Abstract
Introduction: Members of the family Enterobacteriaceae are commonly identified in the clinical laboratory, being responsible for a substantial range of infections. This study aimed to investigate phenotypic and genotypic resistance traits in pathogenic Enterobacteriaceae isolated from outpatients in Cluj-Napoca, Romania. Methods: Pathogenic Enterobacteriaceae were isolated from urinary tract infections, wound infections and persistent diarrhea in a private laboratory from Cluj-Napoca, Romania. Bacterial strains were biochemically identified and subjected to antimicrobial susceptibility testing by disk diffusion. The carriage of antibiotic resistance genes and of class 1 integron were assessed by PCR. Results: E. coli and Enterobacter spp. were the most prevalent pathogens. High levels of resistance were observed against folate pathway inhibitors (74%), fluoroquinolones (49%) and penicillins (44%). The incidence of carbapenem resistance was 3%. The strains displaying phenotypic resistance were able to produce β-lactamase enzymes encoded by blaTEM, blaTEM-1, blaSHV-1 and blaCTX-M, aminoglycoside modifying enzymes due to the carriage of aac(3)-IIIa,aac(6')-II and aac(6’)-Ie-aph(2”), to possess fluoroquinolones resistance due to qnrS DNA gyrase protection proteins and resistance to folate pathway inhibitors due to dihydropteroate synthases encoded by sul1, sul2 and sul3 genes. The high frequency of intI1 integrase was associated to sulphonamide resistance (r = 0.48; p < 0.001) and also to fluoroquinolone resistance (r = 0.27; p = 0.011), but no significant associations in the co-occurrence of specific antibiotic resistance genes and intI1 were found in pathogenic Enterobacteriaceae. Conclusions: An important proportion of pathogenic Enterobacteriaceae were multidrug resistant, due to a wide diversity of mechanisms encoding genetic resistance.
Introduction
Members of the family Enterobacteriaceae are commonly isolated in the clinical laboratory, being responsible for a variety of infections: enteric disease, urinary tract infections, wound and burn infections, bloodstream infections, pneumonia, ocular, ear and sinus infections. Various Escherichia coli serotypes, or strains belonging to Citrobacter, Enterobacter, Hafnia, Klebsiella, Proteus, Providencia, Salmonella, Yersinia and other genera have long been responsible for a substantial percentage of nosocomial and community-acquired infections [1].
Antimicrobial resistant (AMR) bacteria exert a great burden on healthcare systems, management of the infectious diseases becoming progressively complicated. In Romania, the incidence of multidrug resistant (MDR) Enterobacteriaceae reaches one of the highest levels, as indicated by the latest annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). Recently, significant increases were observed for third-generation cephalosporin and aminoglycoside resistance, as well as for combined resistance to fluoroquinolones, aminoglycosides and third-generation cephalosporins [2].
Antibiotic resistance genes (ARGs) can arise and spread rapidly in different bacteria. This way, AMR is more available than ever before to pathogenic organisms, narrowing the options of clinical medicine for drug regimens used to treat bacterial infection. The spread of particular ARG types or AMR strains can widely occur from patient to patient in hospitals, from person to person in the community or between the clinical settings and the environment [3].
Gram-negative bacteria possess naturally occurring resistance mechanisms, such as the chromosomally mediated β-lactamases. TEM-type are the first plasmid-mediated ARGs. The genes blaTEM, blaSHV and blaCTX-M are frequently found in Enterobacteriaceae. A variety of extended-spectrum β-lactamases (ESBLs) arise by mutations of the parent genes and of other common genes. VIM- and NDM-type metallo-β-lactamases, OXA-48 and KPC appear as the most successful enzymes and may threaten the efficacy of carbapenems in the near future [4].
The combination of mutagenesis with the ability of ARGs to transfer as part of integrons, gene cassettes, transposons, or integrative conjugative elements results in the capacity of resistance genes to virtually reach all bacterial types [5]. In Gram-negative bacteria, particularly the Enterobacteriaceae, ARGs and associated mobile elements contained by plasmids often cluster together in large multiresistance regions [6]. More recently, genetic determinants conferring resistance to fluoroquinolones, aminoglycosides and sulphonamides, such as the qnr, aac(6’)-Ib-cr and sul genes were found to be associated with β-lactamase ARGs in complex integron structures, and most frequently with class 1 integron [7,8,9]. Thus, class 1 integrons pose major clinical concern, being among the biggest contributors to the problem of MDR infections. In pathogenic Enterobacteriaceae the cassette arrays can vary, but overall they have certain common, albeit not universal, characteristic features [10].
