Identification and Functional Characterization of the RcFAH12 Promoter from Castor Bean in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
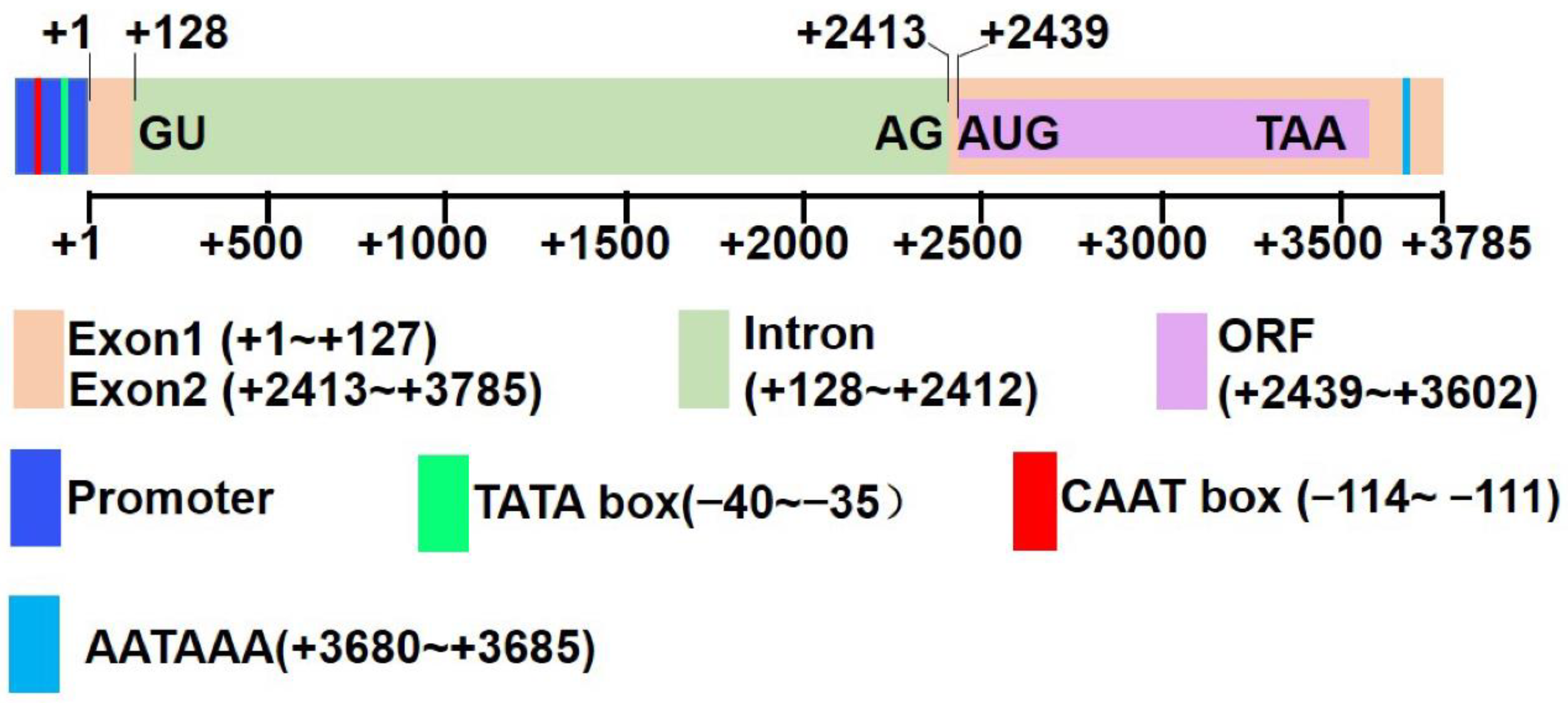
2.1. Sequence Analysis of RcFAH12
2.2. Sequence Analysis of the RcFAH12 Promoter
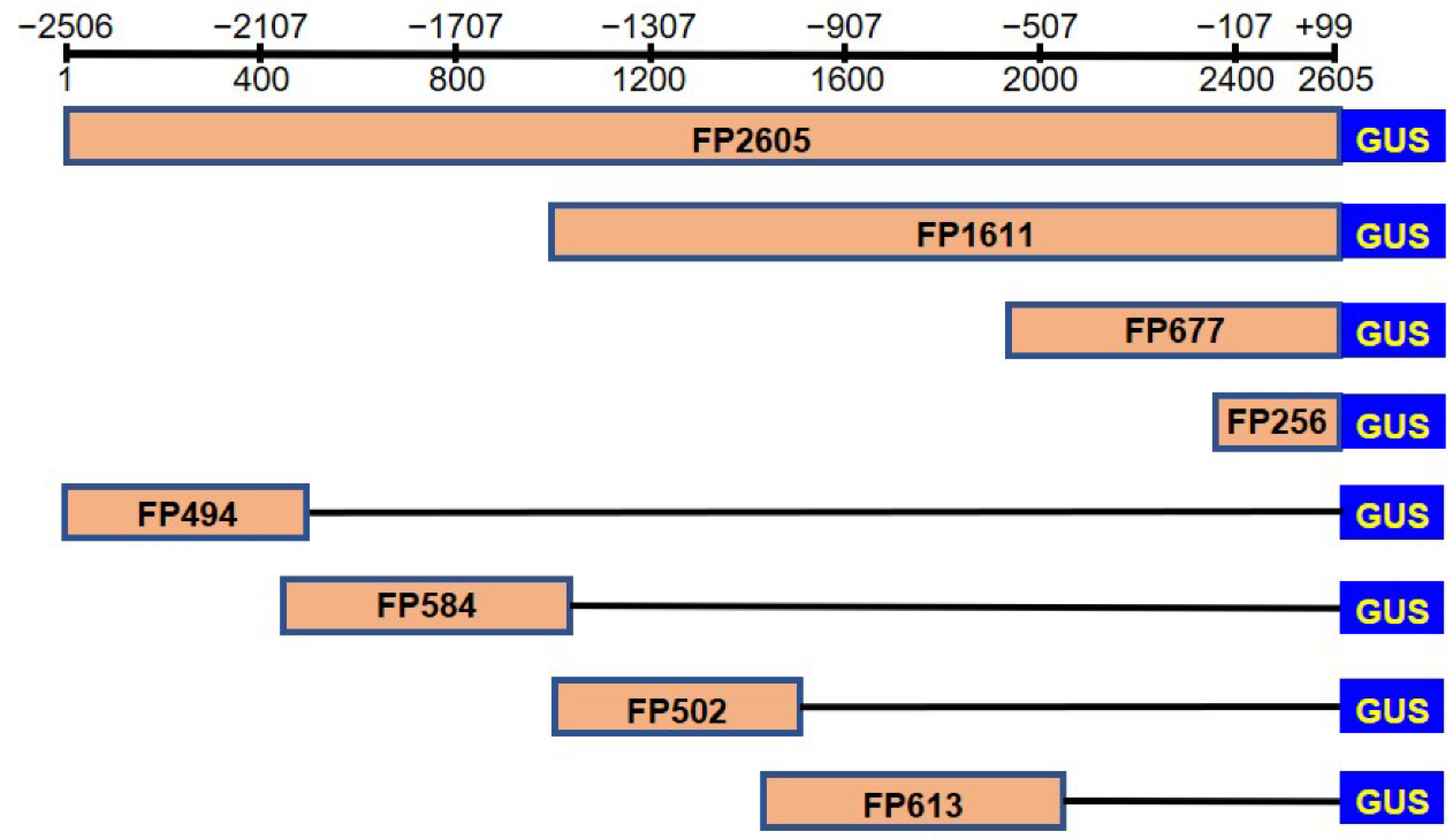
2.3. Deletion Analysis of the Promoter
3. Discussion
3.1. RcFAH12 Has Multiple TSSs
3.2. RcFAH12 Expression Is Subject to Complex Transcriptional Regulation
3.3. RcFAH12 Promoter Can Be Used for Genetic Engineering
4. Materials and Methods
4.1. Plant Material
4.2. Rapid Amplification of cDNA Ends (RACE)
4.2.1. Intermediate Fragment Cloning of RcFAH12 cDNA
4.2.2. Cloning of the 5′ and 3′ Terminal Sequences
4.3. Cloning of the RcFAH12 Promoter and Sequence Analyses
4.4. Deletion Analysis of Cloned RcFAH12 Promoter Fragment
4.5. Construction of a Series of Promoters::GUS Vectors and Transformation of Arabidopsis
4.6. GUS Histochemical Staining
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Da Silva Ramos, L.C.; Tango, J.S.; Savi, A.; Leal, N.R. Variability for oil and fatty acid composition in castorbean varieties. J. Am. Oil Chem. Soc. 1984, 61, 1841–1843. [Google Scholar] [CrossRef]
- Mensah, M.B.; Awudza, J.A.M.; O’Brien, P. Castor oil: A suitable green source of capping agent for nanoparticle syntheses and facile surface functionalization. R. Soc. Open Sci. 2018, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Viana, A.D.; Nobrega, E.T.D.; Moraes, E.P.; Neto, A.O.W.; Menezes, F.G.; Gasparotto, L.H.S. Castor oil derivatives in the environmentally friendly one-pot synthesis of silver nanoparticles: Application in cysteine sensing. Mater. Res. Bull. 2020, 124, 7. [Google Scholar] [CrossRef]
- Muntz, A.; Sandford, E.; Claassen, M.; Curd, L.; Jackson, A.K.; Watters, G.; Wang, M.T.M.; Craig, J.P. Randomized trial of topical periocular castor oil treatment for blepharitis. Ocul. Surface 2021, 19, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Bayoumi, Y.; Alalfy, M.; Sharkawy, M.; Ali, A.; Gouda, H.; Hatem, D.L. Castor oil for labor initiation in women with a previous cesarean section: A double-blind randomized study. J. Matern. Fetal Neonatal Med. 2021, 35, 8945–8951. [Google Scholar] [CrossRef] [PubMed]
- James, A.T.; Hadaway, H.C.; Webb, J.P. The Biosynthesis of Ricinoleic Acid. Biochem. J. 1965, 95, 448–452. [Google Scholar] [CrossRef]
