Impact of a New SARS-CoV-2 Variant on the Population: A Mathematical Modeling Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Mathematical Model
2.2. Parameter Values
2.3. Initial Conditions for the Scenarios
3. Results
Numerical Simulation of Scenarios
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking changes in SARS-CoV-2 Spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef]
- Lemieux, J.E.; Li, J.Z. Uncovering Ways that Emerging SARS-CoV-2 Lineages May Increase Transmissibility. J. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Kupferschmidt, K. Vaccinemakers ponder how to adapt to virus variants. Science 2021, 371, 448–449. [Google Scholar] [CrossRef] [PubMed]
- Le Page, M. Threats from new variants. New Sci. 2021, 249, 8–9. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hall, N.; Ly, H.; Tyler, K.M. COVID-19 evolution during the pandemic–Implications of new SARS-CoV-2 variants on disease control and public health policies. Virulence 2021, 12, 507. [Google Scholar] [CrossRef]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L.; et al. Emergence of SARS-CoV-2 b. 1.1. 7 lineage—United States, 29 December 2020–12 January 2021. Morb. Mortal. Wkly. Rep. 2021, 70, 95. [Google Scholar] [CrossRef]
- Gandhi, M.; Rutherford, G.W. Facial masking for Covid-19—Potential for “variolation” as we await a vaccine. N. Engl. J. Med. 2020, 383, e101. [Google Scholar] [CrossRef] [PubMed]
- Syal, K. COVID-19: Herd immunity and convalescent plasma transfer therapy. J. Med. Virol. 2020, 92, 1380–1382. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.; Lau, E.H.; Wong, J.Y.; et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Sanche, S.; Lin, Y.T.; Xu, C.; Romero-Severson, E.; Hengartner, N.W.; Ke, R. The novel coronavirus, 2019-nCoV, is highly contagious and more infectious than initially estimated. arXiv 2020, arXiv:2002.03268. [Google Scholar]
- Pitlik, S.D. COVID-19 Compared to Other Pandemic Diseases. Rambam Maimonides Med. J. 2020, 11, e0027. [Google Scholar] [CrossRef]
- Tang, K.; Huang, Y.; Chen, M. Novel Coronavirus 2019 (Covid-19) epidemic scale estimation: Topological network-based infection dynamic model. medRxiv 2020. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. 2020. Available online: https://www.cdc.gov/coronavirus/2019-nCoV/index.html (accessed on 26 March 2021).
- Johns Hopkins University and Medicine. 2020. Available online: https://coronavirus.jhu.edu (accessed on 26 March 2021).
- Lovato, A.; de Filippis, C.; Marioni, G. Upper airway symptoms in coronavirus disease 2019 (COVID-19). Am. J. Otolaryngol. 2020. [Google Scholar] [CrossRef]
- Pinky, L.; Dobrovolny, H.M. SARS-CoV-2 coinfections: Could influenza and the common cold be beneficial? J. Med. Virol. 2020, 92, 2623–2630. [Google Scholar] [CrossRef]
- Mandal, S.; Bhatnagar, T.; Arinaminpathy, N.; Agarwal, A.; Chowdhury, A.; Murhekar, M.; Gangakhedkar, R.R.; Sarkar, S. Prudent public health intervention strategies to control the coronavirus disease 2019 transmission in India: A mathematical model-based approach. Indian J. Med. Res. 2020, 151, 190. [Google Scholar]
- Reis, R.F.; de Melo Quintela, B.; de Oliveira Campos, J.; Gomes, J.M.; Rocha, B.M.; Lobosco, M.; dos Santos, R.W. Characterization of the COVID-19 pandemic and the impact of uncertainties, mitigation strategies, and underreporting of cases in South Korea, Italy, and Brazil. Chaos Solitons Fractals 2020, 136, 109888. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pasco, R.F.; Du, Z.; Petty, M.; Fox, S.J.; Galvani, A.P.; Pignone, M.; Johnston, S.C.; Meyers, L.A. Impact of social distancing measures on coronavirus disease healthcare demand, central Texas, USA. Emerg. Infect. Dis. 2020, 26, 2361. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolny, H.M. Modeling the role of asymptomatics in infection spread with application to SARS-CoV-2. PLoS ONE 2020, 15, e0236976. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolny, H.M. Quantifying the effect of remdesivir in rhesus macaques infected with SARS-CoV-2. Virology 2020, 550, 61–69. [Google Scholar] [CrossRef]
- Zhang, X.; Tan, Y.; Ling, Y.; Lu, G.; Liu, F.; Yi, Z.; Jia, X.; Wu, M.; Shi, B.; Xu, S.; et al. Viral and host factors related to the clinical outcome of COVID-19. Nature 2020, 583, 437–440. [Google Scholar] [CrossRef]
- Ran, L.; Chen, X.; Wang, Y.; Wu, W.; Zhang, L.; Tan, X. Risk factors of healthcare workers with corona virus disease 2019: A retrospective cohort study in a designated hospital of Wuhan in China. Clin. Infect. Dis. 2020, 71, 2218–2221. [Google Scholar] [CrossRef]
- Yang, H.; Duan, G. Analysis on the epidemic factors for the corona virus disease. Zhonghua Yu Fang Yi Xue Za Zhi Chin. J. Prev. Med. 2020, 54, E021. [Google Scholar]
- Boris Johnson Says the UK’s Coronavirus Variant May Have up to a 30% Higher Mortality Rate. Available online: https://tinyurl.com/6q3g47b8 (accessed on 26 March 2021).
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Rangarajan, E.S.; Izard, T.; Farzan, M.; Choe, H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Hanage, W.P.; Rasmussen, A.L. Making sense of mutation: What D614G means for the COVID-19 pandemic remains unclear. Cell 2020, 182, 794–795. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2020, 1–6. [Google Scholar] [CrossRef]
- Zhu, W.; Yang, J.; Lu, S.; Lan, R.; Jin, D.; Luo, X.L.; Pu, J.; Wu, S.; Xu, J. Beta-and Novel Delta-Coronaviruses Are Identified from Wild Animals in the Qinghai-Tibetan Plateau, China. Virol. Sin. 2020, 1–10. [Google Scholar] [CrossRef]
- Rahimi, F.; Abadi, A.T.B. Implications of the Emergence of a New Variant of SARS-CoV-2, VUI-202012/01. Arch. Med. Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance 2021, 26, 2002106. [Google Scholar] [CrossRef]
- Fiorentini, S.; Messali, S.; Zani, A.; Caccuri, F.; Giovanetti, M.; Ciccozzi, M.; Caruso, A. First detection of SARS-CoV-2 spike protein N501 mutation in Italy in August 2020. Lancet Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Yi, C.; Sun, X.; Ye, J.; Ding, L.; Liu, M.; Yang, Z.; Lu, X.; Zhang, Y.; Ma, L.; Gu, W.; et al. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020, 17, 621–630. [Google Scholar] [CrossRef]
- Public Health England. 2021. Available online: https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 (accessed on 26 March 2021).
- Reuters. 2020. Available online: https://tinyurl.com/y5fe8q2u (accessed on 26 March 2021).
