The Mass Spectrometry Identification, Antimicrobial Genes Detection, and Proteomics Analysis of Stutzerimonas stutzeri Strain Was Isolated from Industrial Wastewater
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Culture
2.2. Extraction of DNA
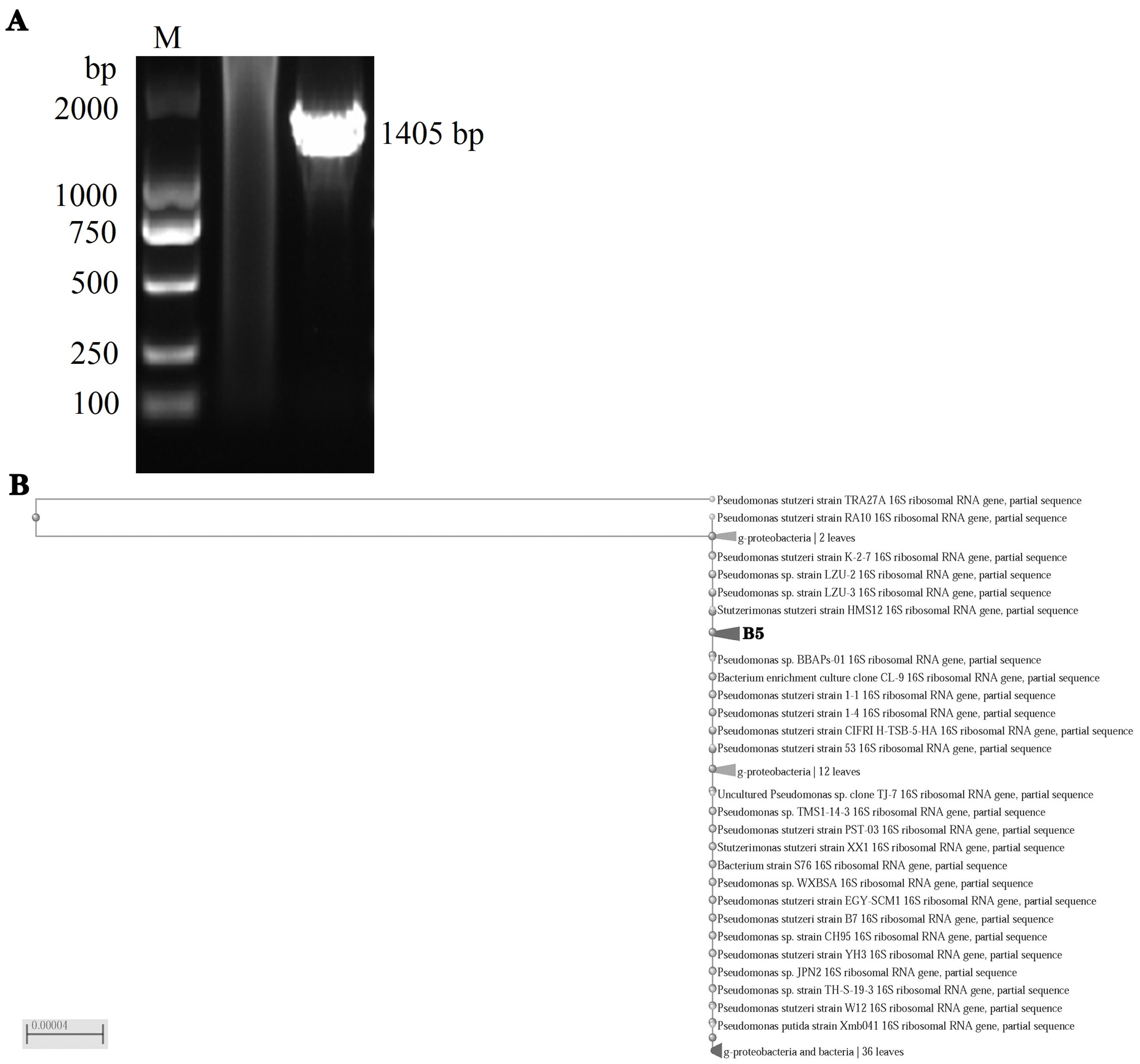
2.3. Construction of Phylogenetic Tree
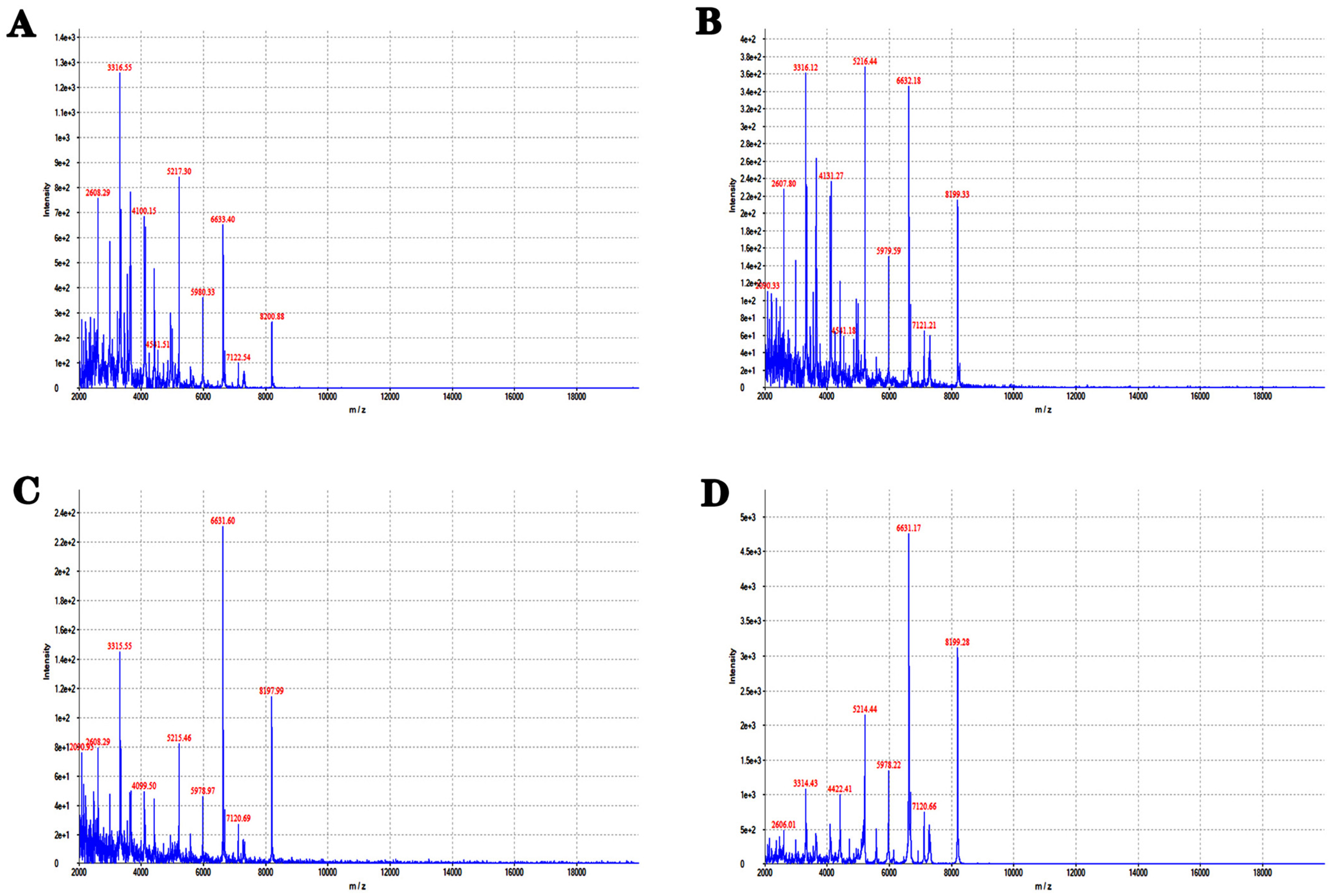
2.4. Identification of Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOFMS)
2.5. Biochemical Identification
2.6. Antibiotic Resistance Analysis
2.7. Detection of Bacterial Growth Curve
2.8. Identification of Drug-Resistant Genes
2.9. Proteomic Analysis
3. Results
3.1. Rapid Identification of Stutzerimonas stutzeri Using MALDI-TOFMS
3.2. Molecular Identification of Stutzerimonas stutzeri
3.3. Biochemical Identification of Stutzerimonas stutzeri
3.4. Antibiotic Susceptibility of Stutzerimonas stutzeri
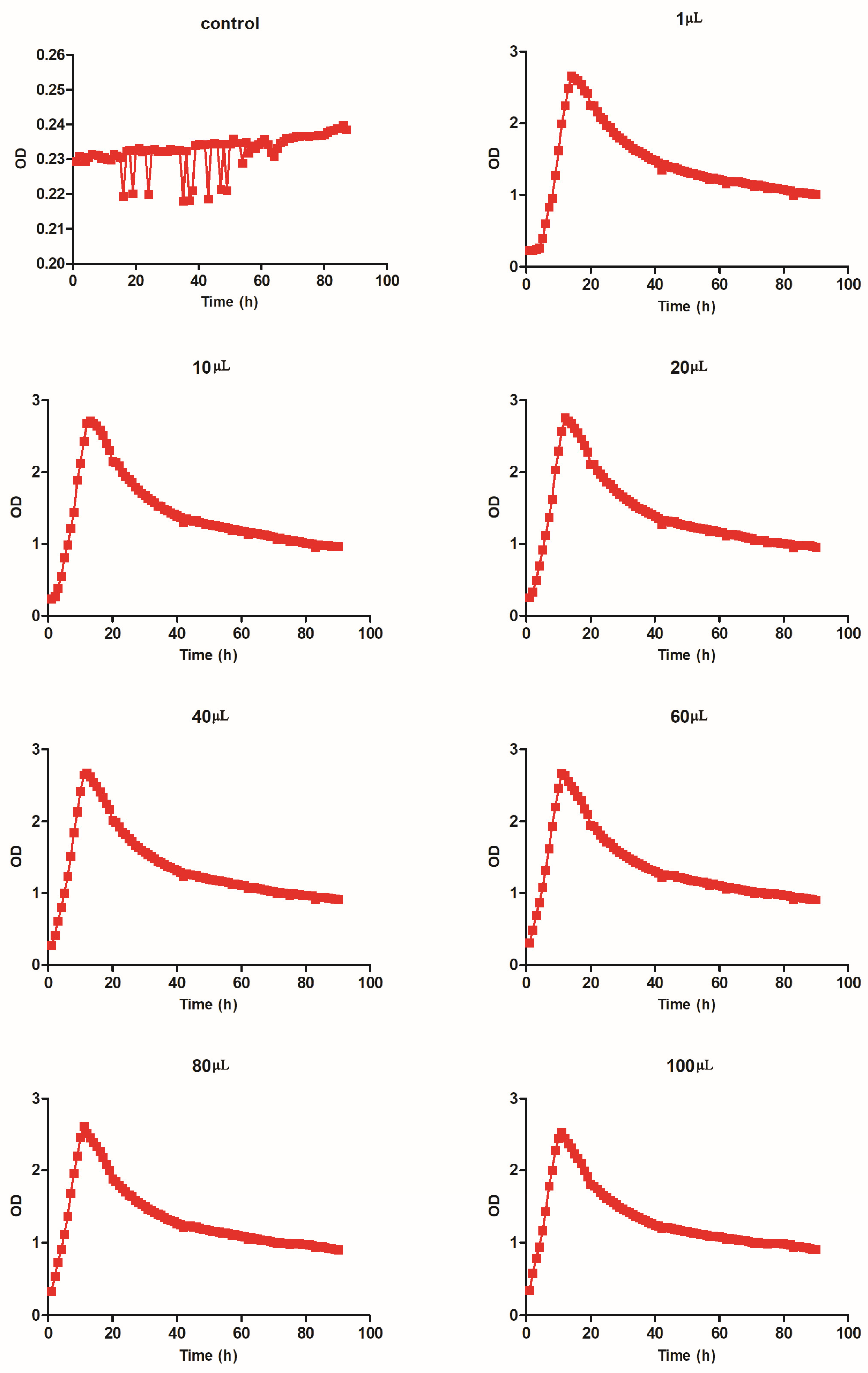
3.5. Growth Test for Stutzerimonas stutzeri
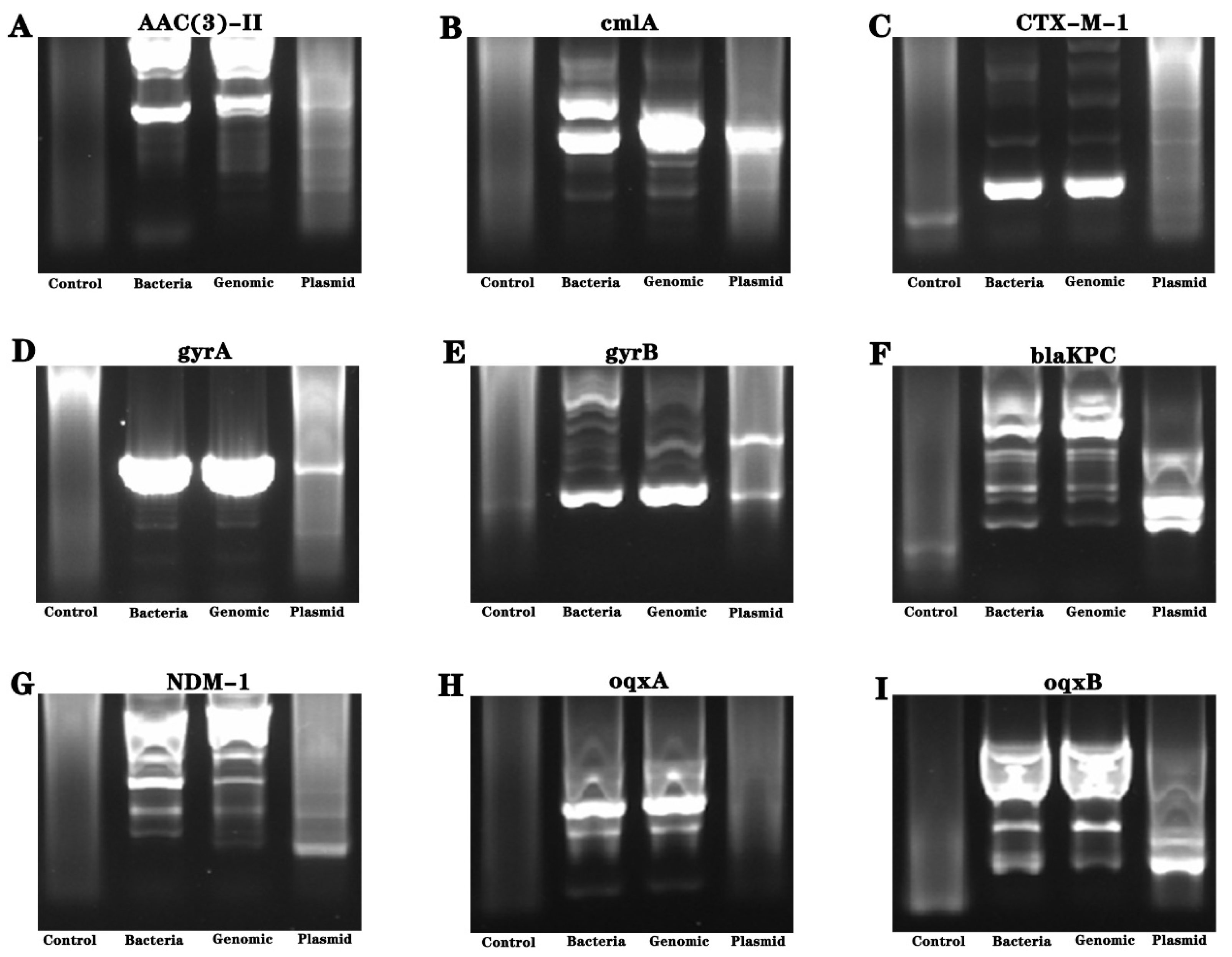
3.6. Detection of Drug-Resistant Genes
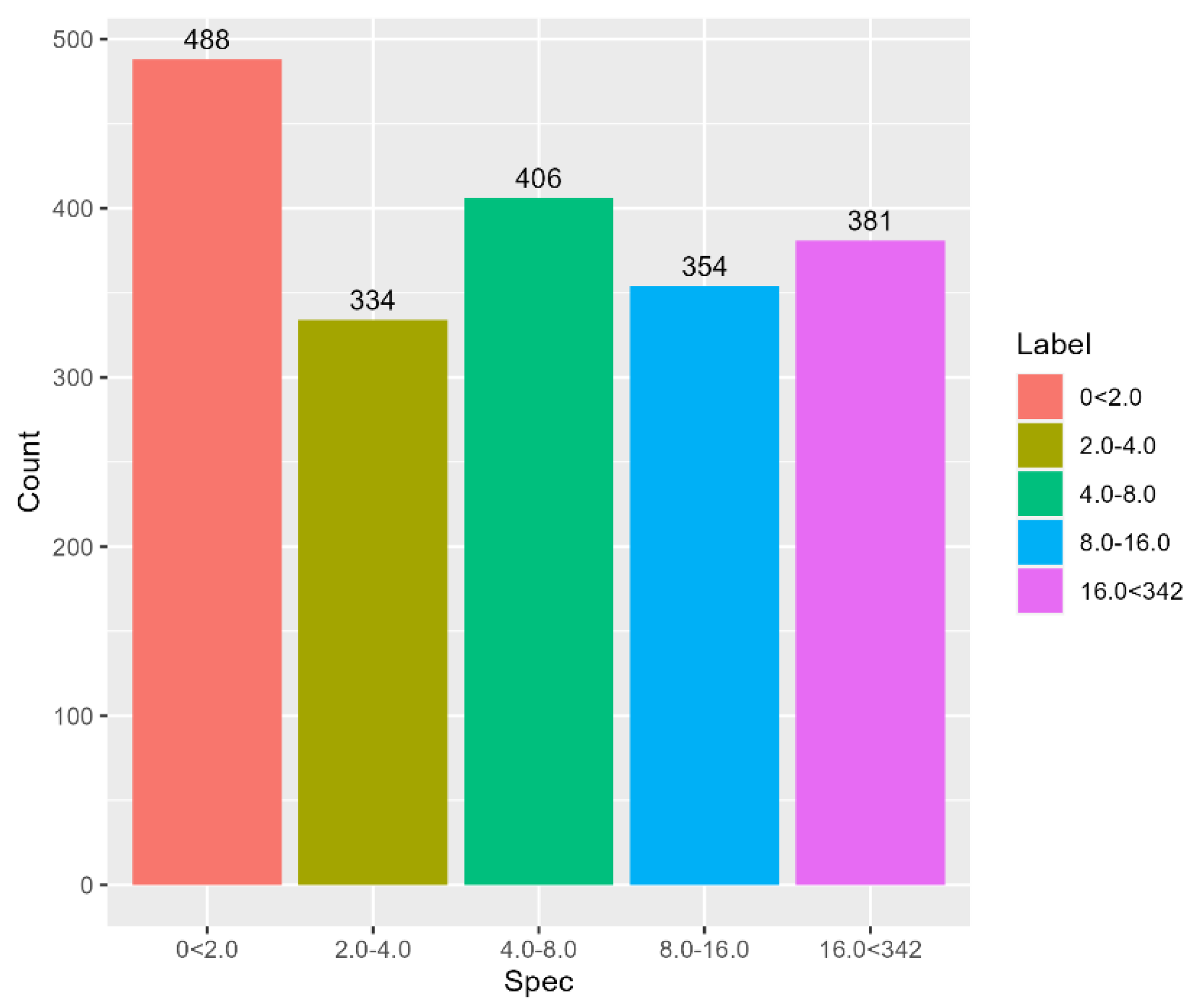
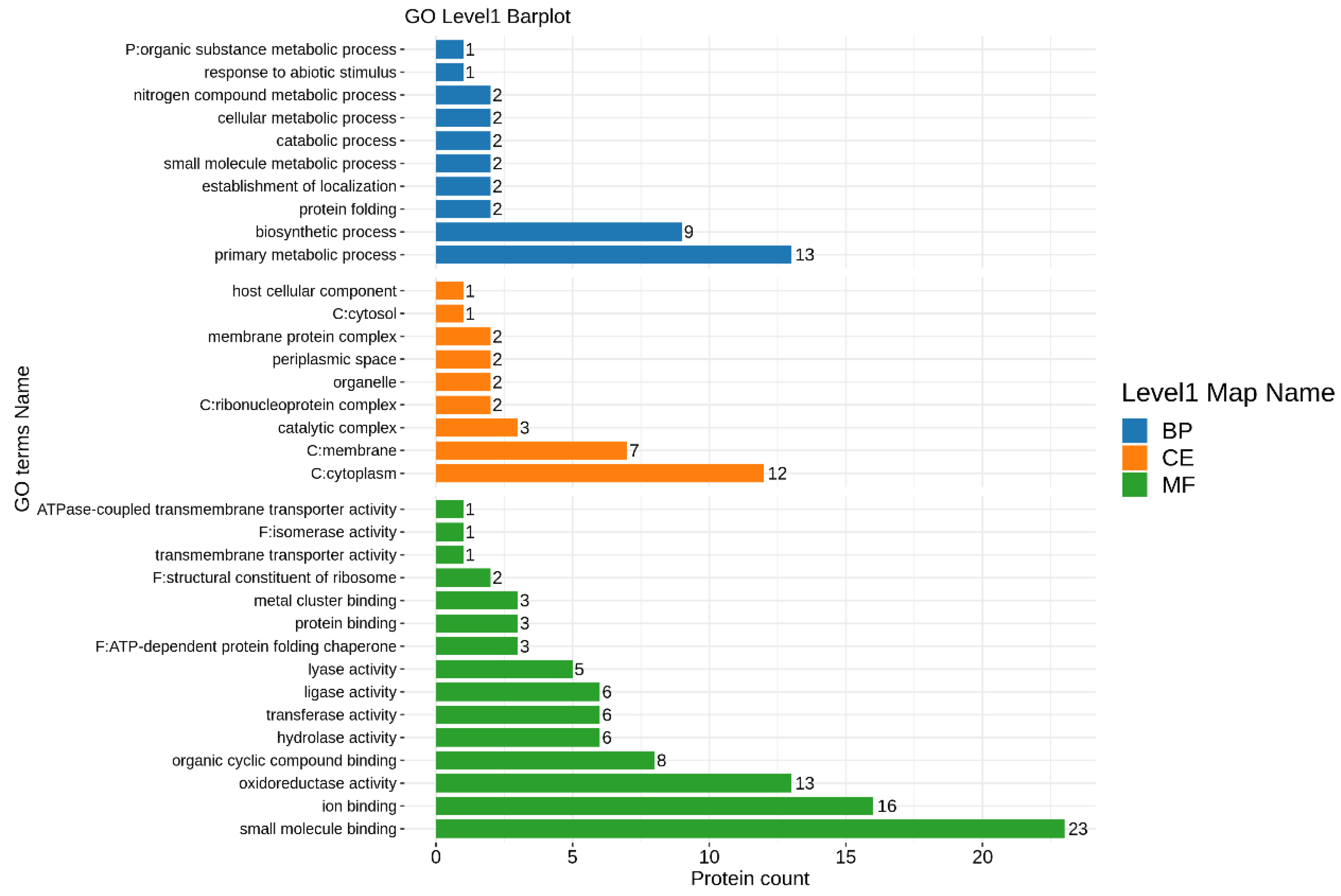
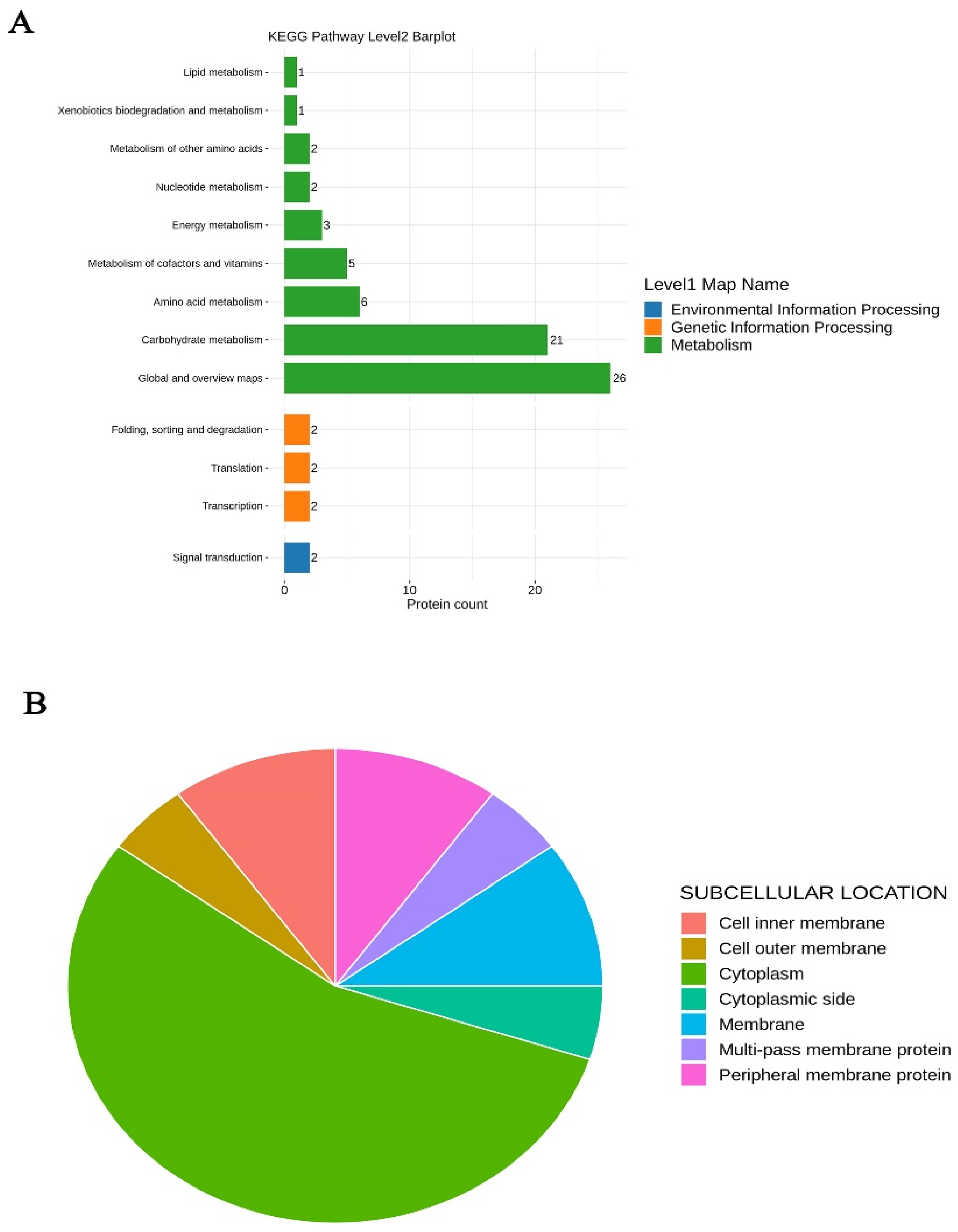
3.7. Proteomic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, J.; Qian, W.; Du, J.; Liu, Y. Effective allocation of resources in water pollution treatment alternatives: A multi-stage gray group decision-making method based on hesitant fuzzy linguistic term sets. Environ. Sci. Pollut. Res. Int. 2020, 27, 3173–3186. [Google Scholar] [CrossRef] [PubMed]
- Torun, M.; Gültekin, O.; Şolpan, D.; Güven, O. Mineralization of paracetamol in aqueous solution with advanced oxidation processes. Environ. Technol. 2014, 36, 970–982. [Google Scholar] [CrossRef]
- Villota, N.; Lomas, J.M.; Camarero, L.M. Study of the paracetamol degradation pathway that generates color and turbidity in oxidized wastewaters by photo-Fenton technology. J. Photochem. Photobiol. A Chem. 2016, 329, 113–119. [Google Scholar] [CrossRef]