Previous Romanian studies of resistance in clinical Enterobacteriaceae were focused on detection of β-lactamases, ESBLs and carbapenemases [11,12,13,14]. Even with a global surveillance, dynamics of antimicrobial resistance occurring both under antibiotic treatment and in the environment are poorly understood. Therefore, a continuous monitoring of a diversity of mechanisms is imperative. Most of the existing information on the antibiotic resistance patterns is based on samples from hospitalized patients and limited information is available regarding outpatient infections.
The aim of the present work was to analyze the antimicrobial susceptibility profiles in pathogenic Enterobacteriaceae from outpatients and to characterize a variety of genetic mechanisms responsible for resistance. Enzymes involved in drug alteration (β-lactamases, aminoglycoside modifying enzymes), catalytic enzymes (in the folic acid biosynthesis) and protecting proteins (for the DNA-enzyme complexes from the action of quinolones) have been investigated. Also, the prevalence of class 1 integron and the association with phenotypic and genotypic resistance was assessed in all Enterobacteriaceae isolates.
Methods
Bacterial isolates
A cross-sectional study was conducted to assess the mechanisms of antibiotic resistance among Enterobacteriaceae isolated from outpatient infections in Cluj-Napoca, Romania. A collection of 68 non-susceptible bacterial strains were investigated for AMR and ARG patterns. From January to March 2016, clinical specimens were collected from individual patients addressing a private laboratory located in the center of the city. After the inoculation of samples into specific culture media including blood agar, MacConkey agar, deoxycholate citrate Leifson agar, Hektoen enteric agar and eosin methylene blue agar and following overnight incubation, typical presumptive Enterobacteriaceae colonies were purified and recultured. The isolates were identified according to standard methods by colony appearance, pigment production, catalase test, oxidase test, Gram staining, triple sugar iron fermentation, motility, indole, urease production, citrate utilization, lysine decarboxylase, phenylalanine deaminase detection and oxidation-fermentation of sugars. For confirmation, isolates were speciated with the API-20E biochemical kits (bioMérieux, Marcy-l'Étoile, France). Enteropathogenic E. coli (EPEC) strains isolated from fecal samples were serogrouped by slide agglutination using O-specific antisera (Bio-Rad, Hercules, USA).
Antimicrobial susceptibility testing
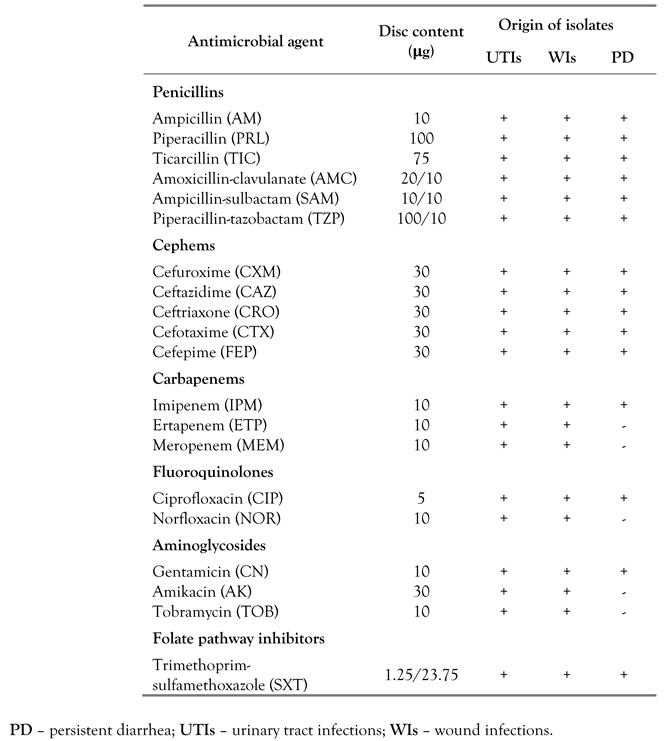
All bacterial strains were subjected to antimicrobial susceptibility testing against six antibiotic classes: penicillins, cephems, carbapenems, fluoroquinolones, aminoglycosides and folate pathway inhibitors. Enterobacteriaceae isolated from urinary tract infections (UTIs) and from wound infections (WIs) were tested for resistance against up to 20 individual drugs. The panel for EPEC strains isolated from patients with persistent diarrhea (PD) included up to 15 antibiotics (Supplementary Table 1). Antibiotic susceptibility testing was performed by disk diffusion in Mueller-Hinton agar plates. Following overnight incubation, the inhibition zone diameters were measured and the results were interpreted according to CLSI guidelines [15]. MDR was defined as non-susceptibility to at least one agent in three or more antimicrobial categories.