- Wang, Y.; Wu, J.; Zhang, L.; Zhang, Y.; Wang, H.; Ding, R.; Nie, J.; Li, Q.; Liu, S.; Yu, Y.; et al. The Infectivity and Antigenicity of Epidemic SARS-CoV-2 Variants in the United Kingdom. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Murray, J.D. Mathematical Biology I: An Introduction, Volume 17 of Interdisciplinary Applied Mathematics; Springer: New York, NY, USA, 2002. [Google Scholar]
- Brauer, F.; Castillo-Chavez, C. Mathematical Models in Population Biology and Epidemiology; Springer: Berlin/Heidelberg, Germany, 2001; Volume 40. [Google Scholar]
- Rios-Doria, D.; Chowell, G. Qualitative analysis of the level of cross-protection between epidemic waves of the 1918–1919 influenza pandemic. J. Theor. Biol. 2009, 261, 584–592. [Google Scholar] [CrossRef]
- González-Parra, G.; Arenas, A.; Diego, F.; Aranda, L.S. Modeling the epidemic waves of AH1N1/09 influenza around the world. Spat. Spatio Temporal Epidemiol. 2011, 2, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Andreasen, V.; Viboud, C.; Simonsen, L. Epidemiologic Characterization of the 1918 Influenza Pandemic Summer Wave in Copenhagen: Implications for Pandemic Control Strategies. J. Infect. Dis. 2008, 197, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.; Tobias, M. Predicting and preventing measles epidemics in New Zealand: Application of a mathematical model. Epidemiol. Infect. 2000, 124, 279–287. [Google Scholar] [CrossRef]
- Legrand, J.; Grais, R.F.; Boelle, P.Y.; Valleron, A.J.; Flahault, A. Understanding the dynamics of Ebola epidemics. Epidemiol. Infect. 2007, 135, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.M.; Duintjer Tebbens, R.J.; Pallansch, M.A. Evaluation of response scenarios to potential polio outbreaks using mathematical models. Risk Anal. 2006, 26, 1541–1556. [Google Scholar] [CrossRef]
- Kim, Y.; Barber, A.V.; Lee, S. Modeling influenza transmission dynamics with media coverage data of the 2009 H1N1 outbreak in Korea. PLoS ONE 2020, 15, e0232580. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Jung, E. Mathematical model of transmission dynamics and optimal control strategies for 2009 A/H1N1 influenza in the Republic of Korea. J. Theor. Biol. 2017, 412, 74–85. [Google Scholar] [CrossRef]
- Roda, W.C.; Varughese, M.B.; Han, D.; Li, M.Y. Why is it difficult to accurately predict the COVID-19 epidemic? Infect. Dis. Model. 2020, 5, 271–281. [Google Scholar] [CrossRef]
- Holmdahl, I.; Buckee, C. Wrong but useful- what covid-19 epidemiologic models can and cannot tell us. N. Engl. J. Med. 2020, 383, 303–305. [Google Scholar] [CrossRef]
- Jewell, N.P.; Lewnard, J.A.; Jewell, B.L. Caution Warranted: Using the Institute for Health Metrics and Evaluation Model for Predicting the Course of the COVID-19 Pandemic. Ann. Intern. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sperrin, M.; Grant, S.W.; Peek, N. Prediction models for diagnosis and prognosis in Covid-19. BMJ 2020, 369. [Google Scholar] [CrossRef]
- Taylor, L. Covid-19: Is Manaus the final nail in the coffin for natural herd immunity? BMJ 2021, 372, n394. [Google Scholar] [CrossRef]
- da Silva, P.C.V.; Velásquez-Rojas, F.; Connaughton, C.; Vazquez, F.; Moreno, Y.; Rodrigues, F.A. Epidemic spreading with awareness and different timescales in multiplex networks. Phys. Rev. E 2019, 100, 032313. [Google Scholar] [CrossRef]
- Abdel-Hamid, T.; Ankel, F.; Battle-Fisher, M.; Gibson, B.; Gonzalez-Parra, G.; Jalali, M.; Kaipainen, K.; Kalupahana, N.; Karanfil, O.; Marathe, A.; et al. Public and health professionals’ misconceptions about the dynamics of body weight gain/loss. Syst. Dyn. Rev. 2014, 30, 58–74. [Google Scholar] [CrossRef]
- Malecki, K.M.; Keating, J.A.; Safdar, N. Crisis communication and public perception of COVID-19 risk in the era of social media. Clin. Infect. Dis. 2021, 72, 697–702. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, C.; Reynolds, T.L.; Bai, Q.; Huang, Y.; Li, C.; Zheng, K.; Chen, Y. Why do people oppose mask wearing? A comprehensive analysis of US tweets during the COVID-19 pandemic. J. Am. Med. Inform. Assoc. 2021. [Google Scholar] [CrossRef]
- Kucharski, A.J.; Russell, T.W.; Diamond, C.; Liu, Y.; Edmunds, J.; Funk, S.; Eggo, R.M.; Sun, F.; Jit, M.; Munday, J.D.; et al. Early dynamics of transmission and control of COVID-19: A mathematical modelling study. Lancet Infect. Dis. 2020, 20, 553–558. [Google Scholar] [CrossRef]
- Stutt, R.O.; Retkute, R.; Bradley, M.; Gilligan, C.A.; Colvin, J. A modelling framework to assess the likely effectiveness of facemasks in combination with lock-down in managing the COVID-19 pandemic. Proc. R. Soc. A 2020, 476, 20200376. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, N.M.; Laydon, D.; Nedjati-Gilani, G.; Imai, N.; Ainslie, K.; Baguelin, M.; Bhatia, S.; Boonyasiri, A.; Cucunubá, Z.; Cuomo-Dannenburg, G.; et al. Impact of Non-Pharmaceutical Interventions (NPIs) to Reduce COVID-19 Mortality and Healthcare Demand; Imperial College: London, UK, 2020. [Google Scholar] [CrossRef]
- Tang, Z.; Li, X.; Li, H. Prediction of New Coronavirus Infection Based on a Modified SEIR Model. medRxiv 2020. [Google Scholar] [CrossRef]
- Kuniya, T. Prediction of the Epidemic Peak of Coronavirus Disease in Japan, 2020. J. Clin. Med. 2020, 9, 789. [Google Scholar] [CrossRef]
- Zhong, L.; Mu, L.; Li, J.; Wang, J.; Yin, Z.; Liu, D. Early Prediction of the 2019 Novel Coronavirus Outbreak in the Mainland China based on Simple Mathematical Model. IEEE Access 2020, 8, 51761–51769. [Google Scholar] [CrossRef] [PubMed]
- Postnikov, E.B. Estimation of COVID-19 dynamics “on a back-of-envelope”: Does the simplest SIR model provide quantitative parameters and predictions? Chaos Solitons Fractals 2020, 135, 109841. [Google Scholar] [CrossRef] [PubMed]
- Barmparis, G.D.; Tsironis, G. Estimating the infection horizon of COVID-19 in eight countries with a data-driven approach. Chaos Solitons Fractals 2020, 135, 109842. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Ahmed, A.; Hossain, K.M.; Haque, T.; Hossain, M.A. Impact of control strategies on COVID-19 pandemic and the SIR model based forecasting in Bangladesh. medRxiv 2020. [Google Scholar] [CrossRef]
- Chaves, L.F.; Hurtado, L.A.; Rojas, M.R.; Friberg, M.D.; Rodríguez, R.M.; Avila-Aguero, M.L. COVID-19 basic reproduction number and assessment of initial suppression policies in Costa Rica. Math. Model. Nat. Phenom. 2020, 15, 32. [Google Scholar] [CrossRef]
