CyVerse Austria—A Local, Collaborative Cyberinfrastructure
Abstract
1. Introduction
2. Materials and Methods
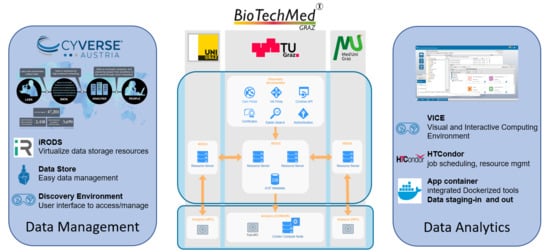
2.1. Data Management
Integrated Rule-Oriented Data Systems (iRODS) Deployment
2.2. Analytics
2.2.1. Discovery Environment
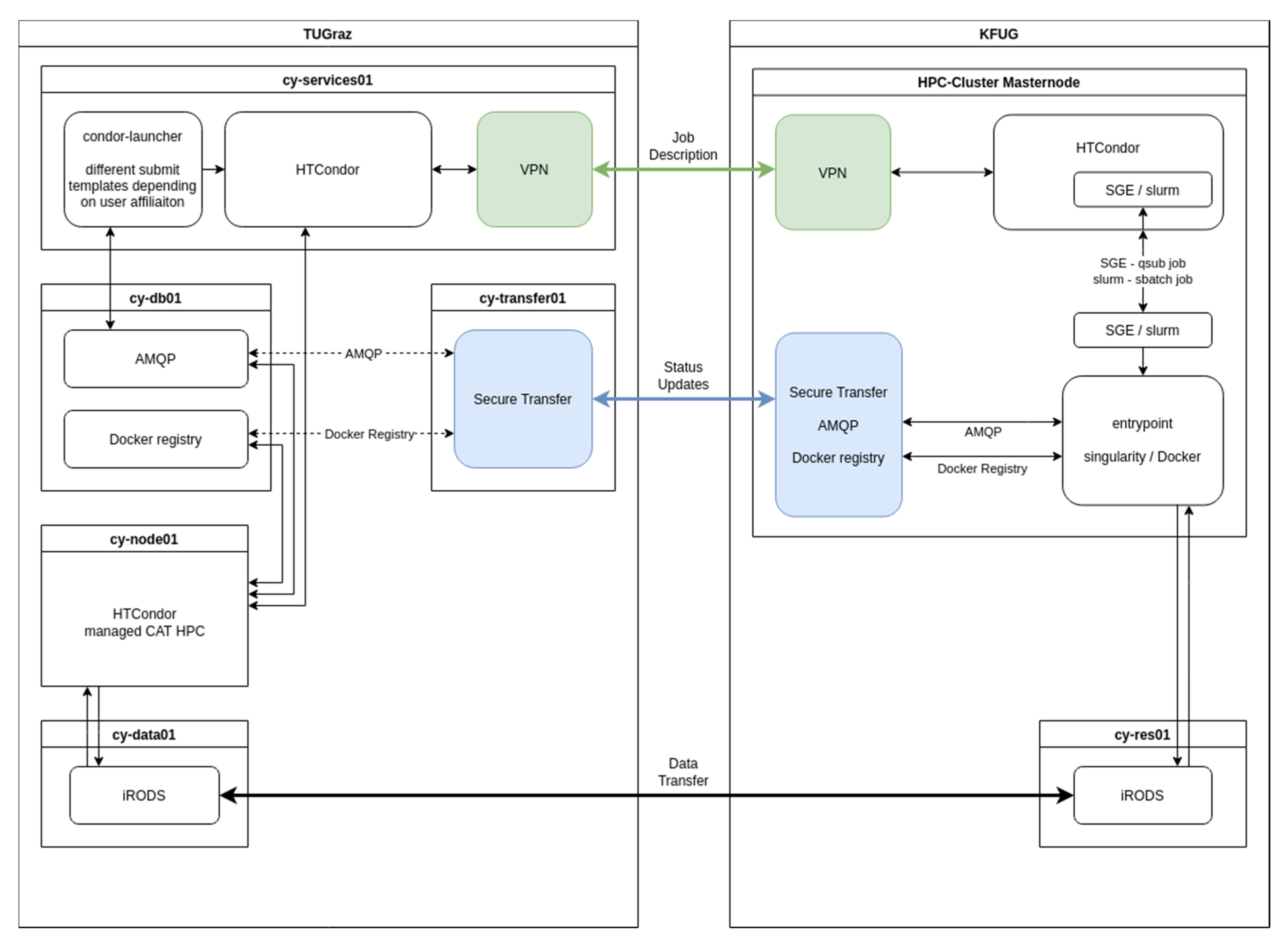
2.2.2. HPC Connection
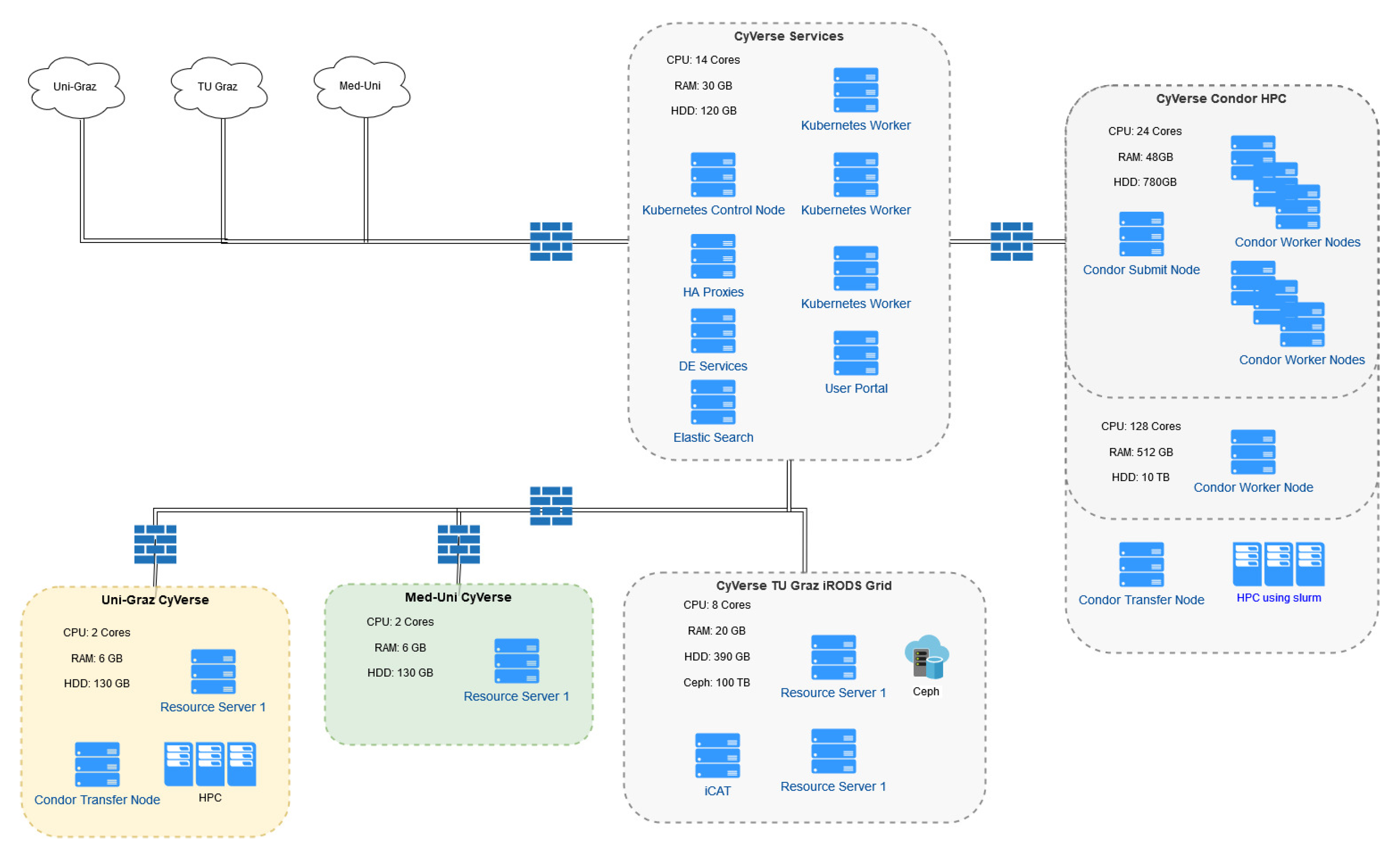
3. Results
3.1. Data Management
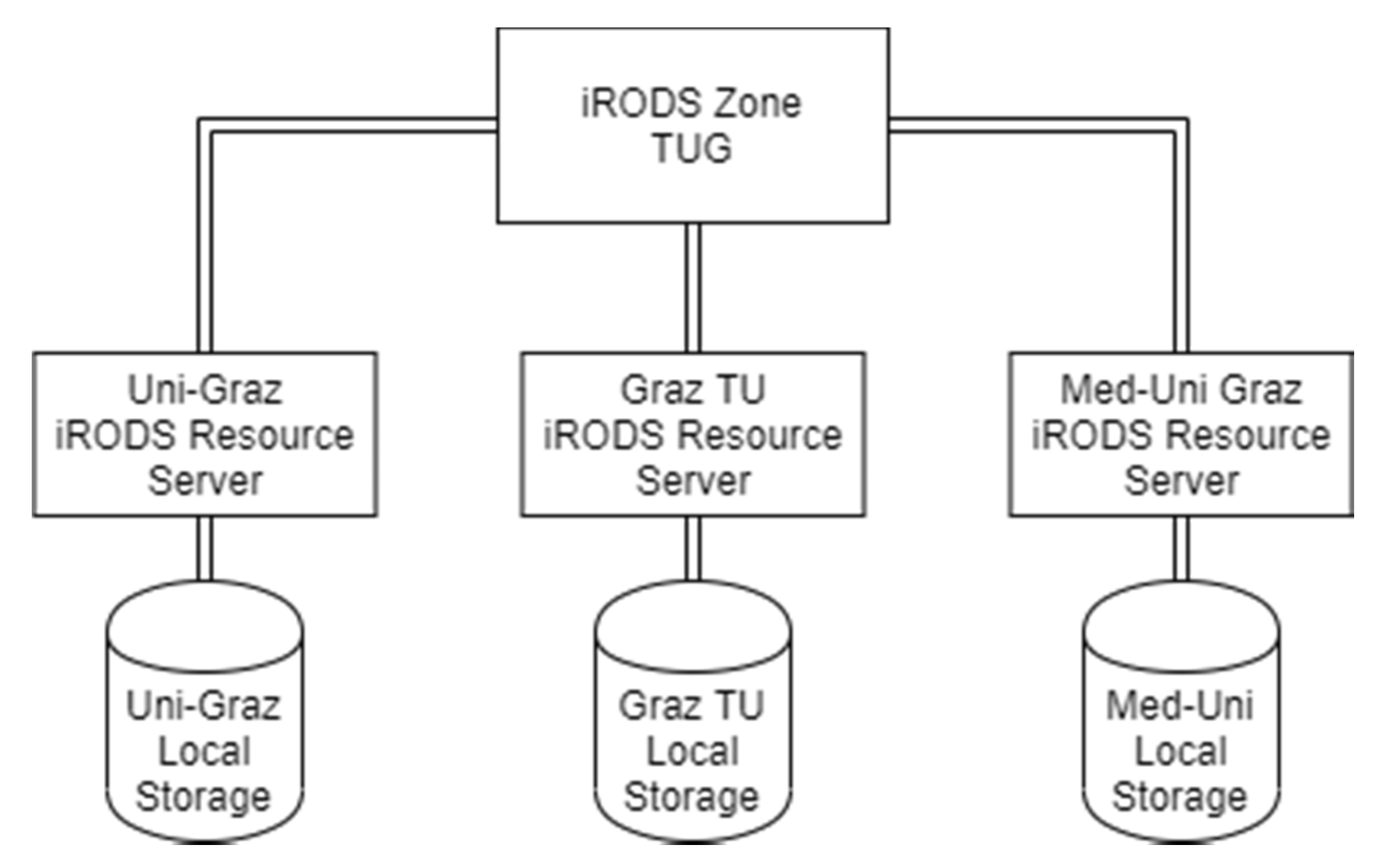
3.1.1. Integrated Rule-Oriented Data Systems (iRODS) Deployment
TUG Main Node
KFUG Node
MUG Node
Summarising
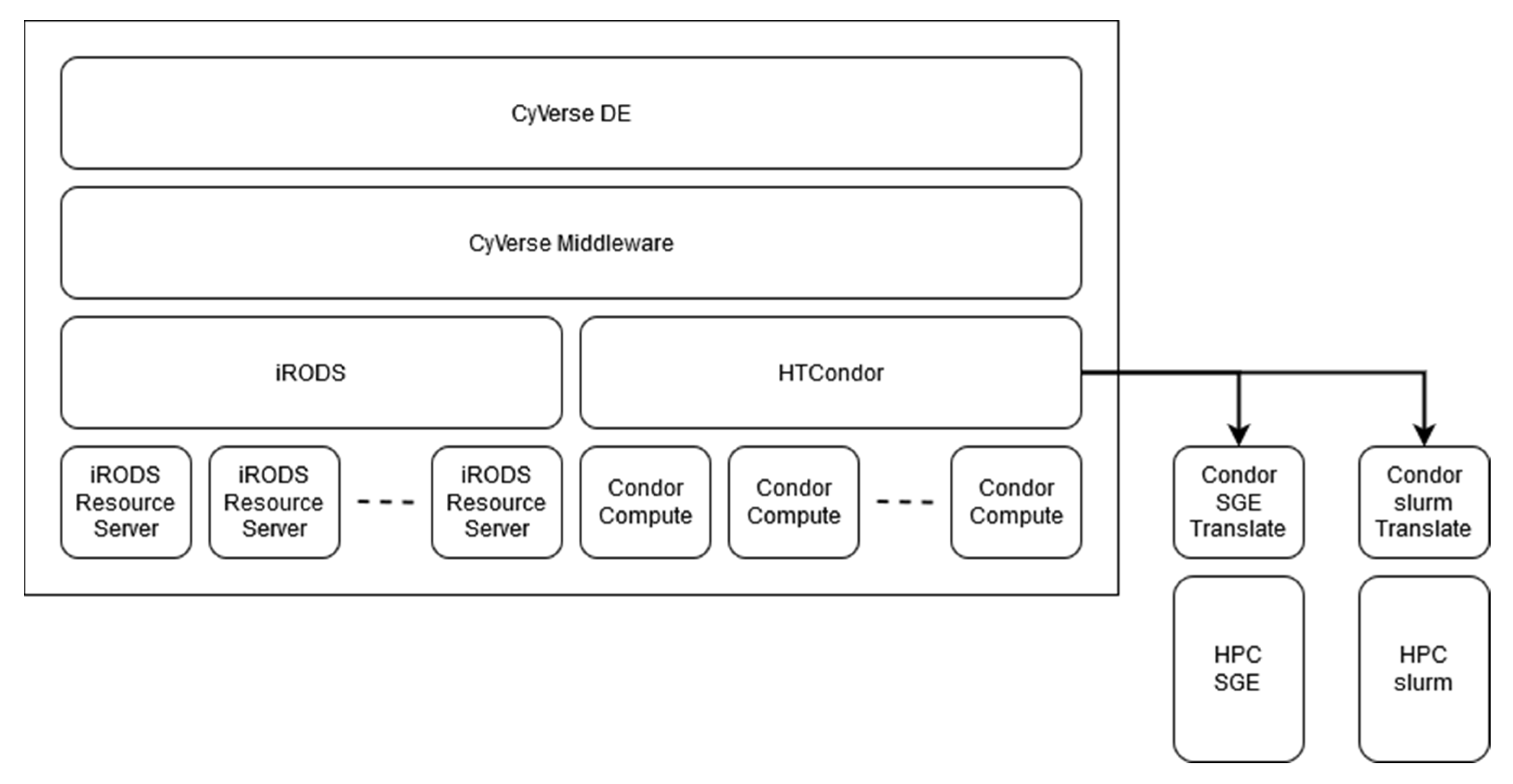
3.2. Analytics
3.2.1. HPC Connection
3.2.2. DE and Docker
3.3. Users
3.3.1. What Does CAT Offer to Researchers?
- Data management: (i) Data transfer: CAT data transfer is enabled by the third-party software tool Cyberduck [51] to drag and drop files between your computer and the Data store. In addition, iCommands, a collection of tools developed by iRODS, enables command line based data transfer. (ii) Data storage in the Data store: data can be managed in file systems comparable to a Windows