- Culebras, M.; Pishnamazi, M.; Walker, G.M.; Collins, M.N. Facile Tailoring of Structures for Controlled Release of Paracetamol from Sustainable Lignin Derived Platforms. Molecules 2021, 26, 1593. [Google Scholar] [CrossRef] [PubMed]
- Gomila, M.; Mulet, M.; García-Valdés, E.; Lalucat, J. Genome-Based Taxonomy of the Genus Stutzerimonas and Proposal of S. frequens sp. nov. and S. degradans sp. nov. and Emended Descriptions of S. perfectomarina and S. chloritidismutans. Microorganisms 2022, 10, 1363. [Google Scholar] [CrossRef]
- Li, B.; Jing, F.; Wu, D.; Xiao, B.; Hu, Z. Simultaneous removal of nitrogen and phosphorus by a novel aerobic denitrifying phosphorus-accumulating bacterium, Pseudomonas stutzeri ADP-19. Bioresour. Technol. 2021, 321, 124445. [Google Scholar] [CrossRef] [PubMed]
- Kusunur, A.B.; Mogilipuri, S.S.; Moturu, D.; Benala, M.; Vaiyapuri, M.; Panda, S.K.; George, J.C.; Badireddy, M.R. Tetracycline resistance potential of heterotrophic bacteria isolated from freshwater fin-fish aquaculture system. J. Appl. Microbiol. 2023, 134, 12. [Google Scholar] [CrossRef]
- Fu, W.-L.; Duan, P.-F.; Wang, Q.; Liao, Y.-X.; Wang, Y.-S.; Xu, M.-J.; Jiang, H.-H.; Zhang, X.; Rao, Z.-M. Transcriptomics reveals the effect of ammonia nitrogen concentration on Pseudomonas stutzeri F2 assimilation and the analysis of amtB function. Synth. Syst. Biotechnol. 2023, 8, 262–272. [Google Scholar] [CrossRef]
- Fu, W.; Wang, Q.; Chen, S.; Wang, Y.; Wang, Y.; Duan, P.; Yi, G.; Liu, C.; Zhang, X.; Rao, Z. Isolation and Identification of an Efficient Aerobic Denitrifying Pseudomonas stutzeri Strain and Characterization of Its Nitrite Degradation. Catalysts 2021, 11, 1214. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Q.; Yan, Y.; Ke, X.; Han, Y.; Wu, S.; Lv, F.; Shao, Y.; Jiang, S.; Lin, M.; et al. Master regulator NtrC controls the utilization of alternative nitrogen sources in Pseudomonas stutzeri A1501. World J. Microbiol. Biotechnol. 2021, 37, 177. [Google Scholar] [CrossRef]