- Grauer, J.; Löwen, H.; Liebchen, B. Strategic spatiotemporal vaccine distribution increases the survival rate in an infectious disease like Covid-19. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Hou, C.; Chen, J.; Zhou, Y.; Hua, L.; Yuan, J.; He, S.; Guo, Y.; Zhang, S.; Jia, Q.; Zhao, C.; et al. The effectiveness of quarantine of Wuhan city against the Corona Virus Disease 2019 (COVID-19): A well-mixed SEIR model analysis. J. Med. Virol. 2020, 92, 841–848. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, X.; Sun, H.; Tick, G.R.; Wei, W.; Jin, B. Applicability of time fractional derivative models for simulating the dynamics and mitigation scenarios of COVID-19. Chaos Solitons Fractals 2020, 138, 109959. [Google Scholar] [CrossRef]
- Matrajt, L.; Eaton, J.; Leung, T.; Brown, E.R. Vaccine optimization for COVID-19, who to vaccinate first? medRxiv 2020. [Google Scholar] [CrossRef]
- Fain, B.; Dobrovolny, H.M. Initial Inoculum and the Severity of COVID-19: A Mathematical Modeling Study of the Dose-Response of SARS-CoV-2 Infections. Epidemiologia 2020, 1, 5–15. [Google Scholar] [CrossRef]
- Thomas, D.M.; Sturdivant, R.; Dhurandhar, N.V.; Debroy, S.; Clark, N. A primer on COVID-19 Mathematical Models. Obesity 2020, 28, 1375–1377. [Google Scholar] [CrossRef]
- Buitrago-Garcia, D.; Egli-Gany, D.; Counotte, M.J.; Hossmann, S.; Imeri, H.; Ipekci, A.M.; Salanti, G.; Low, N. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: A living systematic review and meta-analysis. PLoS Med. 2020, 17, e1003346. [Google Scholar] [CrossRef] [PubMed]
- Pachetti, M.; Marini, B.; Giudici, F.; Benedetti, F.; Angeletti, S.; Ciccozzi, M.; Masciovecchio, C.; Ippodrino, R.; Zella, D. Impact of lockdown on Covid-19 case fatality rate and viral mutations spread in 7 countries in Europe and North America. J. Transl. Med. 2020, 18, 1–7. [Google Scholar] [CrossRef]
- van Dorp, L.; Acman, M.; Richard, D.; Shaw, L.P.; Ford, C.E.; Ormond, L.; Owen, C.J.; Pang, J.; Tan, C.C.; Boshier, F.A.; et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020, 83, 104351. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 2020, 182, 1284–1294. [Google Scholar] [CrossRef]
- Area, I.; Vidal, X.H.; Nieto, J.J.; Hermida, M.J.P. Determination in Galicia of the required beds at Intensive Care Units. Alex. Eng. J. 2021, 60, 559–564. [Google Scholar] [CrossRef]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed asymptomatic carrier transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, X.; Zhang, X.; Wei, Z.; Zhang, L.; Xu, J.; Liang, P.; Xu, Y.; Zhang, C.; Xu, A. Rapid asymptomatic transmission of COVID-19 during the incubation period demonstrating strong infectivity in a cluster of youngsters aged 16–23 years outside Wuhan and characteristics of young patients with COVID-19: A prospective contact-tracing study. J. Infect. 2020, 80, e1–e13. [Google Scholar] [CrossRef]
- Mizumoto, K.; Kagaya, K.; Zarebski, A.; Chowell, G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance 2020, 25, 2000180. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Cornforth, D.M.; Dushoff, J.; Weitz, J.S. The time scale of asymptomatic transmission affects estimates of epidemic potential in the COVID-19 outbreak. Epidemics 2020, 31, 100392. [Google Scholar] [CrossRef]
- Shao, S.; Zhou, D.; He, R.; Li, J.; Zou, S.; Mallery, K.; Kumar, S.; Yang, S.; Hong, J. Risk assessment of airborne transmission of COVID-19 by asymptomatic individuals under different practical settings. J. Aerosol Sci. 2020, 151, 105661. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.; Yokoe, D.S.; Havlir, D.V. Asymptomatic Transmission, the Achilles’ Heel of Current Strategies to Control Covid-19. N. Engl. J. Med. 2020, 382, 2158–2160. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, R.; Anzai, A.; Jung, S.M.; Linton, N.M.; Miyama, T.; Kobayashi, T.; Hayashi, K.; Suzuki, A.; Yang, Y.; Akhmetzhanov, A.R.; et al. Containment, Contact Tracing and Asymptomatic Transmission of Novel Coronavirus Disease (COVID-19): A Modelling Study. J. Clin. Med. 2020, 9, 3125. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Li, R.; Han, Y.; Zhang, R.; Li, J. COVID-19: Insight into the asymptomatic SARS-COV-2 infection and transmission. Int. J. Biol. Sci. 2020, 16, 2803. [Google Scholar] [CrossRef]
- Teixeira, S.C. Mild and asymptomatic cases of COVID-19 are potential threat for faecal–oral transmission. Braz. J. Infect. Dis. 2020, 24, 368. [Google Scholar] [CrossRef]
- Pollán, M.; Pérez-Gómez, B.; Pastor-Barriuso, R.; Oteo, J.; Hernán, M.A.; Pérez-Olmeda, M.; Sanmartín, J.L.; Fernández-García, A.; Cruz, I.; de Larrea, N.F.; et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): A nationwide, population-based seroepidemiological study. Lancet 2020, 396, 535–544. [Google Scholar] [CrossRef]
- Ndaïrou, F.; Area, I.; Nieto, J.J.; Torres, D.F. Mathematical modeling of COVID-19 transmission dynamics with a case study of Wuhan. Chaos Solitons Fractals 2020, 135, 109846. [Google Scholar] [CrossRef]
- Nikolai, L.A.; Meyer, C.G.; Kremsner, P.G.; Velavan, T.P. Asymptomatic SARS Coronavirus 2 infection: Invisible yet invincible. Int. J. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Kronbichler, A.; Kresse, D.; Yoon, S.; Lee, K.H.; Effenberger, M.; Shin, J.I. Asymptomatic patients as a source of COVID-19 infections: A systematic review and meta-analysis. Int. J. Infect. Dis. 2020, 98, 180–186. [Google Scholar] [CrossRef]
- He, J.; Guo, Y.; Mao, R.; Zhang, J. Proportion of asymptomatic coronavirus disease 2019: A systematic review and meta-analysis. J. Med. Virol. 2021, 93, 820–830. [Google Scholar] [CrossRef]
- Johansson, M.A.; Quandelacy, T.M.; Kada, S.; Prasad, P.V.; Steele, M.; Brooks, J.T.; Slayton, R.B.; Biggerstaff, M.; Butler, J.C. SARS-CoV-2 Transmission from People without COVID-19 Symptoms. JAMA Netw. Open 2021, 4, e2035057. [Google Scholar] [CrossRef]
- Silva, C.J.; Cruz, C.; Torres, D.F.; Muñuzuri, A.P.; Carballosa, A.; Area, I.; Nieto, J.J.; Fonseca-Pinto, R.; Passadouro, R.; Dos Santos, E.S.; et al. Optimal control of the COVID-19 pandemic: Controlled sanitary deconfinement in Portugal. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Havers, F.P.; Reed, C.; Lim, T.; Montgomery, J.M.; Klena, J.D.; Hall, A.J.; Fry, A.M.; Cannon, D.L.; Chiang, C.F.; Gibbons, A.; et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, 23 March–12 May 2020. JAMA Intern. Med. 2020, 180, 1576–1586. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B. 1.1. 7 in England. Science 2021. [Google Scholar] [CrossRef]
- İşlier, Z.G.; Güllü, R.; Hörmann, W. An exact and implementable computation of the final outbreak size distribution under Erlang distributed infectious period. Math. Biosci. 2020, 325, 108363. [Google Scholar] [CrossRef] [PubMed]
- González-Parra, G.; Acedo, L.; Villanueva-Micó, R.J.; Arenas, A.J. Modeling the social obesity epidemic with stochastic networks. Phys. A Stat. Mech. Appl. 2010, 389, 3692–3701. [Google Scholar] [CrossRef]