explorer system with folders and subfolders. (iii) Metadata: In order to make data FAIR, one can add metadata to a single file or in bulk to large collections of data in the Data store. (iv) Data Sharing: Research data deposited in CAT can be shared with other CAT users. Users can assign read, write or own rights to the data [52].
- Data analytics: The DE offers a variety of tools for bioinformatics analyses to run as a script in the background, but also as a VICE app, with web-based user interface; check whether your tool is already available in the DE (CAT or others—transfer from CyVerse US tools is straightforward). You can find a more detailed description in reference [50]. In order to implement a new tool of interest to CAT, you need to (i) install Docker and all its dependencies on your local machine and (ii) have a URL for the tool and all its executables from a reliable source (e.g., GitHub).
3.3.2. Who Should Use CAT?
- ‘I would like to increase the efficiency and effectiveness of my day to day work with simplification of data management and publication processes.’
- ‘I want to apply for a European Commission call and I need a data management solution that is EOSC conform.’
- ‘I would like to connect my research data with appropriate standardized metadata to ensure adequate documentation of my data.’
- ‘I am collaborating with researchers at other institutions and we need a way to safely transfer data.’
- ‘I am working in a collaborative project with different research institutions and therefore different storage locations. However, we would like to have a combined data management, where we can see and structure all data, even if they are not stored at the same place.’
- ‘I would like to ensure reproducibility of my bioinformatics analyses also few years later (e.g., preserve analytics tool).’
- ‘I would like to automatize monotonous steps in my daily work, such as metadata extraction.’
- ‘I would like to create an easy-to-use, intuitive and standardized workflow for data analytics of command line-based applications for my PhD students.’
- ‘I would like to work with a platform that uses existing standards for data semantics and community standard practices for data management in order to ensure interoperability.’
