Methods
Study design and population
This cross-sectional study included 302 infants ≤ 1 year of age in the Pediatrics Department, The Jordan University Hospital (JUH) in Amman, Jordan, over a 10-month period from May 2016 through February 2017. A total of 302 fresh fecal samples from soiled diapers were collected randomly using cotton swabs from infants hospitalized in the neonatal intensive care unit (NICU) or examined in the outpatient department (OPD). All fecal specimens were sent within 2-3 h to research microbiology labs. For each sample, the following data were recorded: date of collection, patient number, hospital ward, mode of delivery, infant’s name, sex, age, birth weight, type of feeding, presence of diarrhea, and the recent use of one or more antibiotics. Permission for the study was obtained from the Ethical Review Board (ERB) at the JUH, no. 118/May 2016. Verbal consent was obtained from all mothers of infants after explaining the aim of the study.
Culture and identification
Fresh fecal samples were treated by absolute ethanol (
v/
v) for 1 hour before inoculation onto Perfringens Tryptose-Sulphite-Cycloserine (TSC) agar plates with egg-yolk (EY) (Oxoid, Altrincham, England). Samples were carefully mixed, and 0.1 mL inoculum was directly inoculated on EY-TSC agar plates and incubated in anaerobic jar for 48 hours at 37°C by using Anaerobe bag (AnaeroGen AN25; Oxoid). All suspected colonies resembling
C. perfringens which appeared black and surrounded by egg yolk precipitate were examined by the catalase-negative reaction. Five to ten colonies suspected to be
C. perfringens were picked from each plate and streaked on blood agar plates (Oxoid) supplemented with human blood (5%), and incubated for 48 h at 37°C under anaerobic conditions. All isolates that formed white colonies surrounded by hemolysis and displayed Gram-positive rod shape with subterminal spores were identified primarily as
C. perfringens isolates [
9]
. C. perfringens control strain (CMUL Clo. Per. SL IX-004) was obtained from Prof. Monzer Hamze, Laboratoire de Microbiologie Santé et Environnement (LMSE), Faculty of Public Health, Lebanese University, Tripoli, Lebanon. The control strain contains the toxins alpha, beta and epsilon and was included for quality control in biochemical identification of
C. perfringens isolates, antibiotic susceptibility and as a positive control in genetic identification of
C. perfringens toxins. All 82
C. perfringens isolates were inoculated in 1.8 mL of brain-heart infusion broth (Oxoid) with 20% glycerol and were stored at -70°C for further characterization of the toxin genes.
Antimicrobial susceptibility testing
All
C. perfringens isolates were tested using E-test. The minimum inhibitory concentration (MIC) breakpoints were used to indicate the resistance to each tested drug according to the recommendation of the Clinical and Laboratory Standards Institute (CLSI) [
10].
DNA and plasmid extraction
All C. perfringens isolates stored in cryogenic tubes at -70°C were thawed at room temperature and cultured onto TSC agar. After incubation at 37°C for 48 h, a few colonies (6-7 colonies) were picked from the agar and inoculated into 5 mL Mueller Hinton broth and incubated at 37°C overnight. According to the manufacturer’s protocol, the bacterial DNA was isolated using Wizard Genomic DNA purification kit (Promega, Madison, WI, USA); bacterial plasmid was extracted using EZ-10 Spin Column Plasmid DNA Minipreps Kit (Bio Basic, Markham, ON, Canada).
16S rDNA confirmation for C. perfringens identification
PCR confirmation for
C. perfringens isolates was performed by amplification of the 16S rRNA gene using
C. perfringens specific primers (
Table 1). The PCR assay was performed at the final volume of 20 µL using the following mixture: 4 µL of KAPPA Fast ready mix (5X) master mix (Solis BioDyne, Tartu, Estonia), one pair of primers were used to detect 16S rDNA. The primer concentration used was 10 µL of each primer, 12 µL of nuclease free water, and a volume of 2 µL of the extracted DNA. A PCR mix with DNA from the
C. perfringens control strain was used as positive control, whereas a PCR mix containing nuclease free water was used as negative control. All samples were amplified in a PCR thermal cycler (Bioer XP Cycler, Hangzhou, China), with cycling condition of the initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 53°C for 1 min then extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products, controls and 100 bp ladder marker (Biolabs, Ipswich, MA, USA) were detected in 2.0% agarose gel (Promega) stained with red safe (CONDA Pronadisa, Madrid, Spain).
