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, T.; Sakamoto, M.; Takeuchi, Y.; Maruyama, N.; Ohkuma, M.; Izumi, Y. Comprehensive microbiological findings in peri-implantitis and periodontitis. J. Clin. Periodontol. 2013, 40, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Dabdoub, S.M.; Tsigarida, A.A.; Kumar, P.S. Patient-specific analysis of periodontal and peri-implant microbiomes. J. Dent. Res. 2013, 92, 168S–175S. [Google Scholar] [CrossRef] [PubMed]
- Lafaurie, G.I.; Sabogal, M.A.; Castillo, D.M.; Rincón, M.V.; Gómez, L.A.; Lesmes, Y.A.; Chambrone, L. Microbiome and Microbial Biofilm Profiles of Peri-Implantitis: A Systematic Review. J. Periodontol. 2017, 88, 1066–1089. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahmad, A.; Muzafferiy, F.; Anderson, A.C.; Wölber, J.P.; Ratka-Krüger, P.; Fretwurst, T.; Nelson, K.; Vach, K.; Hellwig, E. Shift of microbial composition of peri-implantitis-associated oral biofilm as revealed by 16S rRNA gene cloning. J. Med. Microbiol. 2018, 67, 332–340. [Google Scholar] [CrossRef]
- Maruyama, N.; Maruyama, F.; Takeuchi, Y.; Aikawa, C.; Izumi, Y.; Nakagawa, I. Intraindividual variation in core microbiota in peri-implantitis and periodontitis. Sci. Rep. 2014, 4, 6602. [Google Scholar] [CrossRef]
- Kumar, P.S.; Mason, M.R.; Brooker, M.R.; O’Brien, K. Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J. Clin. Periodontol. 2012, 39, 425–433. [Google Scholar] [CrossRef]
- Zheng, H.; Xu, L.; Wang, Z.; Li, L.; Zhang, J.; Zhang, Q.; Chen, T.; Lin, J.; Chen, F. Subgingival microbiome in patients with healthy and ailing dental implants. Sci. Rep. 2015, 5, 10948. [Google Scholar] [CrossRef] [PubMed]
- Schaumann, S.; Staufenbiel, I.; Scherer, R.; Schilhabel, M.; Winkel, A.; Stumpp, S.N.; Eberhard, J.; Stiesch, M. Pyrosequencing of supra- and subgingival biofilms from inflamed peri-implant and periodontal sites. BMC Oral Health 2014, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-L.; Chan, Y.; Zhuang, L.; Lai, H.-C.; Lang, N.P.; Keung Leung, W.; Watt, R.M. Intra-oral single-site comparisons of periodontal and peri-implant microbiota in health and disease. Clin. Oral Implant. Res. 2019, 30, 760–776. [Google Scholar] [CrossRef] [PubMed]
- Heuer, W.; Kettenring, A.; Stumpp, S.N.; Eberhard, J.; Gellermann, E.; Winkel, A.; Stiesch, M. Metagenomic analysis of the peri-implant and periodontal microflora in patients with clinical signs of gingivitis or mucositis. Clin. Oral Investig. 2012, 16, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Belibasakis, G.N.; Manoil, D. Microbial Community-Driven Etiopathogenesis of Peri-Implantitis. J. Dent. Res. 2021, 100, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Manoil, D.; Belibasakis, G.N. Microbial Principles of Peri-Implant Infections. In Dental Implants and Oral Microbiome Dysbiosis; Neelakantan, P., Princy Solomon, A., Eds.; Springer: Cham, Switzerland, 2022. [Google Scholar]
- Rakic, M.; Grusovin, M.G.; Canullo, L. The Microbiologic Profile Associated with Peri-Implantitis in Humans: A Systematic Review. Int. J. Oral Maxillofac. Implant. 2016, 31, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Sahrmann, P.; Gilli, F.; Wiedemeier, D.B.; Attin, T.; Schmidlin, P.R.; Karygianni, L. The microbiome of peri-implantitis: A systematic review and meta-analysis. Microorganisms 2020, 8, 661. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association. Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. In Reproductive Health and Human Rights; Oxford University Press: Oxford, UK, 2003; pp. 428–432. [Google Scholar]