- Zhu, Y.; Xu, Q.; Schwarz, S.; Yu, S.; Xia, L.; Xie, F.; Yang, W.; Zhou, L.; Lin, L.; Zhang, W. Identification of a novel MDR plasmid co-harbouring the carbapenem resistance gene bla(VIM-2) and tigecycline resistance gene cluster tmexCD1-toprJ1 in a Pseudomonas stutzeri isolate. J. Antimicrob. Chemother. 2023, 78, 1309–1311. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.X.; Xu, H.; Guo, X.B.; Li, S.; Wang, Q.; Li, Y.; Liu, R.S.; Gou, J.J. Emergence and Genetic Characterization of Plasmid-Encoded VIM-2-Producing Pseudomonas stutzeri with Novel Integron in1998 Isolated from Cerebrospinal Fluid. Infect. Drug Resist. 2021, 14, 3415–3424. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Ordóñez, A.; Aguilar-Romero, I.; Villaverde, J.; Madrid, F.; Morillo, E. Isolation of Novel Bacterial Strains Pseudomonas extremaustralis CSW01 and Stutzerimonas stutzeri CSW02 from Sewage Sludge for Paracetamol Biodegradation. Microorganisms 2023, 11, 196. [Google Scholar] [CrossRef]
- Qiu, L.; Zhao, D.; Zheng, S.; Gong, A.; Liu, Z.; Su, Y.; Liu, Z. Inhibition Effect of Pseudomonas stutzeri on the Corrosion of X70 Pipeline Steel Caused by Sulfate-Reducing Bacteria. Materials 2023, 16, 2896. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, W.; Tan, Y.; Meng, G.; Liu, H.; Cheng, Y.; Liu, H. Characterizations of the biomineralization film caused by marine Pseudomonas stutzeri and its mechanistic effects on X80 pipeline steel corrosion. J. Mater. Sci. Technol. 2022, 125, 15–28. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel Carbapenem-Hydrolyzing β-Lactamase, KPC-1, from a Carbapenem-Resistant Strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef]
- Yu, Y.; Ji, S.; Chen, Y.; Zhou, W.; Wei, Z.; Li, L.; Ma, Y. Resistance of strains producing extended-spectrum β-lactamases and genotype distribution in China. J. Infect. 2007, 54, 53–57. [Google Scholar] [CrossRef]