- Chen, G.Q.; Turner, C.; He, X.; Nguyen, T.; McKeon, T.A.; Laudencia-Chingcuanco, D. Expression profiles of genes involved in fatty acid and triacylglycerol synthesis in castor bean (Ricinus communis L.). Lipids 2007, 42, 263–274. [Google Scholar] [CrossRef]
- Brown, A.P.; Kroon, J.T.M.; Swarbreck, D.; Febrer, M.; Larson, T.R.; Graham, I.A.; Caccamo, M.; Slabas, A.R. Tissue-Specific Whole Transcriptome Sequencing in Castor, Directed at Understanding Triacylglycerol Lipid Biosynthetic Pathways. PLoS ONE 2012, 7, e30100. [Google Scholar] [CrossRef]
- Lin, J.T.; Woodruff, C.L.; Lagouche, O.J.; McKeon, T.A.; Stafford, A.E.; Goodrich-Tanrikulu, M.; Singleton, J.A.; Haney, C.A. Biosynthesis of triacylglycerols containing ricinoleate in castor microsomes using 1-acyl-2-oleoyl-sn-glycero-3-phosphocholine as the substrate of oleoyl-12-hydroxylase. Lipids 1998, 33, 59–69. [Google Scholar] [CrossRef]
- Brown, A.P.; Kroon, J.T.; Topping, J.F.; Robson, J.L.; Simon, W.J.; Slabas, A.R. Components of complex lipid biosynthetic pathways in developing castor (Ricinus communis) seeds identified by MudPIT analysis of enriched endoplasmic reticulum. J. Proteome Res. 2011, 10, 3565–3577. [Google Scholar] [CrossRef]
- Han, B.; Wu, D.; Zhang, Y.; Li, D.-Z.; Xu, W.; Liu, A. Epigenetic regulation of seed-specific gene expression by DNA methylation valleys in castor bean. BMC Biol. 2022, 20, 57. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Li, C.; Yan, C.; Zhao, X.; Wang, J.; Sun, Q.; Shan, S. Isolation and characterization of a novel seed-specific promoter from peanut (Arachis hypogaea L.). Mol. Biol. Rep. 2019, 46, 3183–3191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Sun, R.; Zheng, Y.; Yuan, Y.; Li, D. Isolation and characterization of the EgWRI1 promoter from oil palm (Elaeis guineensis Jacq.) and its response to environmental stress and ethylene. PLoS ONE 2019, 14, e0225115. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhu, H.; Pujiana, D.; Zheng, L.; Chen, L.; Ma, A. Cloning and functional characterization of gpd and alpha-tubulin promoters from Annulohypoxylon stygium, a companion fungus of Tremella fuciformis. Mycoscience 2020, 61, 1–8. [Google Scholar] [CrossRef]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Kim, W.C. A Fruitful Decade Using Synthetic Promoters in the Improvement of Transgenic Plants. Front. Plant Sci. 2019, 10, 1433. [Google Scholar] [CrossRef] [PubMed]