Antibiotic resistance genes screening
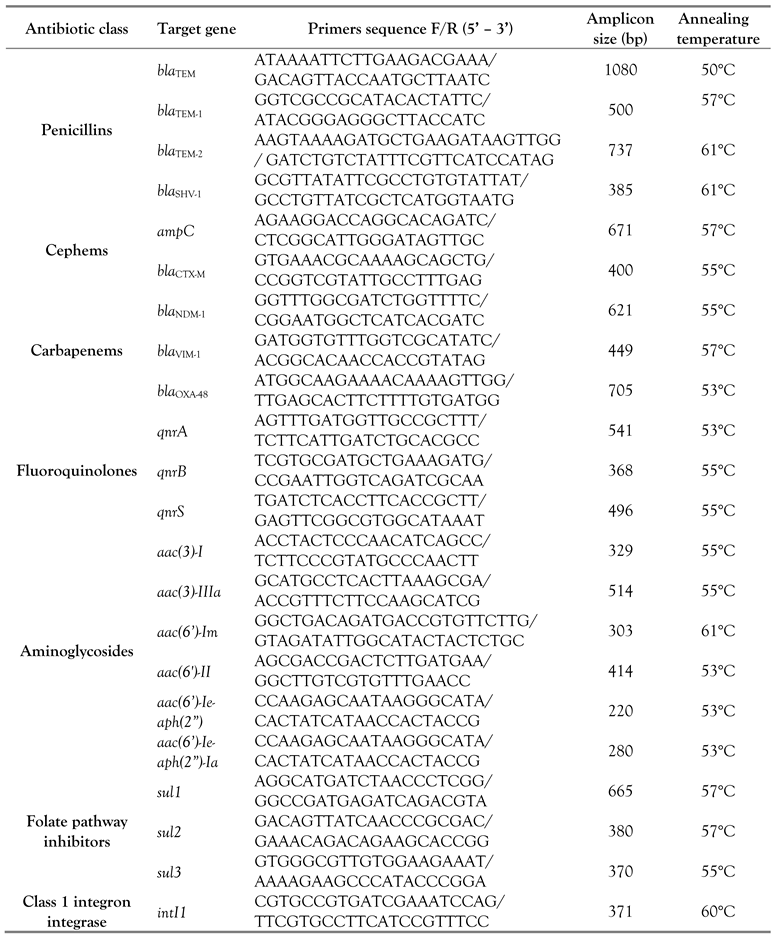
Bacterial strains displaying phenotypic resistance were included in the molecular testing for detection of ARGs conferring resistance to the corresponding antibiotic classes. The carriage of class I integron integrase was screened in all isolates. PCR amplifications were performed in 25 μL reaction mix containing: nuclease-free water, DreamTaq Green PCR master mix, forward and reverse primers to a final concentration of 0.5 μM, and DNA template as fresh bacterial suspensions. The PCR reaction conditions were set up as follows, using a thermocycler TProfessional TRIO (Analytik Jena, Jena, Germany): initial denaturation at 94°C for 5 min; 35-fold repeated cycles for denaturation, annealing (Supplementary Table 2) and synthesis; a final elongation at 72°C for 8 min. Agarose gel electrophoresis was used for separation of DNA amplicons. Data acquisition and analysis were performed using the BDA Digital Compact System and BioDocAnalyze Software (Analytik Jena). Molecular reagents and oligonucleotide primers were purchased from Cleaver Scientific (Rugby, UK), Eurogentec (Seraing, Belgium), Lonza (Basel, Switzerland) and Thermo Fisher Scientific (Massachusets, USA).
Statistical analysis
Inferential statistics methods were used to calculate the proportions of AMR and MDR Enterobacteriaceae as well as the abundances of ARGs and intI1 in bacterial strains displaying clinical resistance. Correlations between the AMR and ARG numbers were explored. Resistant and susceptible phenotypes (with the intermediate resistance phenotype considered as susceptible) and positive/negative genotypes were converted to binary matrices (1-0). It was verified whether the phenotypic or genotypic resistance was associated with the presence of intI1 gene, while linear and logistic regression models were used to assess the degree of relationship. Two-sided significance level was set at 0.05, inferred using the Real Statistics Resource Pack software for Microsoft Excel [16].