- Tonetti, M.S.; Sanz, M. Implementation of the new classification of periodontal diseases: Decision-making algorithms for clinical practice and education. J. Clin. Periodontol. 2019, 46, 398–405. [Google Scholar] [CrossRef]
- Parga, A.; Muras, A.; Otero-Casal, P.; Arredondo, A.; Soler-Ollé, A.; Àlvarez, G.; Alcaraz, L.D.; Mira, A.; Blanc, V.; Otero, A. The quorum quenching enzyme Aii20J modifies in vitro periodontal biofilm formation. Front. Cell. Infect. Microbiol. 2023, 13, 1118630. [Google Scholar] [CrossRef]
- Parga, A.; Balboa, S.; Otero-Casal, P.; Otero, A. New preventive strategy against oral biofilm formation in caries-active children: An in vitro study. Antibiotics 2023, 12, 1263. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Rosier, B.T.; Palazon, C.; Garcia-Esteban, S.; Artacho, A.; Galiana, A.; Mira, A. A Single Dose of Nitrate Increases Resilience Against Acidification Derived From Sugar Fermentation by the Oral Microbiome. Front. Cell. Infect. Microbiol. 2021, 11, 692883. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing, version 4.0.2; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Allaire, J.J.; Horner, J.; Marti, V.; Porte, N. markdown: Markdown Rendering for R, version 0.7.4; 2014. Available online: http://CRAN.R-project.org/package=markdown (accessed on 13 January 2024).
- Xie, Y. Dynamic Documents with R and Knitr; CRC Press (Taylor & Francis Group): Boca Raton, FL, USA, 2014; p. 190. [Google Scholar]
- Boettiger, C. knitcitations: Citations for Knitr Markdown Files, version 1.0.5; 2014. Available online: http://CRAN.R-project.org/package=knitcitations (accessed on 13 January 2024).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package, version 2.4-4; 2017. Available online: https://CRAN.R-project.org/package=vegan (accessed on 13 January 2024).
- Wickham, H. Ggplot2. Elegant Graphics for Data Analysis, 1st ed.; Springer: New York, NY, USA, 2009; p. 213. [Google Scholar]
- Wickham, H. Create Elegant Data Visualisations Using the Grammar of Graphics. Available online: https://ggplot2.tidyverse.org/reference/ggplot2-package.html (accessed on 13 January 2024).
- van der Horst, J.; Buijs, M.J.; Laine, M.L.; Wismeijer, D.; Loos, B.G.; Crielaard, W.; Zaura, E. Sterile paper points as a bacterial DNA-contamination source in microbiome profiles of clinical samples. J. Dent. 2013, 41, 1297–1301. [Google Scholar] [CrossRef] [PubMed]
- Belibasakis, G.N.; Schmidlin, P.R.; Sahrmann, P. Molecular microbiological evaluation of subgingival biofilm sampling by paper point and curette. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2014, 122, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Matić Petrović, S.; Cimbaljević, M.; Radunović, M.; Kuzmanović Pfićer, J.; Jotić, A.; Pucar, A. Detection and sampling methods for isolation of Candidaspp. from oral cavities in diabetics and non-diabetics. Braz. Oral Res. 2015, 29, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Chaparro, P.J.; Duarte, P.M.; Pannuti, C.M.; Figueiredo, L.C.; Mestnik, M.J.; Gonçalves, C.P.S.; Faveri, M.; Feres, M. Evaluation of human and microbial DNA content in subgingival plaque samples collected by paper points or curette. J. Microbiol. Methods 2015, 111, 19–20. [Google Scholar] [CrossRef]