- ‘I would like to use the HPC cluster at our university for computation and I need a safe way to transfer my data.’
- ‘I would like to keep up motivation and productivity of my PhD students by reducing time dedicated to administrative data management processes.’
- ‘I would like to stay up-to-date with the ongoing research in my local network in Graz.’
- ‘I would like to become part of a global network with a strong discipline-specific community developing tools for state-of-the-art research.’
3.3.3. CAT Use Cases
- Use Case 1: A researcher in structural biology has an analysis pipeline which needs several command line-based tools for protein structure prediction. The output of the first tool feeds as input file into the next tool and so on. In order to ensure that tools are properly combined and correct input files are chosen by different users in the lab, creation of an automated workflow to combine the analysis is crucial to ensure reproducibility. CAT was used to import all pieces of the analysis pipeline as docker images and a workflow was defined. Now, the whole team can use the workflow in a reproducible manner.
- Use Case 2: A researcher in computational bioinformatics is teaching a course with 20 students. To ensure that all students work with a shared dataset and regardless of their own hardware, with the same analytics tools and without having any problems with underlying dependencies, he chose CAT. All students receive a CAT account and do their analyses on the same platform using the data shared on the same platform.
- Use Case 3: A researcher in biology performs genomic data analysis requiring high computational power. Since institutes usually cannot serve such requirements, he uses the institutional HPC cluster. Therefore, he has to send his locally stored data to the HPC cluster and to ensure that his tool is available as a Singularity container for analysis on the HPC cluster. By using CAT, he stores his data and analysis tools on the platform which has a connection to the HPC cluster. Thereby, data and tools will be directly transferred and output is stored again in CAT.