Results
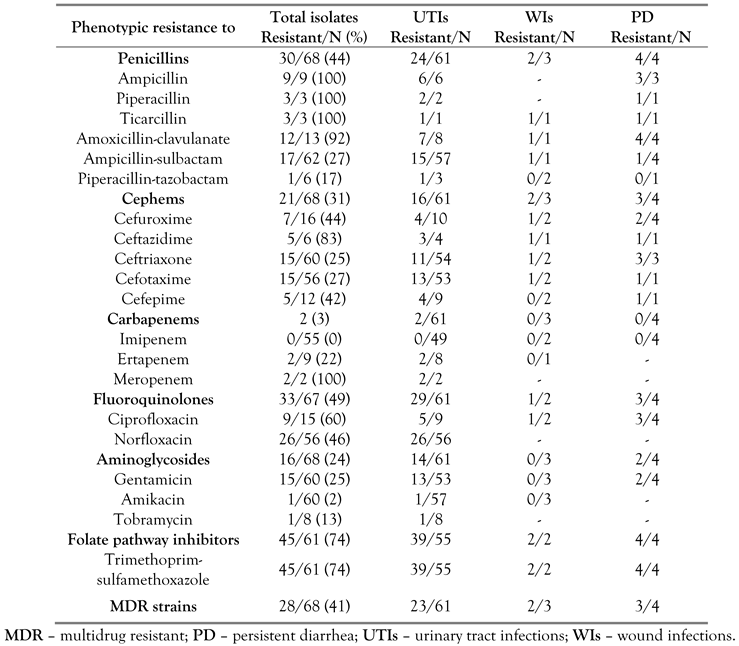
A total of 68 non-susceptible bacterial strains were analyzed during this study: 61 isolates from UTIs (90%), three isolates from WIs (6%) and four isolates from patients with PD (4%). Biochemical identification confirmed 47 E. coli strains (of which 4 were EPEC), 13 Enterobacter spp., 5 Proteus spp., 2 Citrobacter spp. and 1 Providencia spp. Multiple resistance up to 14 individual drugs belonging to maximum six antimicrobial categories was observed (Figure 1). High levels of resistance were displayed for folate pathway inhibitors (74%), fluoroquinolones (49%) and penicillins (44%). Carbapenem resistance was observed in 3% of the isolates. A proportion of 41% of the resistant strains were MDR (Table 1).
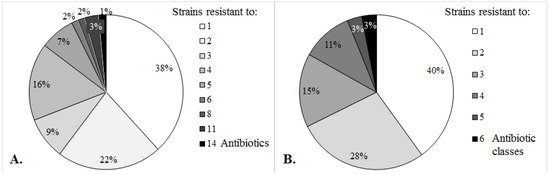
Figure 1.
Antimicrobial resistance in clinical Enterobacteriaceae.

Table 1.
Frequencies of resistant and multidrug resistant Enterobacteriaceae.