- Kim, H.B.; Wang, M.; Park, C.H.; Kim, E.-C.; Jacoby, G.A.; Hooper, D.C. oqxAB Encoding a Multidrug Efflux Pump in Human Clinical Isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 3582–3584. [Google Scholar] [CrossRef]
- Liu, J.-H.; Deng, Y.-T.; Zeng, Z.-L.; Gao, J.-H.; Chen, L.; Arakawa, Y.; Chen, Z.-L. Coprevalence of Plasmid-Mediated Quinolone Resistance Determinants QepA, Qnr, and AAC(6′)-Ib-cr among 16S rRNA Methylase RmtB-Producing Escherichia coli Isolates from Pigs. Antimicrob. Agents Chemother. 2008, 52, 2992–2993. [Google Scholar] [CrossRef]
- Mulet, M.; Gomila, M.; Lalucat, J.; Bosch, R.; Rossello-Mora, R.; García-Valdés, E. Stutzerimonas decontaminans sp. nov. isolated from marine polluted sediments. Syst. Appl. Microbiol. 2023, 46, 126400. [Google Scholar] [CrossRef]
- Fu, W.; Duan, P.; Wang, Q.; Song, J.; Wang, Y.; Zhang, Z.; Wang, P.; Jiang, H.; Zhang, X.; Song, G.; et al. Effect of Pseudomonas stutzeri F2 on rearing water quality and growth, innate immunity, visceral morphology and gut microbiota structure of juvenile spotted seabass (Lateolabrax maculatus). Aquac. Rep. 2023, 30, 12. [Google Scholar] [CrossRef]
- Wei, F.; Xu, R.; Rao, Q.; Zhang, S.; Ma, Z.; Ma, Y. Biodegradation of asphaltenes by an indigenous bioemulsifier-producing Pseudomonas stutzeri YWX-1 from shale oil in the Ordos Basin: Biochemical characterization and complete genome analysis. Ecotoxicol. Environ. Saf. 2023, 251, 114551. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Cheng, D.Y.; Tan, P.; An, Q.; Guo, J.S.J. Characterization of an aerobic denitrifier Pseudomonas stutzeri strain XL-2 to achieve efficient nitrate removal. Bioresour. Technol. 2018, 250, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Yuan, P.; Ding, J.; Wu, H.; Wang, L.; Chen, K.; Jiang, N.; Dai, Y. Novel biodegradation pathway of insecticide flonicamid mediated by an amidase and its unusual substrate spectrum. J. Hazard. Mater. 2023, 441, 129952. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Wu, Y.; He, P.; Hu, S.; Li, Q.; Chen, S. Assembled denitrifying consortia for efficient nitrate removal under low-COD/N conditions. Chem. Eng. J. 2023, 460, 10. [Google Scholar] [CrossRef]