- Roberts, M.; Andreasen, V.; Lloyd, A.; Pellis, L. Nine challenges for deterministic epidemic models. Epidemics 2015, 10, 49–53. [Google Scholar] [CrossRef]
- González-Parra, G.; Dobrovolny, H.M. Assessing uncertainty in A2 respiratory syncytial virus viral dynamics. Comput. Math. Methods Med. 2015, 2015, 567589. [Google Scholar] [CrossRef]
- Wearing, H.J.; Rohani, P.; Keeling, M.J. Appropriate models for the management of infectious diseases. PLoS Med. 2005, 2, e174. [Google Scholar] [CrossRef]
- Faes, C.; Abrams, S.; Van Beckhoven, D.; Meyfroidt, G.; Vlieghe, E.; Hens, N. Time between Symptom Onset, Hospitalisation and Recovery or Death: Statistical Analysis of Belgian COVID-19 Patients. Int. J. Environ. Res. Public Health 2020, 17, 7560. [Google Scholar] [CrossRef]
- Faust, J.S.; del Rio, C. Assessment of Deaths From COVID-19 and From Seasonal Influenza. JAMA Intern. Med. 2020. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Walsh, F. Loss and resilience in the time of COVID-19: Meaning making, hope, and transcendence. Fam. Process 2020, 59, 898–911. [Google Scholar] [CrossRef]
- Wiegand, B.B.; Wiegand, S.d.C.B. Losses in times of a pandemic: Bioethical perceptions about mourning caused by COVID-19 in Brazil. Voices Bioeth. 2020, 6. [Google Scholar] [CrossRef]
- Zhai, Y.; Du, X. Loss and grief amidst COVID-19: A path to adaptation and resilience. Brain Behav. Immun. 2020, 87, 80–81. [Google Scholar] [CrossRef]
- Paltiel, A.D.; Schwartz, J.L.; Zheng, A.; Walensky, R.P. Clinical outcomes of a COVID-19 vaccine: Implementation over efficacy: Study examines how definitions and thresholds of vaccine efficacy, coupled with different levels of implementation effectiveness and background epidemic severity, translate into outcomes. Health Aff. 2020, 10–1377. [Google Scholar] [CrossRef]
- Ehrhardt, M.; Gašper, J.; Kilianová, S. SIR-based mathematical modeling of infectious diseases with vaccination and waning immunity. J. Comput. Sci. 2019, 37, 101027. [Google Scholar] [CrossRef]
- Iboi, E.A.; Ngonghala, C.N.; Gumel, A.B. Will an imperfect vaccine curtail the COVID-19 pandemic in the US? Infect. Dis. Model. 2020, 5, 510–524. [Google Scholar] [PubMed]
- Eikenberry, S.E.; Mancuso, M.; Iboi, E.; Phan, T.; Eikenberry, K.; Kuang, Y.; Kostelich, E.; Gumel, A.B. To mask or not to mask: Modeling the potential for face mask use by the general public to curtail the COVID-19 pandemic. Infect. Dis. Model. 2020, 5, 293–308. [Google Scholar] [CrossRef]
- Team, I.C.F. Modeling COVID-19 scenarios for the United States. Nat. Med. 2020. [Google Scholar] [CrossRef]
- Law, K.B.; Peariasamy, K.M.; Gill, B.S.; Singh, S.; Sundram, B.M.; Rajendran, K.; Dass, S.C.; Lee, Y.L.; Goh, P.P.; Ibrahim, H.; et al. Tracking the early depleting transmission dynamics of COVID-19 with a time-varying SIR model. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hethcote, H.W. Mathematics of infectious diseases. SIAM Rev. 2005, 42, 599–653. [Google Scholar] [CrossRef]
- van den Driessche, P.; Watmough, J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 2002, 180, 29–48. [Google Scholar] [CrossRef]
- Van den Driessche, P.; Watmough, J. Further Notes on the Basic Reproduction Number; Springer: Berlin/Heidelberg, Germany, 2008; pp. 159–178. [Google Scholar]
- Xia, S.; Duan, K.; Zhang, Y.; Zhao, D.; Zhang, H.; Xie, Z.; Li, X.; Peng, C.; Zhang, Y.; Zhang, W.; et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: Interim analysis of 2 randomized clinical trials. JAMA 2020, 324, 951–960. [Google Scholar] [CrossRef]
- Martinez-Rodriguez, D.; Gonzalez-Parra, G.; Villanueva-Mico, R.J. Analysis of key factors of a SARS-CoV-2 vaccination program: A mathematical modeling approach. medRxiv 2021. [Google Scholar] [CrossRef]
- Kim, L.; Garg, S.; O’Halloran, A.; Whitaker, M.; Pham, H.; Anderson, E.J.; Armistead, I.; Bennett, N.M.; Billing, L.; Como-Sabetti, K.; et al. Risk factors for intensive care unit admission and in-hospital mortality among hospitalized adults identified through the US coronavirus disease 2019 (COVID-19)-associated hospitalization surveillance network (COVID-NET). Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Yehia, B.R.; Winegar, A.; Fogel, R.; Fakih, M.; Ottenbacher, A.; Jesser, C.; Bufalino, A.; Huang, R.H.; Cacchione, J. Association of race with mortality among patients hospitalized with coronavirus disease 2019 (COVID-19) at 92 US hospitals. JAMA Netw. Open 2020, 3, e2018039. [Google Scholar] [CrossRef]
- Mukandavire, Z.; Nyabadza, F.; Malunguza, N.J.; Cuadros, D.F.; Shiri, T.; Musuka, G. Quantifying early COVID-19 outbreak transmission in South Africa and exploring vaccine efficacy scenarios. PLoS ONE 2020, 15, e0236003. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, A.; Yi, B.; Ding, K.; Wang, H.; Wang, J.; Xu, G. The epidemiological characteristics of infection in close contacts of COVID-19 in Ningbo city. Chin. J. Epidemiol. 2020, 41, 668–672. [Google Scholar]
- Mc Evoy, D.; McAloon, C.G.; Collins, A.B.; Hunt, K.; Butler, F.; Byrne, A.W.; Casey, M.; Barber, A.; Griffin, J.M.; Lane, E.A.; et al. The relative infectiousness of asymptomatic SARS-CoV-2 infected persons compared with symptomatic individuals: A rapid scoping review. medRxiv 2020. [Google Scholar] [CrossRef]
- Orenes-Piñero, E.; Baño, F.; Navas-Carrillo, D.; Moreno-Docón, A.; Marín, J.M.; Misiego, R.; Ramírez, P. Evidences of SARS-CoV-2 virus air transmission indoors using several untouched surfaces: A pilot study. Sci. Total Environ. 2021, 751, 142317. [Google Scholar] [CrossRef]
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection: A Narrative Review. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef]
- Zhao, H.J.; Lu, X.X.; Deng, Y.B.; Tang, Y.J.; Lu, J.C. COVID-19: Asymptomatic carrier transmission is an underestimated problem. Epidemiol. Infect. 2020, 148, 1–7. [Google Scholar] [CrossRef]
- Lauer, S.A.; Grantz, K.H.; Bi, Q.; Jones, F.K.; Zheng, Q.; Meredith, H.R.; Azman, A.S.; Reich, N.G.; Lessler, J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: Estimation and application. Ann. Intern. Med. 2020, 172, 577–582. [Google Scholar] [CrossRef]
- Hoseinpour Dehkordi, A.; Alizadeh, M.; Derakhshan, P.; Babazadeh, P.; Jahandideh, A. Understanding epidemic data and statistics: A case study of COVID-19. J. Med. Virol. 2020, 92, 868–882. [Google Scholar] [CrossRef] [PubMed]
- Quah, P.; Li, A.; Phua, J. Mortality rates of patients with COVID-19 in the intensive care unit: A systematic review of the emerging literature. Crit. Care 2020, 24, 1–4. [Google Scholar] [CrossRef]
- The World Bank. 2021. Available online: https://data.worldbank.org/ (accessed on 26 March 2021).