- Use Case 4: A researcher from KFUG is leading a project with a consortium including TUG and MUG. As a coordinator of the project, the researcher has to ensure that data management of the whole project is done according to FAIR principles. Therefore, it is essential for them to have a central platform where all research data from the project is findable. By using CAT, they ensured that all data are either stored on the CAT storage resources or registered in iRODS making data findable.
3.3.4. Experiences/Lessons Learned
- Importance of support from CyVerse US. CAT is a local, independent instance of CyVerse; however, it is part of a global community with a lot of experience and know-how. The CAT team is steadily exchanging knowledge with the team from CyVerse US. There are a lot of joint development tasks in specific areas (e.g., HPC connections) ongoing. Moreover, tools for data analysis are easily exchanged between CAT and CyVerse US, showing the international level of collaborative science and the importance of FAIR.
- Data protection is a very important topic, mainly in the contexts of Medical Universities and industry collaborations. During the deployment of CAT and the connection of the MUG to our network, we learned that research data sharing, mainly in the clinical context, is an extremely sensitive topic. This leads to the necessity of tight regulations to ensure that it is clear which data can be shared in which format.
- Docker containerisation is a very nice, convenient and innovative tool for light conservation of data analytics software. However, due to superuser privileges that come with Docker containers, it is essential to use Docker-to-Singularity conversion to be able to work with the tools on HPC clusters.
- Although new hardware is permanently acquired for CAT, storage capacity and computing power will always remain a main limiting factor for researchers due to the accelerating progress in the development of big data disciplines, and therefore, also the need for high computational power. Therefore, the access to shared HPC resources is essential. With this, institutions can ensure that enough resources are available for researchers. Researchers can scale up their analyses if needed, but, in case they do not need the resources, computing power is available for other users.
- Motivation to invest time in learning the new technologies offered in CAT is essential. CAT offers many advantages for researchers; however, it requires some effort to learn how to use the platform and to reorganise existing data to the CAT Data Store. Hence, a certain commitment and motivation on the part of researchers is needed. Furthermore, the crucial aspect on maintaining user motivation is the investment in social aspects like build-up and care of a community and training possibilities. It is not sufficient just to provide the technical infrastructure; the human aspect has to be supported too. In addition, know-how for user support in CAT is required. CAT does not only imply new technologies for researchers, it does this for the IT staff too. It is of vital importance that there are adequate training possibilities offered to ensure a competent support staff keeping up-to-date with the fast-evolving progress of this field.
4. Discussion
4.1. Benefits of CAT for LS and HPC
4.2. Future Direction of CAT
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- ANR. Available online: https://anr.fr/en/latest-news/read/news/the-anr-introduces-a-data-management-plan-for-projects-funded-in-2019-onwards/ (accessed on 22 June 2020).
- NIH. Available online: https://grants.nih.gov/grants/policy/data_sharing/data_sharing_guidance.htm (accessed on 22 June 2020).
- Wellcome. Available online: https://wellcome.ac.uk/funding/guidance/how-complete-outputs-management-plan (accessed on 22 June 2020).
- FWF. Available online: https://www.fwf.ac.at/en/research-funding/open-access-policy/research-data-management/ (accessed on 22 June 2020).
- Boeckhout, M.; Zielhuis, G.A.; Bredenoord, A.L. The FAIR guiding principles for data stewardship: Fair enough? Eur. J. Hum. Genet. 2018, 26, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Guiding Principles for Findable, Accessible, Interoperable and Re-usable Data Publishing Version B1.0. Available online: https://www.force11.org/node/6062 (accessed on 22 June 2020).
- How do the FAIR principles translate to different disciplines? Available online: https://digitalscholarshipleiden.nl/articles/how-do-the-fair-principles-translate-to-different-disciplines (accessed on 22 June 2020).
- RDM policy at University of Graz. Available online: https://static.uni-graz.at/fileadmin/strategische-entwicklung/Dateien/FDM-Policy_EN_FINAL_Layout.pdf (accessed on 22 June 2020).
- RDM policy at TU Graz. Available online: https://www.tugraz.at/sites/research-data-management-rdm/policy/rdm-policy-at-tu-graz/ (accessed on 22 June 2020).