- Guo, J.; Xu, S.; Liu, Y.; Zhang, C.; Hou, S. Complete Genome Sequence of Stutzerimonas stutzeri Strain SOCE 002, a Marine Bacterium Isolated from the Surface Seawater of Dapeng Bay. Microbiol. Resour. Ann. 2023, 12, e0015023. [Google Scholar] [CrossRef]
- Wood, H.N.; Venken, T.; Willems, H.; Jacobs, A.; Reis, A.J.; da Silva, P.E.A.; Homolka, S.; Niemann, S.; Rohde, K.H.; Hooyberghs, J. Molecular drug susceptibility testing and strain typing of tuberculosis by DNA hybridization. PLoS ONE 2019, 14, e0212064. [Google Scholar] [CrossRef]
- Lalucat, J.; Bennasar, A.; Bosch, R.; García-Valdés, E.; Palleroni, N.J. Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 2006, 70, 510–547. [Google Scholar] [CrossRef]
- Kuroda, M.; Sei, K.; Yamashita, M.; Ike, M. Draft Genome Sequence of Stutzerimonas stutzeri NT-I, Which Reduces Selenium Oxyanions into Elemental Selenium and Volatile Selenium Species. Genome Announc. 2022, 11, e0101622. [Google Scholar] [CrossRef]
- Ma, N.; Cai, R.; Sun, C. Threonine dehydratase enhances bacterial cadmium resistance via driving cysteine desulfuration and biomineralization of cadmium sulfide nanocrystals. J. Hazard. Mater. 2021, 417, 126102. [Google Scholar] [CrossRef]
- Lee, H.; Rhee, S. Structural and mutational analyses of the bifunctional arginine dihydrolase and ornithine cyclodeaminase AgrE from the cyanobacterium Anabaena. J. Biol. Chem. 2020, 295, 5751–5760. [Google Scholar] [CrossRef] [PubMed]
- Damin, B.I.S.; Kovalski, F.C.; Fischer, J.; Piccin, J.S.; Dettmer, A. Challenges and perspectives of the β-galactosidase enzyme. Appl. Microbiol. Biotechnol. 2021, 105, 5281–5298. [Google Scholar] [CrossRef] [PubMed]
- Martinho, N.; Pires, R.F.; Zloh, M.; Bonifácio, V.D.B. Intrinsic acetamide brush-off by polyurea biodendrimers. J. Mater. Chem. B 2021, 9, 3371–3376. [Google Scholar] [CrossRef]