- Beyer, K.; Brandt, B.W.; Buijs, M.J.; Brun, J.G.; Crielaard, W.; Bolstad, A.I.; Zaura, E. Comparison of paperpoint and curette sampling of subgingival microbiome composition as analyzed by 16S rRNA gene amplicon sequencing. J. Oral Microbiol. 2017, 9, 1325260. [Google Scholar] [CrossRef][Green Version]
- Loomer, P.M. Microbiological diagnostic testing in the treatment of periodontal diseases. Periodontology 2000 2004, 34, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Gazil, V.; Bandiaky, O.N.; Renard, E.; Idiri, K.; Struillou, X.; Soueidan, A. Current Data on Oral Peri-Implant and Periodontal Microbiota and Its Pathological Changes: A Systematic Review. Microorganisms 2022, 10, 2466. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ahn, D.H.; Yu, Y.; Han, H.; Kim, S.Y.; Joo, J.Y.; Chung, J.; Na, H.S.; Lee, J.Y. Microbial profiling of peri-implantitis compared to the periodontal microbiota in health and disease using 16S rRNA sequencing. J. Periodontal Implant Sci. 2023, 53, 69–84. [Google Scholar] [CrossRef]
- Kotsakis, G.A.; Olmedo, D.G. Peri-implantitis is not periodontitis: Scientific discoveries shed light on microbiome-biomaterial interactions that may determine disease phenotype. Periodontology 2000 2021, 86, 231–240. [Google Scholar] [CrossRef]
| Sample No. | Sex | Age | Smoker | Sprinkler | Antibiotic Uptake | CHX | Bleeding on Probing | Implant Connection | Implant Time | PI Time |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | 47 | Yes | Yes | No | No | No | External | 10 years | 1 year |
| 2 | Male | 67 | Yes | Yes | No | No | No | External | 6 years | 2 years |
| 3 | Female | 54 | No (4 years) | Yes | No | No | Yes | External | 8 years | 3 years |
| 4 | Female | 71 | No | Yes | No | No | No | Internal | 8 years | 2 years |
| 5 | Female | 59 | Yes | Yes | Yes | Yes | No | Internal | 12 years | 1 month |
| 6 | Female | 54 | Yes | Yes | No | No | Yes | Internal | 6 years | 2 years |
| 7 | Male | 45 | No (6 years) | No | Yes | No | Yes | Internal | 1 year | 1 month |
| 8 | Female | 47 | N/A | Yes | No | Yes | Yes | External | 5 years | 6 months |
| 9 | Male | 65 | No | Yes | No | No | No | Internal | 4 years | 2 months |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parga, A.; Pose-Rodríguez, J.M.; Muras, A.; Baus-Domínguez, M.; Otero-Casal, P.; Ortega-Quintana, M.L.; Torres-Lagares, D.; Otero, A. Do Concurrent Peri-Implantitis and Periodontitis Share Their Microbiotas? A Pilot Study. Dent. J. 2024, 12, 113. https://doi.org/10.3390/dj12040113
Parga A, Pose-Rodríguez JM, Muras A, Baus-Domínguez M, Otero-Casal P, Ortega-Quintana ML, Torres-Lagares D, Otero A. Do Concurrent Peri-Implantitis and Periodontitis Share Their Microbiotas? A Pilot Study. Dentistry Journal. 2024; 12(4):113. https://doi.org/10.3390/dj12040113
Chicago/Turabian StyleParga, Ana, José Manuel Pose-Rodríguez, Andrea Muras, María Baus-Domínguez, Paz Otero-Casal, Marcos Luis Ortega-Quintana, Daniel Torres-Lagares, and Ana Otero. 2024. "Do Concurrent Peri-Implantitis and Periodontitis Share Their Microbiotas? A Pilot Study" Dentistry Journal 12, no. 4: 113. https://doi.org/10.3390/dj12040113
APA StyleParga, A., Pose-Rodríguez, J. M., Muras, A., Baus-Domínguez, M., Otero-Casal, P., Ortega-Quintana, M. L., Torres-Lagares, D., & Otero, A. (2024). Do Concurrent Peri-Implantitis and Periodontitis Share Their Microbiotas? A Pilot Study. Dentistry Journal, 12(4), 113. https://doi.org/10.3390/dj12040113