- Santarpia, J.L.; Rivera, D.N.; Herrera, V.; Morwitzer, M.J.; Creager, H.; Santarpia, G.W.; Crown, K.K.; Brett-Major, D.; Schnaubelt, E.; Broadhurst, M.J.; et al. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci. Rep. 2020, 110, 12732. [Google Scholar] [CrossRef] [PubMed]
- Hasanoglu, I.; Korukluoglu, G.; Asilturk, D.; Cosgun, Y.; Kalem, A.K.; Altas, A.B.; Kayaaslan, B.; Eser, F.; Kuzucu, E.A.; Guner, R. Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection 2020, 49, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Bragazzi, N.L.; Li, Q.; Tang, S.; Xiao, Y.; Wu, J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov). Infect. Dis. Model. 2020, 5, 248–255. [Google Scholar] [CrossRef]
- Surkova, E.; Nikolayevskyy, V.; Drobniewski, F. False-positive COVID-19 results: Hidden problems and costs. Lancet Respir. Med. 2020, 8, 1167–1168. [Google Scholar] [CrossRef]
- Sharfstein, J.M.; Becker, S.J.; Mello, M.M. Diagnostic testing for the novel coronavirus. JAMA 2020, 323, 1437–1438. [Google Scholar] [CrossRef]
- Raffle, A.E.; Pollock, A.M.; Harding-Edgar, L. Covid-19 mass testing programmes. BMJ 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Bisoffi, Z.; Pomari, E.; Deiana, M.; Piubelli, C.; Ronzoni, N.; Beltrame, A.; Bertoli, G.; Riccardi, N.; Perandin, F.; Formenti, F.; et al. Sensitivity, specificity and predictive values of molecular and serological tests for COVID-19: A longitudinal study in emergency room. Diagnostics 2020, 10, 669. [Google Scholar] [CrossRef] [PubMed]
- Wikipedia. 2020. Available online: https://en.wikipedia.org/wiki/List_of_countries_by_population_(United_Nations) (accessed on 26 March 2021).
- Ministerio Salud Colombia. 2021. Available online: https://www.minsalud.gov.co/portada-covid-19.html (accessed on 26 March 2021).
- Byambasuren, O.; Cardona, M.; Bell, K.; Clark, J.; McLaws, M.L.; Glasziou, P. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: Systematic review and meta-analysis. Off. J. Assoc. Med. Microbiol. Infect. Dis. Can. 2020, 5, 223–234. [Google Scholar]
- Al-Qahtani, M.; AlAli, S.; AbdulRahman, A.; Alsayyad, A.S.; Otoom, S.; Atkin, S.L. The prevalence of asymptomatic and symptomatic COVID-19 in a cohort of quarantined subjects. Int. J. Infect. Dis. 2020, 102, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Cullenbine, C.A.; Rohrer, J.W.; Almand, E.A.; Steel, J.J.; Davis, M.T.; Carson, C.M.; Hasstedt, S.; Sitko, J.C.; Wickert, D.P. Fizzle Testing: An Equation Utilizing Random Surveillance to Help Reduce COVID-19 Risks. Math. Comput. Appl. 2021, 26, 16. [Google Scholar]
- Menachemi, N.; Yiannoutsos, C.T.; Dixon, B.E.; Duszynski, T.J.; Fadel, W.F.; Wools-Kaloustian, K.K.; Needleman, N.U.; Box, K.; Caine, V.; Norwood, C.; et al. Population point prevalence of SARS-CoV-2 infection based on a statewide random sample—Indiana, 25–29 April 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 960. [Google Scholar] [CrossRef]
- Shakiba, M.; Nazemipour, M.; Heidarzadeh, A.; Mansournia, M. Prevalence of asymptomatic COVID-19 infection using a seroepidemiological survey. Epidemiol. Infect. 2020, 148, e300. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.; Prendecki, M.; Dhutia, A.; Ali, M.A.; Sajjad, H.; Shivakumar, O.; Lightstone, L.; Kelleher, P.; Pickering, M.C.; Thomas, D.; et al. High prevalence of asymptomatic COVID-19 infection in hemodialysis patients detected using serologic screening. J. Am. Soc. Nephrol. 2020, 31, 1969–1975. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Chen, J.; Li, L.; Jia, N.; Jiangtulu, B.; Xue, T.; Zhang, L.; Li, Z.; Ye, R.; Wang, B. Modeling the prevalence of asymptomatic COVID-19 infections in the Chinese mainland. Innovation 2020, 1, 100026. [Google Scholar] [CrossRef]
- Sartor, G.; Del Riccio, M.; Dal Poz, I.; Bonanni, P.; Bonaccorsi, G. COVID-19 in Italy: Considerations on official data. Int. J. Infect. Dis. 2020, 98, 188–190. [Google Scholar] [CrossRef]
- Ghaffarzadegan, N.; Rahmandad, H. Simulation-based estimation of the early spread of COVID-19 in Iran: Actual versus confirmed cases. Syst. Dyn. Rev. 2020, 36, 101–129. [Google Scholar] [CrossRef]
- Nature. 2021. Available online: https://www.nature.com/articles/d41586-021-00728-2 (accessed on 26 March 2021).