- Padhi, L.; Panda, S.K.; Mohapatra, P.P.; Sahoo, G. Antibiotic susceptibility of cultivable aerobic microbiota from the oral cavity of Echis carinatus from Odisha (India). Microb. Pathog. 2020, 143, 104121. [Google Scholar] [CrossRef]
- Collin, F.; Karkare, S.; Maxwell, A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl. Microbiol. Biotechnol. 2011, 92, 479–497. [Google Scholar] [CrossRef]
- Zidar, N.; Tomašič, T.; Macut, H.; Sirc, A.; Brvar, M.; Montalvão, S.; Tammela, P.; Ilaš, J.; Kikelj, D. New N-phenyl-4,5-dibromopyrrolamides and N-Phenylindolamides as ATPase inhibitors of DNA gyrase. Eur. J. Med. Chem. 2016, 117, 197–211. [Google Scholar] [CrossRef]
- Kaul, M.; Mark, L.; Zhang, Y.; Parhi, A.K.; Lyu, Y.L.; Pawlak, J.; Saravolatz, S.; Saravolatz, L.D.; Weinstein, M.P.; LaVoie, E.J.; et al. TXA709, an FtsZ-Targeting Benzamide Prodrug with Improved Pharmacokinetics and Enhanced In Vivo Efficacy against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2015, 59, 4845–4855. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhang, Z.; Li, X.; Song, Y.; Kang, J.; Yin, D.; Gao, Y.; Shi, N.; Duan, J. Mutations in gyrB play an important role in ciprofloxacin-resistant Pseudomonas aeruginosa. Infect. Drug Resist. 2019, 12, 261–272. [Google Scholar] [CrossRef]
- Cacciotto, P.; Basciu, A.; Oliva, F.; Malloci, G.; Zacharias, M.; Ruggerone, P.; Vargiu, A.V. Molecular rationale for the impairment of the MexAB-OprM efflux pump by a single mutation in MexA. Comput. Struct. Biotechnol. J. 2022, 20, 252–260. [Google Scholar] [CrossRef]
- Barlow, V.L.; Lai, S.-J.; Chen, C.-Y.; Tsai, C.-H.; Wu, S.-H.; Tsai, Y.-H. Effect of membrane fusion protein AdeT1 on the antimicrobial resistance of Escherichia coli. Sci. Rep. 2020, 10, 20464. [Google Scholar] [CrossRef]
- Fabre, L.; Ntreh, A.T.; Yazidi, A.; Leus, I.V.; Weeks, J.W.; Bhattacharyya, S.; Ruickoldt, J.; Rouiller, I.; Zgurskaya, H.I.; Sygusch, J. A “Drug Sweeping” State of the TriABC Triclosan Efflux Pump from Pseudomonas aeruginosa. Structure 2021, 29, 261–274.e6. [Google Scholar] [CrossRef] [PubMed]