- Shahmuradov, I.A.; Umarov, R.K.; Solovyev, V.V. TSSPlant: A new tool for prediction of plant Pol II promoters. Nucleic Acids Res. 2017, 45, e65. [Google Scholar] [CrossRef]
- Sanchez-Alvarez, A.; Ruiz-Lopez, N.; Moreno-Perez, A.J.; Martinez-Force, E.; Garces, R.; Salas, J.J. Agrobacterium-Mediated Transient Gene Expression in Developing Ricinus communis Seeds: A First Step in Making the Castor Oil Plant a Chemical Biofactory. Front. Plant Sci. 2019, 10, 11. [Google Scholar] [CrossRef]
- Sharif, Y.; Chen, H.; Deng, Y.; Ali, N.; Khan, S.; Zhang, C.; Xie, W.; Chen, K.; Cai, T.; Yang, Q.; et al. Cloning and Functional Characterization of a Pericarp Abundant Expression Promoter From Peanut (Arachis hypogaea L.). Front. Genet. 2021, 12, 821281. [Google Scholar] [CrossRef]
- Danino, Y.M.; Even, D.; Ideses, D.; Juven-Gershon, T. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta 2015, 1849, 1116–1131. [Google Scholar] [CrossRef]
- Kumar, A.; Chan, J.; Taguchi, M.; Kono, H. Interplay among transacting factors around promoter in the initial phases of transcription. Curr. Opin. Struct. Biol. 2021, 71, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Wang, G.; Abou-Elwafa, S.F.; Fu, J.; Liu, Z.; Zhang, P.; Xie, X.; Ku, L.; Ma, Y.; Guan, X.; et al. Genome-wide identification of HD-ZIP transcription factors in maize and their regulatory roles in promoting drought tolerance. Physiol. Mol. Biol. Plants 2022, 28, 425–437. [Google Scholar] [CrossRef]
- Thomas, M.; Chiang, C. The general transcription machinery and general cofactors. Crit. Rev. Biochem. 2006, 41, 105–178. [Google Scholar] [CrossRef]
- Mejia-Guerra, M.K.; Li, W.; Doseff, A.I.; Grotewold, E. Genome-wide TSS identification in maize. In Plant Transcription Factors: Methods and Protocols; Yamaguchi, N., Ed.; Springer: New York, NY, USA, 2018; pp. 239–256. [Google Scholar] [CrossRef]