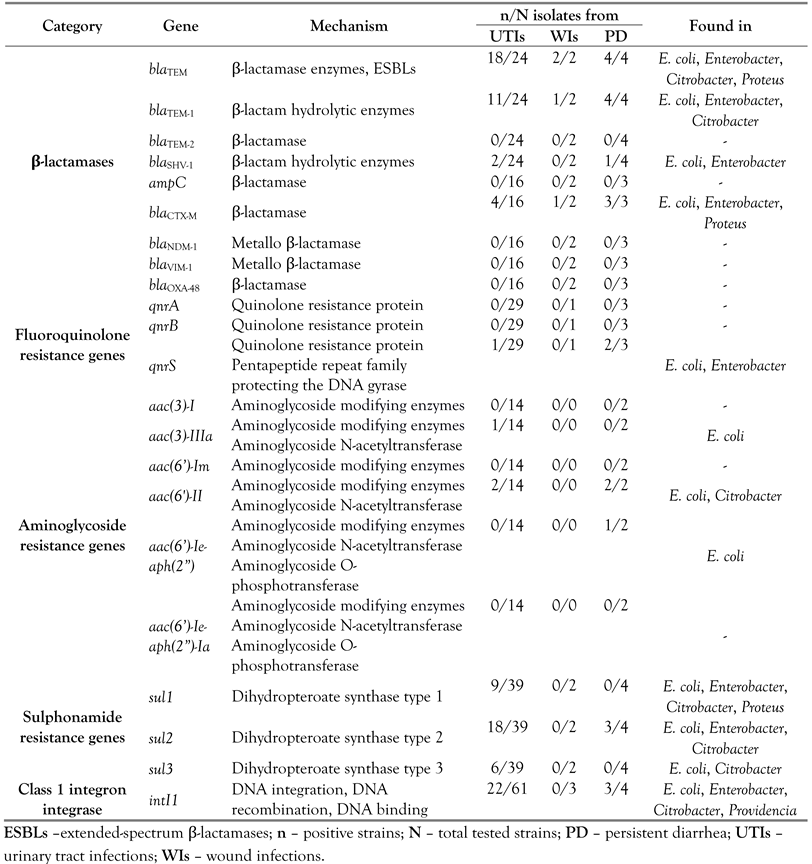
Nine β-lactamase genes were screened during this study, and only four were detected: blaTEM, blaTEM-1, blaSHV-1 and blaCTX-M. The most frequent ARGs were blaTEM, found in 24 out of 30 strains resistant to penicillins. blaTEM-1 encoding the parent enzyme of TEM-type ESBLs was found in 16 isolates. blaCTX-M, the most influential mechanism providing cephalosporin resistance in Enterobacteriaceae, was present in 8 out of 21 bacterial strains. Three genes responsible for resistance to fluoroquinolones (qnrA, qnrB and qnrS) were investigated, qnrS genes being detected in three out of 33 isolates resistant to ciprofloxacin or norfloxacin. Six ARGs encoding aminoglycoside modifying enzymes were screened by PCR. aac(3)-IIIa, aac(6’)-II and aac(6’)-Ie-aph(2”) were detected in 5 out of 16 isolates phenotypically resistant to gentamicin. Resistance to folate inhibitors pathway was encoded by sul1, sul2, and sul3 genes in 25 of the 45 isolates resistant to trimethoprim-sulfamethoxazole (Figure 2). Three isolates contained only the sul1 gene, 15 other isolates contained only the sul2 gene and one isolate contained only the sul3 gene. sul1, sul2 and sul3 genes occurred simultaneously in one strain, both sul1 and sul2 were found in 5 isolates, while both sul2 and sul3 were found in five strains. sul1 and sul3 genes were not observed concomitantly. A significant proportion of pathogenic Enterobacteriaceae harbored class 1 integrons, the intI1 gene being detected in 40 strains (59%) (Figure 2).
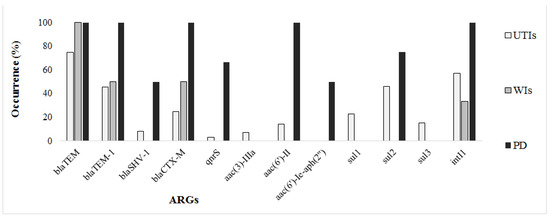
Figure 2.
Genetic resistance in clinical Enterobacteriaceae.
With regard of the sample origin, the blaTEM, blaTEM-1 and blaCTX-M genes were detected in samples from various infections. The blaSHV-1 β-lactamase as well as the genes conferring resistance to fluoroquinolones and aminoglycosides were not observed in samples from WIs, being found frequently in EPEC strains from PD samples. Sulphonamide resistance genes were mostly associated with UTIs, and were not detected in WIs. Regarding the taxonomic incidence, ARGs particularly occurred in Citrobacter and E. coli (particularly EPEC), were less frequent in Enterobacter and Proteus species, and were not found in Providencia isolates (Table 2).

Table 2.
Antibiotic resistance genes and class 1 integron integrase detected in Enterobacteriaceae isolated from clinical samples.