- Yan, P.; Chowell, G. Beyond the Initial Phase: Compartment Models for Disease Transmission. In Quantitative Methods for Investigating Infectious Disease Outbreaks; Springer: Berlin/Heidelberg, Germany, 2019; pp. 135–182. [Google Scholar]
- Clemente-Suárez, V.J.; Hormeño-Holgado, A.; Jiménez, M.; Benitez-Agudelo, J.C.; Navarro-Jiménez, E.; Perez-Palencia, N.; Maestre-Serrano, R.; Laborde-Cárdenas, C.C.; Tornero-Aguilera, J.F. Dynamics of population immunity due to the herd effect in the COVID-19 pandemic. Vaccines 2020, 8, 236. [Google Scholar] [CrossRef] [PubMed]
- Petrosillo, N. SARS-CoV-2, “Common Cold” Coronaviruses’ Cross-Reactivity and “Herd Immunity”: The Razor of Ockham (1285–1347)? Infect. Dis. Rep. 2020, 12, 22–23. [Google Scholar] [CrossRef]
- Köhler, J.M. Infection-Immunity Competition: A Simple Model for Illustrating the Background of Individual Response on Herd Immunity. Appl. Sci. 2020, 10, 3078. [Google Scholar] [CrossRef]
- Bartsch, S.M.; O’Shea, K.J.; Ferguson, M.C.; Bottazzi, M.E.; Wedlock, P.T.; Strych, U.; McKinnell, J.A.; Siegmund, S.S.; Cox, S.N.; Hotez, P.J.; et al. Vaccine efficacy needed for a COVID-19 coronavirus vaccine to prevent or stop an epidemic as the sole intervention. Am. J. Prev. Med. 2020, 59, 493–503. [Google Scholar] [CrossRef]
- Dinleyici, E.C.; Borrow, R.; Safadi, M.A.P.; van Damme, P.; Munoz, F.M. Vaccines and routine immunization strategies during the COVID-19 pandemic. Hum. Vaccines Immunother. 2020, 17, 400–407. [Google Scholar] [CrossRef]
- Knoll, M.D.; Wonodi, C. Oxford—AstraZeneca COVID-19 vaccine efficacy. Lancet 2020, 397, 72–74. [Google Scholar] [CrossRef]
- Krause, P.; Fleming, T.R.; Longini, I.; Henao-Restrepo, A.M.; Peto, R.; Dean, N.; Halloran, M.; Huang, Y.; Fleming, T.; Gilbert, P.; et al. COVID-19 vaccine trials should seek worthwhile efficacy. Lancet 2020, 396, 741–743. [Google Scholar] [CrossRef]
- Verger, P.; Dubé, E. Restoring confidence in vaccines in the COVID-19 era. Expert Rev. Vaccines 2020, 1–3. [Google Scholar] [CrossRef]
- Yamey, G.; Schäferhoff, M.; Hatchett, R.; Pate, M.; Zhao, F.; McDade, K.K. Ensuring global access to COVID-19 vaccines. Lancet 2020, 395, 1405–1406. [Google Scholar] [CrossRef]
- Chowell, G.; Ammon, C.; Hengartner, N.; Hyman, J. Transmission dynamics of the great influenza pandemic of 1918 in Geneva, Switzerland: Assessing the effects of hypothetical interventions. J. Theor. Biol. 2006, 241, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, O.; Heesterbeek, J.; Roberts, M. The construction of next-generation matrices for compartmental epidemic models. J. R. Soc. Interface 2009, 7, 873–885. [Google Scholar] [CrossRef]
- Britton, T.; Ball, F.; Trapman, P. A mathematical model reveals the influence of population heterogeneity on herd immunity to SARS-CoV-2. Science 2020, 369, 846–849. [Google Scholar] [CrossRef] [PubMed]
- Raue, A.; Kreutz, C.; Maiwald, T.; Bachmann, J.; Schilling, M.; Klingmüller, U.; Timmer, J. Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 2009, 25, 1923–1929. [Google Scholar] [CrossRef] [PubMed]
- Roosa, K.; Chowell, G. Assessing parameter identifiability in compartmental dynamic models using a computational approach: Application to infectious disease transmission models. Theor. Biol. Med. Model. 2019, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- González-Parra, G.; Benincasa, T. Mathematical modeling and numerical simulations of Zika in Colombia considering mutation. Math. Comput. Simul. 2019, 163, 1–18. [Google Scholar]
- World Health Organization. COVID-19 Weekly Epidemiological Update; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Washington, N.L.; Gangavarapu, K.; Zeller, M.; Bolze, A.; Cirulli, E.T.; Barrett, K.M.S.; Larsen, B.B.; Anderson, C.; White, S.; Cassens, T.; et al. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B. 1.1. 7 in the United States. medRxiv 2021. [Google Scholar] [CrossRef]
- Davies, N.G.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.; Pearson, C.A.; Russell, T.W.; Tully, D.C.; Abbott, S.; Gimma, A.; et al. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv 2020. [Google Scholar] [CrossRef]
- Iacobucci, G. Covid-19: New UK variant may be linked to increased death rate, early data indicate. BMJ 2021, 372, n230. [Google Scholar] [CrossRef] [PubMed]
- ICNARC; Group, Q. Hazard ratio of ICU admission for VOC infected individuals compared non-VOC infected individuals. Unpublished.
- Cheyne, C.; Burnside, G.; Hughes, D.; Turtle, L.G.F.M.; Semple, C. Analysis on recent hospital case fatality and variant of concern VOC2020-01 using CO-CIN data (10th February 2021). Unpublished.
- Public Health England. 2021. Available online: https://www.gov.uk/government/organisations/public-health-england (accessed on 26 March 2021).