- Drechsel, G.; Kahles, A.; Kesarwani, A.; Stauffer, E.; Behr, J.; Drewe, P.; Rätsch, G.; Wachter, A. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 2013, 25, 3726–3742. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, L.; Drewe-Boß, P.; Wießner, T.; Wagner, G.; Geue, S.; Lee, H.; Obermüller, D.; Kahles, A.; Behr, J.; Sinz, F.; et al. Alternative Splicing Substantially Diversifies the Transcriptome during Early Photomorphogenesis and Correlates with the Energy Availability in Arabidopsis. Plant Cell Online 2016, 28, 2715–2734. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Huang, S.; Wise, A.; Castanon, R.; Nery, J.; Chen, H.; Watanabe, M.; Thomas, J.; Bar-Joseph, Z.; Ecker, J. A transcription factor hierarchy defines an environmental stress response network. Science 2016, 354, aag1550. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, Y.; Makita, Y.; Kawashima, M.; Fujita, T.; Iwasaki, S.; Matsui, M. Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 7831–7836. [Google Scholar] [CrossRef]
- Policastro, R.A.; Zentner, G.E. Global approaches for profiling transcription initiation. Cell Rep. Methods 2021, 1, 100081. [Google Scholar] [CrossRef]
- Borlin, C.S.; Cvetesic, N.; Holland, P.; Bergenholm, D.; Siewers, V.; Lenhard, B.; Nielsen, J. Saccharomyces cerevisiae displays a stable transcription start site landscape in multiple conditions. FEMS Yeast Res. 2019, 19, foy128. [Google Scholar] [CrossRef]
- Van de Loo, F.J.; Broun, P.; Turner, S.; Somerville, C. An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc. Natl. Acad. Sci. USA 1995, 92, 6743–6747. [Google Scholar] [CrossRef]
- Chan, C.-B.; Tang, W.-K.; Cheng, C.H.K.; Fong, W.-P. Cloning of the black seabream (Acanthopagrus schlegeli) antiquitin gene and functional characterization of its promoter region. Mol. Cell. Biochem. 2007, 297, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Xiong, J.; Li, X.; Qin, X. A 43-bp A/T-rich element upstream of the kinesin gene AtKP1 promoter functions as a silencer in Arabidopsis. Plant Cell Rep. 2009, 28, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Foster, R.; Nakajima, M.; Chua, S.N.H. The rice bZIP transcriptional activator RITA-1 is highly expressed during seed development. Plant Cell 1994, 6, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Ellerström, M.; Stålberg, K.; Ezcurra, I.; Rask, L. Functional dissection of a napin gene promoter: Identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol. Biol. 1996, 32, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, L.F.; Carrie, C.; Andersson, C.R.; Sivasithamparam, K.; Whelan, J.; Singh, K.B. Differential Gene Expression and Subcellular Targeting of Arabidopsis Glutathione S-Transferase F8 Is Achieved through Alternative Transcription Start Sites. J. Biol. Chem. 2007, 282, 28915–28928. [Google Scholar] [CrossRef]