16S rDNA confirmation for C. perfringens identification
PCR confirmation for
C. perfringens isolates was performed by amplification of the 16S rRNA gene using
C. perfringens specific primers (
Table 1). The PCR assay was performed at the final volume of 20 µL using the following mixture: 4 µL of KAPPA Fast ready mix (5X) master mix (Solis BioDyne, Tartu, Estonia), one pair of primers were used to detect 16S rDNA. The primer concentration used was 10 µL of each primer, 12 µL of nuclease free water, and a volume of 2 µL of the extracted DNA. A PCR mix with DNA from the
C. perfringens control strain was used as positive control, whereas a PCR mix containing nuclease free water was used as negative control. All samples were amplified in a PCR thermal cycler (Bioer XP Cycler, Hangzhou, China), with cycling condition of the initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 53°C for 1 min then extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products, controls and 100 bp ladder marker (Biolabs, Ipswich, MA, USA) were detected in 2.0% agarose gel (Promega) stained with red safe (CONDA Pronadisa, Madrid, Spain).
Statistical analysis
Data generated from the study were tabulated with Microsoft Excel and uploaded to Statistical Package for Social Sciences (SPSS, version 20, IBM Corp, Armonk, NY, USA).
Frequency and percentage were calculated for the categorical data, and Pearson’s Chi-squared test or Fisher’s exact test were applied to determine potential factors associated with C. perfingens and to determine whether there are any statistical differences between groups. The level of significance was set at a p<0.05 to test the hypothesis of no association. Fisher’s exact test replaced the Chi-squared test when the minimum expected count was less than five.
Discussion
Detection of
C. perfringens and its various toxins became straightforward after introducing the PCR typing method, which was first developed by Yoo et al [
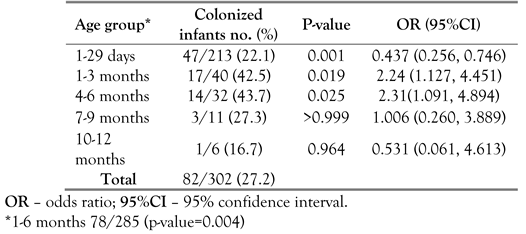
12]. This study shows that intestinal colonization with
Clostridium perfringens was detected in 27.2% of feces from infants aged ≤ 1 year at JUH. Infants aged ≤ 6 months had a higher (p<0.004) colonization rate than older infants. A similar study from Japan found
C. perfringens in the feces of 18.2% healthy infants approximately 30 days old [
13]. A three-year follow up study of 89 healthy infants in Japan showed that the incidence of early life intestinal colonization with α-toxigenic and enterotoxigenic
C. perfringens was found only in 3% of infants at one week of age, and was later increased to 33-39% after 1-3 months [
14]. This increased incidence probably occurred because the infant’s gut is first colonized primarily by facultative anaerobes that create a reduced environment and contribute to the establishment of strict anaerobic bacteria in the intestine, including
Clostridium species [
14].
The present study shows that the
C. perfringens colonization rate was lower but not significantly in breast-fed compared to formula-fed infants (23.7% versus 28.8%). We found also that infants born by caesarean delivery were significantly (p=0.001) more frequently colonized by toxigenic
C. perfringens than those born by vaginal delivery (
Table 2). These results are in agreement with a recent Japanese study, which has also reported that caesarean-born infants had a higher carriage of
C. perfringens compared to vaginally-born infants [
14]. However, the Japanese study demonstrated that the rate of colonization by
C. perfringens was much lower in breast-fed (17%) than in formula-fed infants (43%). It appears that the composition of
Clostridium species in the fecal microbiota of infants can be affected by the type of feeding during the first month of birth. Higher carriage of toxigenic
C. perfringens in caesarean-born infants needs further investigation of its sources and potential clinical significance.
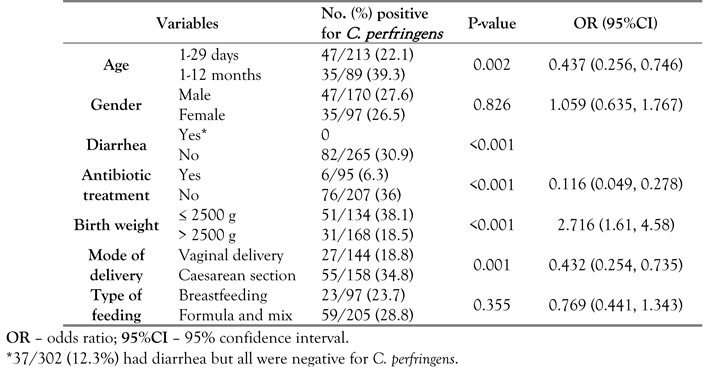
This study shows that there was a significant association (p<0.05) between C. perfringens colonization and certain neonatal condition related to age, birth weight, mode of delivery, and hospital ward, while the presence of diarrhea, antibiotic treatment, type of feeding and gender were not significantly associated with C. perfringens colonization (p>0.05).