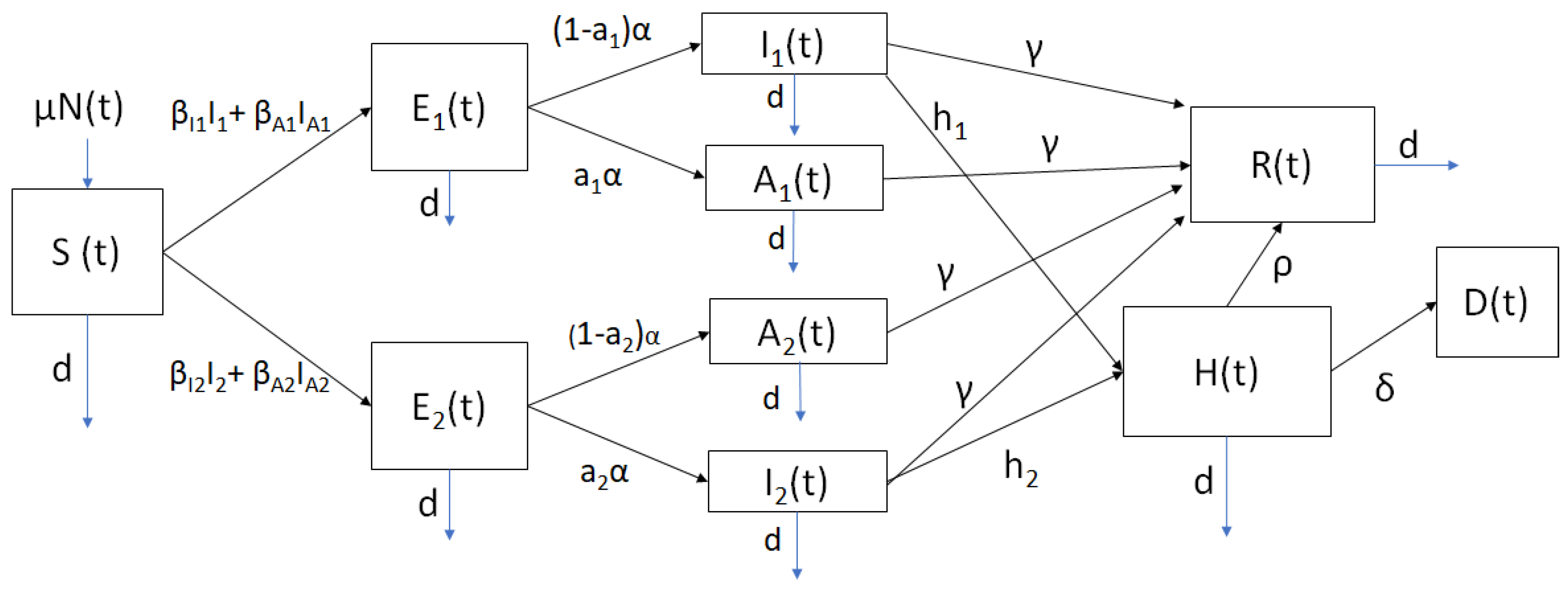
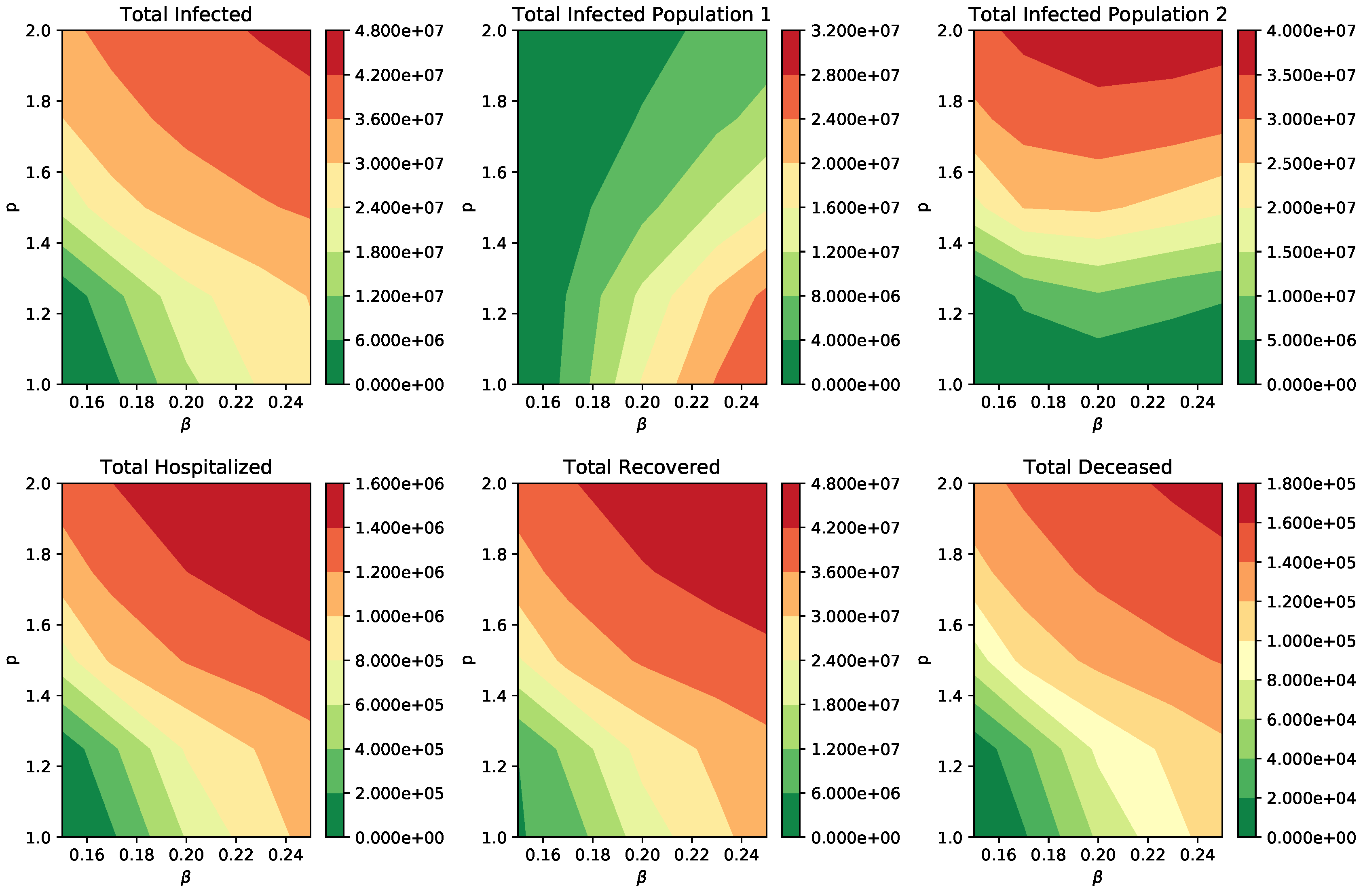
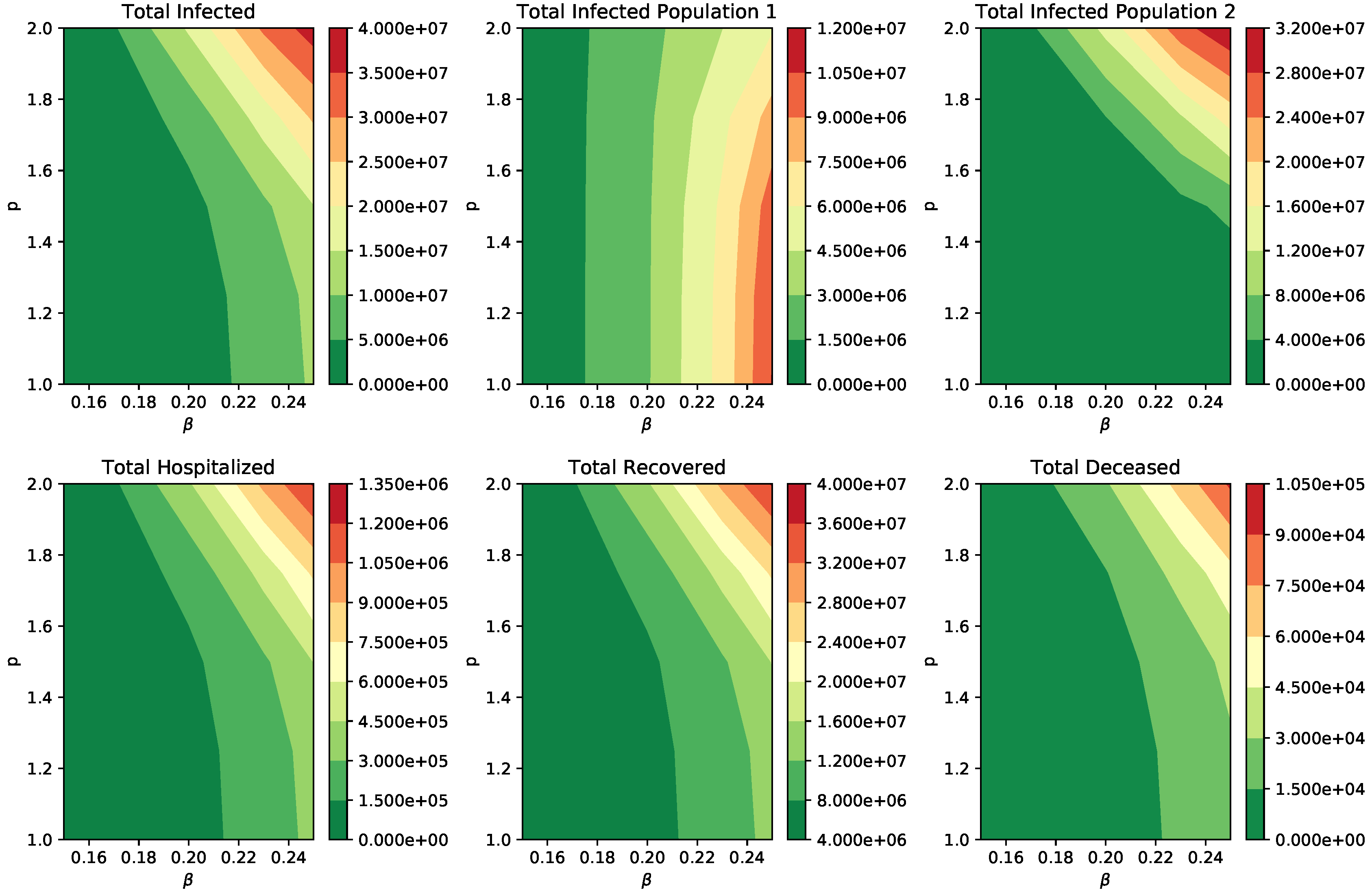
| Parameter | Symbol | Value or Range |
|---|---|---|
| Mean latent period | 5.2 days [9,110,125] | |
| Mean infectious period | 7 days [9] | |
| Rate progression from to H | 3.5 days × [9,58,126] | |
| Rate progression from to R | 10.4 × days [9,58,126] | |
| Death rate (hospitalized) | 10.4 days × [106,127] | |
| Probability of being asymptomatic | a | [13,123] |
| Birth rate | 0.0000407726 days [128] | |
| Death rate | d | 0.00001523835 days [128] |
| Parameter | Symbol | Value |
|---|---|---|
| Infected 1 (symptomatic) | ||
| Infected 2 (symptomatic) | 5 | |
| Latent 1 | ||
| Latent 2 | 5 | |
| Asymptomatic 1 | ||
| Asymptomatic 2 | 5 | |
| Hospitalized | 2589 | |
| Recovered | ||
| Susceptible | ||
| Total population |
| = 0.15 | = 0.17 | = 0.2 | = 0.23 | = 0.25 | |
|---|---|---|---|---|---|
| p = 1.0 | 1.271551 | 4.608728 | 1.653471 | 2.468029 | 2.847209 |
| p = 1.25 | 1.879443 | 1.007416 | 2.223573 | 2.740237 | 3.023723 |
| p = 1.5 | 1.968806 | 2.783918 | 3.267448 | 3.547061 | 3.684327 |
| p = 1.75 | 2.987208 | 3.394945 | 3.770218 | 3.995417 | 4.099287 |
| p = 2.0 | 3.456039 | 3.768143 | 4.058944 | 4.232105 | 4.310245 |
| = 0.15 | = 0.17 | = 0.2 | = 0.23 | = 0.25 | |
|---|---|---|---|---|---|
| p = 1.0 | 1.531743 | 5.600301 | 2.024257 | 3.038535 | 3.516916 |
| p = 1.25 | 6.311486 | 5.972911 | 9.397759 | 6.610649 | 5.402739 |
| p = 1.5 | 1.853126 | 2.515835 | 2.581866 | 2.330687 | 2.117589 |
| p = 1.75 | 2.885323 | 3.199643 | 3.349876 | 3.281885 | 3.175012 |
| p = 2.0 | 3.365184 | 3.612819 | 3.763003 | 3.758772 | 3.708030 |
| = 0.15 | = 0.17 | = 0.2 | = 0.23 | = 0.25 | |
|---|---|---|---|---|---|
| p = 1.0 | 4.957405 | 1.711537 | 6.127556 | 9.164529 | 1.057023 |
| p = 1.25 | 7.065646 | 3.612623 | 8.224247 | 1.017144 | 1.122371 |