- Hollowel, C.; Barnett, J.; Caramarcu, C.; Stercker-Kellogg, W.; Wong, A.; Zaytsev, A. Mixing HTC and HPC Workloads with HTCondor and Slurm. J. Phys. Conf. Ser. 2017, 898. [Google Scholar] [CrossRef]
- Bloom, K.; Gerber, R. Computing Frontier: Distributed Computing and Facility Infrastructures. In Proceedings of the 2013 Community Summer Study on the Future of U.S. Particle Physics: Snowmass on the Mississippi (CSS2013), Minneapolis, MN, USA, 29 July–6 August 2013. [Google Scholar]
- Stewart, C.; Simms, S.; Plale, B.; Link, M.; Hancock, D.; Fox, G. What is Cyberinfrastructure? In Proceedings of the 38th Annual ACM SIGUCCS Fall Conference: Navigation and Discovery, Norfolk, VA, USA, 24–27 October 2010. [Google Scholar] [CrossRef]
- Wikpedia: Dependency Hell. Available online: https://en.wikipedia.org/wiki/Dependency_hell (accessed on 22 June 2020).
- Empowering App Development for Developers | Docker. Available online: https://www.docker.com/ (accessed on 22 June 2020).
- CyVerse US. Available online: https://cyverse.org (accessed on 22 June 2020).
- CyVerse US Code. Available online: https://github.com/cyverse/ (accessed on 22 June 2020).
- CyVerse Austria Code. Available online: https://github.com/cyverse-at/ (accessed on 22 June 2020).
- GoogleWebToolkit. Available online: http://www.gwtproject.org/ (accessed on 22 June 2020).
- OpenLDAP, Main Page. Available online: https://www.openldap.org/ (accessed on 22 June 2020).
- CAS | Apereo. Available online: https://www.apereo.org/projects/cas (accessed on 22 June 2020).
- iRODS. Available online: https://irods.org/ (accessed on 22 June 2020).
- HTCondor—Home. Available online: https://research.cs.wisc.edu/htcondor/ (accessed on 22 June 2020).
- Shibboleth Consortium—Privacy Preserving Identity Management. Available online: https://www.shibboleth.net/ (accessed on 22 June 2020).
- Open Source Search: The Creators of Elasticsearch, ELK Stack & Kibana | Elastic. Available online: https://www.elastic.co/ (accessed on 22 June 2020).
- CyVerse US DE Code. Available online: https://github.com/cyverse-de (accessed on 22 June 2020).
- CyVerse Austria DE Code. Available online: https://github.com/cyverse-at (accessed on 22 June 2020).
- Lenhardt, W.C.; Conway, M.; Scott, E.; Blanton, B.; Krishnamurthy, A.; Hadzikadic, M.; Vouk, M.; Wilson, A. Cross-institutional research cyberinfrastructure for data intensive science. In Proceedings of the 2016 IEEE High Performance Extreme Computing Conference (HPEC), Waltham, MA, USA, 13–15 September 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Merchant, N.; Lyons, E.; Goff, S.; Vaughn, M.; Ware, D.; Micklos, D.; Antin, P. The iPlant Collaborative: Cyberinfrastructure for Enabling Data to Discovery for the Life Sciences. PLOS Biol. 2016, 14, e1002342. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.A.; Vaughn, M.; McKay, S.; Lyons, E.; Stapleton, A.E.; Gessler, D.; Matasci, N.; Wang, L.; Hanlon, M.; Lenards, A.; et al. The iPlant Collaborative: Cyberinfrastructure for Plant Biology. Front. Plant Sci. 2011, 2, 34. [Google Scholar] [CrossRef] [PubMed]
- European Open Science Cloud (EOSC). Available online: https://ec.europa.eu/research/openscience/index.cfm?pg=open-science-cloud (accessed on 22 June 2020).
- B2SAFE. Available online: https://eudat.eu/services/userdoc/eudat-primer#EUDATPrimer-HowtojoinEUDAT (accessed on 22 June 2020).
- EOSC Marketplace. Available online: https://marketplace.eosc-portal.eu/services/b2safe (accessed on 22 June 2020).