The pathogenicity of
C. perfringens is related to the potential presence of its lethal toxin genes. The pattern of potential toxin genes is different depending on the
C. perfringens toxigenic type prevalent in each community, and it is still unknown why such difference occurs and which conditions activate these genes [
5,
6,
15].
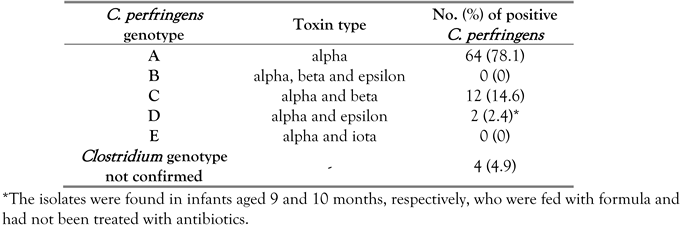
The present study indicates that almost all
C. perfringens isolates were positive for potential presence of the a-toxin gene (95.1%), followed by β2-toxin gene (69.5%), whereas the other toxin genes were detected in few samples, and no single isolate was positive for iota or enterotoxin genes (
Table 3). All our
C. perfringens isolates were obtained from infants without diarrhea, and this might explain the absence of enterotoxin-producing isolates in their intestine. It has been reported that α-toxin, encoded by the
cpa gene, is common to all 5
C. perfringens types (A–E) [
5], and especially
C. perfringens type A can cause fatal diseases in human and several animal species [
5,
6].
This study shows also that 78.1% of
C. perfringens isolates can be classified for epidemiological purposes as genotype A and
cpe-negative (PCR positive for the alpha toxin gene but negative for genes encoding beta, epsilon, iota
, and enterotoxins) as reported by Badagliacca et al. [
15], whereas the other B to E genotypes were found less commonly (0-14.6%) in our isolates (
Table 4).
It is well documented that enterotoxigenic
C. perfringens is one of the most common causes of food-poisoning in humans worldwide [
4,
5,
6]. A study in Jordan reported that no enterotoxin-positive
C. perfringens found in clinical cases was associated with necrotic enteritis in commercial poultry using multiplex PCR, since all their isolates were classified as enterotoxin negative type A [
16]. It would be highly important epidemiologically to initiate a large follow up study to search for the incidence of toxin types among
C. perfringens isolates from food sources and food-poisoning cases in Jordan.
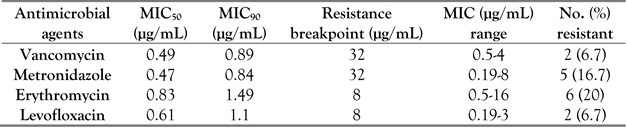
Most published antimicrobial susceptibility studies tested isolates from food items or animals. This study demonstrates that most
C. perfringens isolates from fecal samples of infants were susceptible to vancomycin, levofloxacin, metronidazole, and erythromycin (
Table 5). The minimum inhibitory concentrations for 50% of isolates (MIC
50) for vancomycin, levofloxacin, metronidazole, and erythromycin were 0.49, 0.61, 0.47, and 0.83 µg/mL, respectively. Only 22% of
C. perfringens isolates were resistant to erythromycin. A case study of clindamycin-resistant
C. perfringens human cellulitis was reported by Khanna in England, while it is well known that
C. perfringens is highly susceptible to all penicillin drugs [
17].
A previous study done in Jordan found that all
C. perfringens isolates from animal sources showed a very high minimal inhibitory concentration (MIC
50> 256 mg/mL) for lincomycin, erythromycin, and tilmicosin [
16]. A recent study has reported that all
C. perfringens isolates from cooked beef sold in Côte d’Ivoire were vancomycin susceptible, and 80% and 45% of the isolates were resistant to norfloxacin and erythromycin, respectively [
18]. A recent study done also in Brazil found that all
C. perfringens isolates from insects (
Tinamidae,
Cracidae and
Ramphastidaespecies) were susceptible to vancomycin and metronidazole, while 22.2% were resistant to erythromycin [
19]. Additionally, a recent study from Thailand reported that 54.9% of
C. perfringens isolates from diarrheal newborn piglets were resistant to erythromycin and exhibited a high MIC
50value of 128 μg/mL [
20]. A study done in Egypt found that 34%, 46%, 58%, 67%, 94% and 98% of
C. perfringens isolated from broiler chickens were resistant to rifampicin, chloramphenicol, colistin, ciprofloxacin, norfloxacin, and doxycycline, respectively [
21]. Overall, studies from various countries showed generally an elevated rate of antibiotic resistance among
C. perfringens and among other bacterial pathogens isolated from poultry [
21,
22].