- Tatip, S.; Taggart, J.; Wang, Y.; MacDiarmid, C.; Eide, D.J. Changes in transcription start sites of Zap1-regulated genes during zinc deficiency: Implications for HNT1 gene regulation. Mol. Microbiol. 2020, 113, 285–296. [Google Scholar] [CrossRef]
- Xiao, G.; Zhang, Z.Q.; Yin, C.F.; Liu, R.Y.; Wu, X.M.; Tan, T.L.; Chen, S.Y.; Lu, C.M.; Guan, C.Y. Characterization of the promoter and 5 ’-UTR intron of oleic acid desaturase (FAD2) gene in Brassica.napus. Gene. 2014, 545, 45–55. [Google Scholar] [CrossRef]
- Salimonti, A.; Carbone, F.; Romano, E.; Pellegrino, M.; Benincasa, C.; Micali, S.; Tondelli, A.; Conforti, F.; Perri, E.; Ienco, A.; et al. FAD2-2Association Study of the 5’UTR Intron of the Gene with Oleic and Linoleic Acid Content in Olea europaea L. Front. Plant Sci. 2020, 11, 66. [Google Scholar] [CrossRef]
- Hong, J.K.; Suh, E.J.; Kwon, S.-J.; Lee, S.B.; Kim, J.A.; Lee, S.I.; Lee, Y.-H. Promoter of chrysanthemum actin confers high-level constitutive gene expression in Arabidopsis and chrysanthemum. Sci. Hortic. 2016, 211, 8–18. [Google Scholar] [CrossRef]
- Timerbaev, V.; Dolgov, S. Functional characterization of a strong promoter of the early light-inducible protein gene from tomato. Planta 2019, 250, 1307–1323. [Google Scholar] [CrossRef]
- Sun, M.-L.; Shi, T.-Q.; Lin, L.; Ledesma-Amaro, R.; Ji, X.-J. Advancing Yarrowia lipolytica as a superior biomanufacturing platform by tuning gene expression using promoter engineering. Bioresour. Technol. 2022, 347, 126717. [Google Scholar] [CrossRef] [PubMed]
- Zavallo, D.; Lopez Bilbao, M.; Esteban Hopp, H.; Heinz, R. Isolation and functional characterization of two novel seed-specific promoters from sunflower (Helianthus annuus L.). Plant Cell Rep. 2010, 29, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Sunkara, S.; Bhatnagar-Mathur, P.; Sharma, K.K. Isolation and Functional Characterization of a Novel Seed-Specific Promoter Region from Peanut. Appl. Biochem. Biotechnol. 2014, 172, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhu, Z.; Wang, H.; Wang, L.; Cheng, L.; Yuan, Y.; Zheng, Y.; Li, D. Characterization and functional analysis of a plastidial FAD6 gene and its promoter in the mesocarp of oil palm (Elaeis guineensis). Sci. Hortic. 2018, 239, 163–170. [Google Scholar] [CrossRef]