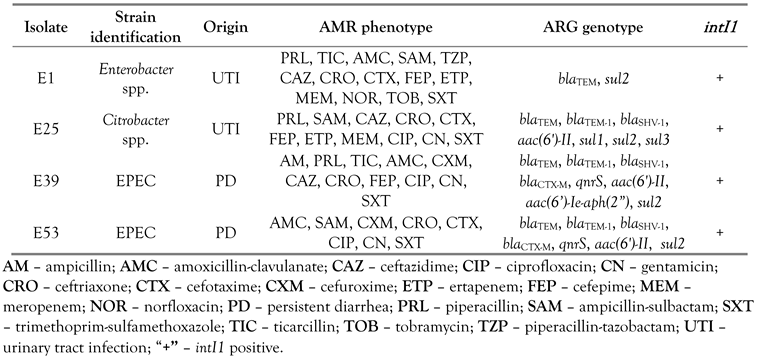
Phenotypic and genotypic profiles of some bacterial strains require special attention. Despite the wide AMR pattern to 14 drugs, Enterobacter spp. E1 was found to carry only two resistance genes. Citrobacter spp. E25, extensively resistant to eleven antibiotics, expressed seven ARGs. Two E. coli strains, EPEC E39 and E53, resistant to eleven and eight drugs, were found to possess a plethora of ARGs, of 9 and 7 genes, respectively. All these 4 strains were positive for intI1. In addition, the simultaneous presence of sul1 and sul3 genes in strain E25 suggests the coexistence of multiple integrons (Table 3).

Table 3.
Characteristics of multidrug resistant strains with exceptional resistance profiles.
Statistically significant correlations were observed between the levels of phenotypic and genotypic resistance (r=0.60; p<0.001). Despite the variable resistance patterns overall, two significant correlations between the integron and the phenotypic resistance were found. The presence of intI1 was correlated to trimethoprim-sulfamethoxazole resistance (r=0.48; p<0.001) and also to fluoroquinolone resistance (r=0.27; p=0.011). ARG abundances were linked to intI1 carriage (r=0.33; p=0.006) but no significant associations in the co-occurrence of specific ARGs and intI1 were found in pathogenic Enterobacteriaceae.
Discussion
Most of the resistant Enterobacteriaceae were isolated from UTIs, E. coli and Enterobacter spp. prevailing in clinical infections. Klebsiella species were not detected in this study, although other Romanian papers reported them among the most frequent pathogens [12,13,14,17]. The frequency of MDR Enterobacteriaceae causing infections in patients from North-Western Romania was similar to that reported in the South-Eastern part of the country [18]. Carbapenems are still valid options for the treatment of MDR strains of Enterobacteriaceae, and imipenem was found to be the most effective drug.
The TEM-1 β-lactamase is able to hydrolyze ampicillin at a higher rate than that for carbenicillin, oxacillin, or cephalothin, and displays negligible activity against extended-spectrum cephalosporins. The SHV-1 enzyme has the same hydrolytic profile as TEM-1, conferring resistance to broad-spectrum penicillins such as ampicillin, ticarcillin and piperacillin but not to oxyimino-substituted cephalosporins. In this screening, the presence of the blaTEM-1 gene was highly related to phenotypic resistance to ampicillin, while the carriage of blaSHV-1 was additionally associated to third and fourth generation cephalosporins resistance. In the past, TEM-1 β-lactamases were described as susceptible to inhibitors such as clavulanic acid and sulbactam. The present study reveals that all the blaTEM-1-carying bacteria were resistant to antibiotic combinations such as amoxicillin-clavulanate or ampicillin-sulbactam. This mechanism was attributed to the TEM-1 β-lactamase hyperproduction [19]. TEM-1 and TEM-2 are not ESBLs. Therefore, detection of strains carrying the blaTEM but not the blaTEM-1or blaTEM-2 genes suggests that approximately 16% of the penicillin-resistant strains carry blaTEM-type ESBL derivatives.
The ampC gene was not detected in study strains, while the blaCTX-M group was present. ampC plasmid genes are derived from the chromosomal ampC genes of various Enterobacteriaceae and ancestors of the blaCTX-M group of β-lactamases [6]. Most of CTX-M enzymes exhibit powerful activity against cefotaxime and ceftriaxone but not ceftazidime, and the presence of the blaCTX-M gene was associated to ceftriaxone resistance. blaNDM, blaVIM and blaOXA-type genes, a significant cause of carbapenem resistance in Enterobacteriaceae, recently found in bacterial strains from hospitalized patients in Romania [11,12,13], were not detected during this study. Qnr proteins, encoded by ARGs such as the qnrS gene, belong to the pentapeptide repeat family capable of protecting DNA gyrase from drug inhibition, being responsible for resistance in the presence of quinolones at therapeutic levels.