- Bialvaei, A.Z.; Rahbar, M.; Hamidi-Farahani, R.; Asgari, A.; Esmailkhani, A.; Dashti, Y.M.; Soleiman-Meigooni, S. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb. Pathog. 2021, 153, 104789. [Google Scholar] [CrossRef] [PubMed]

| Name | Abbreviation | Results |
|---|---|---|
| Arginine dihydrolase | ADH | P |
| Anaerobic glucose fermentation | GLUf | P |
| Hydrogen sulfide production | H2S | N |
| Nitrate reduction | NIT | P |
| Ornithine decarboxylase | ODC | P |
| Amino acid control | C | N |
| Acid production of aerobic glucose | GLU | P |
| Aescin hydrolysis | ESC | N |
| Urease | URE | N |
| Lysine decarboxylase | LDC | P |
| Production of indole | IND | N |
| Acid production of mannose | MNE | P |
| Malic acid utilization | MTE | P |
| Acid production of maltose | MAL | P |
| Gelatin hydrolysis | GEL | N |
| Acid production of sucrose | SAC | P |
| Acid production of lactose | LAC | N |
| Citrate utilization | CIT | P |
| Acid production of xylose | XYL | P |
| Galactosidase | ONPG | P |
| Acid production of fructose | FRU | N |
| Malonic acid salt utilization | MLT | P |
| Acetamide | ACE | P |
| Acid production of mannitol | MAN | N |
| Drug Name | Abbreviation | MIC Value | Results |
|---|---|---|---|
| Gentamicin | GEN | ≤2 | S |
| Tobramycin | TOB | ≤1 | S |
| Ceftazidime | CAZ | ≤1 | S |
| Ciprofloxacin | CIP | ≤1 | S |
| Imipenem | IPM | ≤1 | S |
| MeropeneM | MRP | ≤1 | S |
| Cefepime | FEP | ≤2 | S |
| Levofloxacin | LEV | ≤2 | S |
| Piperacillin/Tazobactam | P/T | ≤4/4 | S |
| Aztreonam | ATM | ≤4 | S |
| Amikacin | AMK | ≤4 | S |
| Compound xinnuomin | SXT | ≤2/38 | S |
| Ceftriaxone | CRO | =2 | S |
| Cefotaxime | CTX | =4 | S |
| Chloramphenicol | CHL | =32 | R |
| Polymyxin B | PB | ≤2 | / |
| Polymyxin E | CT | ≤2 | / |
| Cefoperazone/Sulbactam | CPS | ≤4/2 | S |