- Ceph Homepage—Ceph. Available online: https://ceph.io/ (accessed on 22 June 2020).
- VSC: Home. Available online: http://vsc.ac.at/home/ (accessed on 22 June 2020).
- ZID Services TU Graz. Available online: https://www.tugraz.at/en/research/research-at-tu-graz/services-fuer-forschende/more-services-for-research/ (accessed on 22 June 2020).
- Son of Grid Engine. Available online: https://arc.liv.ac.uk/trac/SGE (accessed on 22 June 2020).
- Singularity. Available online: https://singularity.lbl.gov/ (accessed on 22 June 2020).
- MedBioNode. Available online: https://forschungsinfrastruktur.bmbwf.gv.at/en/institution/medical-university-of-graz_5?id=4107 (accessed on 22 June 2020).
- Slurm Workload Manager—Documentation. Available online: https://slurm.schedmd.com/ (accessed on 22 June 2020).
- Galaxy. Available online: https://usegalaxy.org/ (accessed on 22 June 2020).
- Galaxy Community Hub. Available online: https://galaxyproject.org/ (accessed on 22 June 2020).
- Conda—Conda documentation. Available online: https://docs.conda.io/en/latest/ (accessed on 22 June 2020).
- AMQP. Available online: https://www.amqp.org/ (accessed on 22 June 2020).
- Project Jupyter. Available online: https://www.jupyter.org (accessed on 22 June 2020).
- RStudio. Available online: https://rstudio.com/ (accessed on 22 June 2020).
- Shiny. Available online: https://shiny.rstudio.com/ (accessed on 22 June 2020).
- Randles, B.M.; Pasquetto, I.V.; Golshan, M.S.; Borgman, C.L. Using the Jupyter Notebook as a Tool for Open Science: An Empirical Study. In Proceedings of the 2017 ACM/IEEE Joint Conference on Digital Libraries (JCDL), Toronto, ON, Canada, 19–23 June 2017; pp. 1–2. [Google Scholar] [CrossRef]
- Mendez, K.M.; Pritchard, L.; Reinke, S.N.; Broadhurst, D.I. Toward collaborative open data science in metabolomics using Jupyter Notebooks and cloud computing. Metabolomics 2019, 15, 125. [Google Scholar] [CrossRef] [PubMed]
- Devisetty, U.K.; Kennedy, K.; Sarando, P.; Merchant, N.; Lyons, E. Bringing your tools to CyVerse Discovery Environment using Docker. F1000Research 2016, 5, 1442. [Google Scholar] [CrossRef] [PubMed]
- Cyberduck. Available online: https://cyberduck.io/ (accessed on 22 June 2020).
- CyVerse US Data Store. Available online: https://cyverse-data-store-guide.readthedocs-hosted.com/en/latest/index.html (accessed on 22 June 2020).
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lang, K.; Stryeck, S.; Bodruzic, D.; Stepponat, M.; Trajanoski, S.; Winkler, U.; Lindstaedt, S. CyVerse Austria—A Local, Collaborative Cyberinfrastructure. Math. Comput. Appl. 2020, 25, 38. https://doi.org/10.3390/mca25020038
Lang K, Stryeck S, Bodruzic D, Stepponat M, Trajanoski S, Winkler U, Lindstaedt S. CyVerse Austria—A Local, Collaborative Cyberinfrastructure. Mathematical and Computational Applications. 2020; 25(2):38. https://doi.org/10.3390/mca25020038
Chicago/Turabian StyleLang, Konrad, Sarah Stryeck, David Bodruzic, Manfred Stepponat, Slave Trajanoski, Ursula Winkler, and Stefanie Lindstaedt. 2020. "CyVerse Austria—A Local, Collaborative Cyberinfrastructure" Mathematical and Computational Applications 25, no. 2: 38. https://doi.org/10.3390/mca25020038
APA StyleLang, K., Stryeck, S., Bodruzic, D., Stepponat, M., Trajanoski, S., Winkler, U., & Lindstaedt, S. (2020). CyVerse Austria—A Local, Collaborative Cyberinfrastructure. Mathematical and Computational Applications, 25(2), 38. https://doi.org/10.3390/mca25020038