| p = 1.5 | 7.127291 | 1.031755 | 1.212608 | 1.316221 | 1.367059 |
| p = 1.75 | 1.108271 | 1.259865 | 1.398868 | 1.482267 | 1.520734 |
| p = 2.0 | 1.282502 | 1.398100 | 1.505794 | 1.569921 | 1.598859 |
| = 0.15 | = 0.17 | = 0.2 | = 0.23 | = 0.25 | |
|---|---|---|---|---|---|
| p = 1.0 | 5345.659907 | 17,520.293341 | 62,924.410368 | 94,605.190134 | 109,116.091892 |
| p = 1.25 | 7301.442961 | 35,135.420382 | 84,220.953642 | 104,951.330011 | 115,841.614890 |
| p = 1.5 | 69,942.596753 | 106,053.317619 | 125,125.617692 | 135,814.208170 | 141,050.970076 |
| p = 1.75 | 114,227.805690 | 130,003.038329 | 144,326.962036 | 152,915.947908 | 156,877.382131 |
| p = 2.0 | 132,337.901616 | 144,247.831334 | 155,338.785818 | 161,942.866232 | 164,922.963036 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Parra, G.; Martínez-Rodríguez, D.; Villanueva-Micó, R.J. Impact of a New SARS-CoV-2 Variant on the Population: A Mathematical Modeling Approach. Math. Comput. Appl. 2021, 26, 25. https://doi.org/10.3390/mca26020025
Gonzalez-Parra G, Martínez-Rodríguez D, Villanueva-Micó RJ. Impact of a New SARS-CoV-2 Variant on the Population: A Mathematical Modeling Approach. Mathematical and Computational Applications. 2021; 26(2):25. https://doi.org/10.3390/mca26020025
Chicago/Turabian StyleGonzalez-Parra, Gilberto, David Martínez-Rodríguez, and Rafael J. Villanueva-Micó. 2021. "Impact of a New SARS-CoV-2 Variant on the Population: A Mathematical Modeling Approach" Mathematical and Computational Applications 26, no. 2: 25. https://doi.org/10.3390/mca26020025
APA StyleGonzalez-Parra, G., Martínez-Rodríguez, D., & Villanueva-Micó, R. J. (2021). Impact of a New SARS-CoV-2 Variant on the Population: A Mathematical Modeling Approach. Mathematical and Computational Applications, 26(2), 25. https://doi.org/10.3390/mca26020025