The resistance mechanism of aac(3)-IIIa and aac(6’)-II genes relies on aminoglycoside N-acetyltransferase, which modifies aminoglycosides by acetylation. In addition, the bifunctional aac(6')-Ie-aph(2″) gene modifies aminoglycosides by phosphorylation, being responsible for high-level aminoglycoside resistance. These ARGs are not common for Enterobacteriaceae. They were previously observed in Pseudomonas, Enterococcus or Staphylococcus species [20,21]. Other aminoglycoside ARGs, such as aac(3)-I, aac(6')-Im or aac(6’)-Ie-aph(2”)-Ia, known as frequently present in Enterobacteriaceae, were not detected during this study.
The sul genes encode for dihydropteroate synthases that are not inhibited by sulphonamides, catalyzing the formation of the immediate precursor of folic acid. The more frequent dissemination of sul2 gene, as observed in this study, in 47% of the isolates, could be attributed to the efficacy and stability of type II enzyme. The gene frequency distribution sul2 > sul1 > sul3 is characteristic for pathogenic E. coli in Europe and Canada [22], while the pattern of increased sul1 frequency over sul2 was observed in Australia and Korea [23,24]. Distribution of sul genes in the 3’-conserved segment provides evidence on class 1 integrons diversity: classical integrons contain the sul1 gene while non-classical integrons lack the sul1 cassette, and the sul3 gene is present.
Horizontal gene transfer adds an important dimension to infectious pathology, particularly when an ARG can transfer resistance to multiple otherwise unrelated pathogens [25]. A variety of penicillin, aminoglycoside and sulphonamide resistance determinants are often found as part of transferrable gene cassettes in integrons [6,10,20], and therefore the high frequency of class 1 integrons in pathogenic Enterobacteriaceae displaying AMR.
This study has several limitations. First, a small number of bacterial isolates were sampled in a single facility, over a period of only 3 months. Therefore, the results of the survey may not be generalizable for the entire population. Nevertheless, it brings relevant data on the magnitude of resistance in non-hospitalized patients. Second, the PCR technique is unable to detect the whole range of ARGs and subsequent variants. From hundreds of genes known by far, molecular detection was limited to a selection of ARGs. This set of genes was chosen to cover a diversity of resistance mechanisms with wide occurrence and/or clinical significance for pathogenic Enterobacteriaceae.
Conclusions
Pathogenic Enterobacteriaceae isolated in Cluj-Napoca displayed high levels of phenotypic resistance against folate pathway inhibitors, fluoroquinolones and penicillins. The incidence of carbapenem-resistance was very low. Forty-one percent of the strains were MDR.
A wide diversity of mechanisms encoding genetic resistance were found to be involved in AMR. The strains displaying phenotypic resistance were able to produce β-lactamase enzymes, aminoglycoside modifying enzymes, fluoroquinolone protection proteins as well as enzymes able to confer resistance to sulphonamides. Class 1 integron integrase was detected in 59% of the resistant isolates and significant associations were found between the carriage of intI1 gene and the levels of phenotypic and genotypic resistance. Correlations between the presence of intI1 and resistance to sulphonamides and also regarding the concomitant carriage of intI1 and sul2 genes were found.
Supplementary

Supplementary Table 1.
Antibiotics used in antimicrobial susceptibility testing for pathogenic Enterobacteriaceae.
Supplementary Table 1.
Antibiotics used in antimicrobial susceptibility testing for pathogenic Enterobacteriaceae.
 |

Supplementary Table 2.
Primers and PCR conditions used in this study.
Supplementary Table 2.
Primers and PCR conditions used in this study.
 |
Author Contributions
AF designed and supervised the study, analyzed the data and drafted the manuscript. ET and ABK carried out the laboratory work and reviewed the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This work was partially supported by the European Economic Area 2009-2014 Financial Mechanism under the RO04 programme [Grant number 3499/20.05.2015].
Conflicts of interest
All authors—none to disclose.