- Yuan, M.; Zhu, J.; Gong, L.; He, L.; Lee, C.; Han, S.; Chen, C.; He, G. Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnol. 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, Y.; Sun, M.; Wei, X.; Huo, H.; Yu, L.; Zhang, J. Dynamic transcriptome profiling revealed key genes and pathways associated with cold stress in castor (Ricinus communis L.). Ind Crops Prod. 2022, 178, 114610. [Google Scholar] [CrossRef]
- Jianjun, D.; Qingbo, Z.; Yingfei, S.; Na, Y.; Qinglong, Y.; Yongchun, W.; Jixing, Z. Comparison of Extraction Methods for Total RNA from Castor Seeds. Jiangsu Agric. Sci. 2013, 41, 33–34. [Google Scholar] [CrossRef]
- Lozinsky, S.; Yang, H.; Forseille, L.; Cook, G.R.; Ramirez-Erosa, I.; Smith, M.A. Characterization of an oleate 12-desaturase from Physaria fendleri and identification of 5 ‘ UTR introns in divergent FAD2 family genes. Plant Physiol. Biochem. 2014, 75, 114–122. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di, J.; Li, G.; Wang, X.; Huang, F.; Chen, Y.; Wang, Y.; Sun, J.; Zhang, C.; Zhang, Q.; Wang, G.; et al. Identification and Functional Characterization of the RcFAH12 Promoter from Castor Bean in Arabidopsis thaliana. Separations 2023, 10, 2. https://doi.org/10.3390/separations10010002
Di J, Li G, Wang X, Huang F, Chen Y, Wang Y, Sun J, Zhang C, Zhang Q, Wang G, et al. Identification and Functional Characterization of the RcFAH12 Promoter from Castor Bean in Arabidopsis thaliana. Separations. 2023; 10(1):2. https://doi.org/10.3390/separations10010002
Chicago/Turabian StyleDi, Jianjun, Guorui Li, Xiaoyu Wang, Fenglan Huang, Yongsheng Chen, Yue Wang, Jiaxin Sun, Chunlin Zhang, Qingbo Zhang, Gang Wang, and et al. 2023. "Identification and Functional Characterization of the RcFAH12 Promoter from Castor Bean in Arabidopsis thaliana" Separations 10, no. 1: 2. https://doi.org/10.3390/separations10010002
APA StyleDi, J., Li, G., Wang, X., Huang, F., Chen, Y., Wang, Y., Sun, J., Zhang, C., Zhang, Q., Wang, G., & Zhang, L. (2023). Identification and Functional Characterization of the RcFAH12 Promoter from Castor Bean in Arabidopsis thaliana. Separations, 10(1), 2. https://doi.org/10.3390/separations10010002