| Ampicillin/Sulbactam | AMS | =16/8 | / |
| Doxycycline | DOX | ≤4 | S |
| Piperacillin | PIP | ≤8 | S |
| Minocycline | MIN | =8 | I |
| Ticarcillin/Clavulanic acid | TIM | ≤8/2 | S |
| Tetracycline | TET | ≤4 | S |
| Test Group | Time (h) | Peak Point (OD) |
|---|---|---|
| Stutzerimonas stutzeri-1 μL | 14 | 2.66 |
| Stutzerimonas stutzeri-10 μL | 13 | 2.72 |
| Stutzerimonas stutzeri-20 μL | 12 | 2.76 |
| Stutzerimonas stutzeri-40 μL | 12 | 2.67 |
| Stutzerimonas stutzeri-60 μL | 11 | 2.67 |
| Stutzerimonas stutzeri-80 μL | 11 | 2.61 |
| Stutzerimonas stutzeri-100 μL | 11 | 2.53 |
| Item | Proteomic Analysis |
|---|---|
| 1 | Multidrug/solvent RND membrane fusion protein |
| 2 | RND multidrug efflux membrane fusion protein, MexE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Sun, X.; Chen, X.; Wang, H.; He, H. The Mass Spectrometry Identification, Antimicrobial Genes Detection, and Proteomics Analysis of Stutzerimonas stutzeri Strain Was Isolated from Industrial Wastewater. Separations 2023, 10, 461. https://doi.org/10.3390/separations10090461
Wang Z, Sun X, Chen X, Wang H, He H. The Mass Spectrometry Identification, Antimicrobial Genes Detection, and Proteomics Analysis of Stutzerimonas stutzeri Strain Was Isolated from Industrial Wastewater. Separations. 2023; 10(9):461. https://doi.org/10.3390/separations10090461
Chicago/Turabian StyleWang, Zongwu, Xiaoyan Sun, Xing Chen, Haifeng Wang, and Hongxuan He. 2023. "The Mass Spectrometry Identification, Antimicrobial Genes Detection, and Proteomics Analysis of Stutzerimonas stutzeri Strain Was Isolated from Industrial Wastewater" Separations 10, no. 9: 461. https://doi.org/10.3390/separations10090461
APA StyleWang, Z., Sun, X., Chen, X., Wang, H., & He, H. (2023). The Mass Spectrometry Identification, Antimicrobial Genes Detection, and Proteomics Analysis of Stutzerimonas stutzeri Strain Was Isolated from Industrial Wastewater. Separations, 10(9), 461. https://doi.org/10.3390/separations10090461