References
- Brenner, D.J.; Farmer, J.J.I.I.I. Family Enterobacteriaceae. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.R., Garrity, G.M., Eds.; Springer: New York, 2005; pp. 587–606. [Google Scholar]
- European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe 2016; Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net); ECDC: Stockholm, Sweden, 2017. [Google Scholar]
- Szekeres, E.; Baricz, A.; Chiriac, C.M.; et al. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ Pollut 2017, 225, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Iredell, J.; Brown, J.; Tagg, K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ 2016, 352, h6420. [Google Scholar] [CrossRef] [PubMed]
- Tolmasky, M.E. Overview of dissemination mechanisms of genes coding for resistance to antibiotics. In Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition; Bonomo, R.A., Tolmasky, M.E., Eds.; ASM Press: Washington DC, 2007; pp. 267–720. [Google Scholar]
- Partridge, S.R. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 2011, 35, 820–855. [Google Scholar] [CrossRef] [PubMed]
- de Toro, M.; Sáenz, Y.; Cercenado, E.; etal., *!!! REPLACE !!!*. Genetic characterization of the mechanisms of resistance to amoxicillin/clavulanate and third-generation cephalosporins in Salmonella enterica from three Spanish hospitals. Int Microbiol 2011, 14, 173–181. [Google Scholar] [PubMed]
- Kiiru, J.; Butaye, P.; Goddeeris, B.M.; Kariuki, S. Analysis for prevalence and physical linkages amongst integrons, ISEcp1, ISCR1, Tn21 and Tn7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992–2011). BMC Microbiol 2013, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Rajpara, N.; Kutar, B.M.; Sinha, R.; et al. Role of integrons, plasmids and SXT elements in multidrug resistance of Vibrio cholerae and Providencia vermicola obtained from a clinical isolate of diarrhea. Front Microbiol 2015, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Márquez, C.; Labbate, M.; Raymondo, C.; et al. Urinary tract infections in a South American population: dynamic spread of class 1 integrons and multidrug resistance by homologous and site-specific recombination. J Clin Microbiol 2008, 46, 3417–3425. [Google Scholar] [CrossRef] [PubMed]
- Székely, E.; Damjanova, I.; Jánvári, L.; et al. First description of blaNDM-1, blaOXA-48, blaOXA-181 producing Enterobacteriaceae strains in Romania. Int J Med Microbiol 2013, 303, 697–700. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Flonta, M.; Boudehen, Y.M.; et al. Dissemination of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa in Romania. Antimicrob Agents Chemother 2015, 59, 7100–7103. [Google Scholar] [CrossRef] [PubMed]
- Lixandru, B.E.; Cotar, A.I.; Straut, M.; et al. Carbapenemase-producing Klebsiella pneumoniae in Romania: a six-month survey. PloS One 2015, 10, e0143214. [Google Scholar] [CrossRef] [PubMed]
- Axente, C.; Muntean, D.; Baditoiu, L.; et al. Beta-lactam resistance mechanisms in pathogens isolated from patients admitted to intensive care unit. Rev Chim 2017, 68, 1223–1226. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-fifth informational supplement M100-S25; CLSI: Wayne, 2015. [Google Scholar]
- Zaiontz, C. Real Statistics Using Excel (Release 5.4). 2018. Available online: www.real-statistics.com (accessed on 01 December 2018).
- Pricop, C.; Suditu, N.; Vrinceanu, R.; et al. Multidrug resistant urinary tract infections in Moldova, Romania: focusing on uropathogens and their antibiotic susceptibility. Can we do more? Nobel Med 2015, 11, 42–49. [Google Scholar]
- Arbune, M.; Decusara, M.; Macovei, L.A.; et al. Surveillance of antibiotic resistance among Enterobacteriaceae strains isolated in an infectious diseases hospital from Romania. Rev Chim 2018, 69, 1240–1243. [Google Scholar] [CrossRef]
- Waltner-Toews, R.I.; Paterson, D.L.; Qureshi, Z.A.; et al. Clinical characteristics of bloodstream infections due to ampicillin-sulbactam-resistant, non-extended-spectrum-β-lactamase-producing Escherichia coli and the role of TEM-1 hyperproduction. Antimicrob Agents Chemother 2011, 55, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist Updat 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Medchemcomm 2016, 7, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Blahna, M.T.; Zalewski, C.A.; Reuer, J.; Kahlmeter, G.; Foxman, B.; Marrs, C.F. The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J Antimicrob Chemother 2006, 57, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Gündoğdu, A.; Long, Y.B.; Vollmerhausen, T.L.; Katouli, M. Antimicrobial resistance and distribution of sul genes and integron-associated intI genes among uropathogenic Escherichia coli in Queensland, Australia. J Med Microbiol 2011, 60 Pt 11, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.W.; Lim, J.; Kim, S.; Kim, J.; Kwon, G.C.; Koo, S.H. Characterization of trimethoprim-sulfamethoxazole resistance genes and their relatedness to class 1 integron and insertion sequence common region in Gram-negative bacilli. J Microbiol Biotechnol 2015, 25, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can J Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2019.