Abstract
We evaluated the gene prevalence of the icaADBC operon, its correlation with biofilm formation and antibiotic resistance through a global meta-analysis. We searched for articles that reported the prevalence of icaADBC operon, biofilm, and antibiotic resistance in S. aureus from 2000 up to 1st March 2024. The search was done in scientific databases such as PubMed, Scopus, Google Scholar, EMBASE, and Web of Science. The MESH keywords were: icaADBC operon, biofilm, methicillin-resistant Staphylococcus aureus, antibiotic resistance. Comprehensive Meta-Analysis Software was used for data analysis. The estimation of the combined prevalence of each desired variable was performed by depicting a forest plot through the random-effects model with a 95% confidence interval. Data heterogeneity was estimated by Q and I2 indices, and p-value <0.05 was reflected as statistically significant heterogeneity. Fifteen articles were eligible for inclusion. The prevalence of ica operon genes varied between 28–51.5%. The prevalence of total ica operon genes in S. aureus was reported at 42.4% (95%CI: 29.4–56.5). Biofilm formation prevalence of S. aureus isolates in different studies was reported between 10-100%. The rate of total biofilm in S. aureus was 95.8%. The rate of total strong, moderate, and weak biofilm in S. aureus was reported at 35.4%, 35.3%, and 23.9%, respectively. Most reviewed studies reported a correlation between ica genes and biofilm. We found that many studies reported a correlation between the high prevalence of ica operon genes, phenotypic biofilm production, and antibiotic resistance. Also, regardless of whether the strains were MRSA or not, the high biofilm formation ability was reported at 95.8% by most studies.
Introduction
Staphylococcus aureus is known as an important pathogen in hospitals. The spread of infections caused by methicillin-resistant Staphylococcus aureus (MRSA) strains in all health-treatment centers of the world, especially vital units such as the intensive care unit (ICU) and the respiratory unit, has been reported [1]. Today, widespread and increasing resistance to the antibiotics available to treat infections caused by this microorganism has reached its peak, which depicts a difficult future for infections caused by MRSA strains [2].
From the pathogenic aspects of this microorganism, we can mention the extraordinary ability to form biofilm outside and inside the human body in different infections [3]. This high ability to produce biofilm has led to the spread of resistance against current antibiotics and disinfectants [4,5].
Attachment to host cells is the primary stage necessary for the initiation and establishment of infection. The organization of Staphylococcus aureus is facilitated by the expression of multiple proteins and adhesins, notably the microbial surface components that identify adhesive matrix molecules (MSCRAMMs), enabling binding to laminin, fibrinogen, fibronectin, and collagen [6]. Once the initial attachment is established, the biofilm forms and expands through proliferation and extracellular matrix production. Extracellular matrix components such as polysaccharide extracellular adhesions, extracellular DNA, proteins, and teichoic acids contribute to biofilm formation [7]. Also, S. aureus provides biofilm thickening through the synthesis of polysuccinyl glucosamine. All these steps are regulated and controlled by icaADBC operon specifically icaD gene [8]. The icaA gene encodes the enzyme N-acetyl-glucosaminyl transferase, which plays a crucial role in the synthesis of N-acetyl-glucosamine oligomers derived from UDP-N-acetyl-glucosamine. The complete phenotypic manifestation of the capsular polysaccharide is associated with the icaD gene [9]. The produced polysaccharide wraps around the bacterial cells, functioning as a shield against the immune system of the host and the action of antimicrobial agents [10].
The involvement of the icaADBC operon in biofilm development and its link to antibiotic resistance has been convincingly demonstrated in various research efforts. However, there is currently no extensive review that integrates all these foundational studies. This global systematic review and meta-analysis intends to fill this gap by thoroughly examining the relationship between the icaADBC operon, biofilm production, and antibiotic resistance.
Methods
Search plan
The search plan was based on articles that reported the prevalence of the icaADBC operon, biofilm, and antibiotic resistance in S. aureus from 2000 up to 1st March 2024. The search was done in scientific databases such as PubMed, Scopus, Google Scholar, EMBASE, and Web of Science. The MESH keywords were: icaADBC operon, biofilm, biofilm formation, S. aureus, and methicillin-resistant Staphylococcus aureus, S. aureus, MRSA, antibiotic resistance, antimicrobial resistance, ica genes.
Eligibility criteria
The studies that reported the prevalence of biofilm-related genes, biofilm, and the relationship with microbial resistance, from clinical samples between 2000 up to 1st March 2024, were included in our analysis. Studies on non-clinical strains (environmental and animal) were excluded from our study. Systematic review studies, case controls, and clinical trials were excluded from the current review.
Quality assessment
To determine the quality of the studies, a checklist of Appraisal tools for Cross-Sectional Studies (AXIS) was used [11], in which 20 different questions were asked about varied parts of the study, from the title to the conclusion, and finally, studies characterized by weaknesses were omitted (Supplementary S1).
Data extraction
In this review, our authors independently used a series of designed forms that contained information such as first author, publication time, first name, year of study, year of publication, type of study, location, sample size, S. aureus isolates, ica gene prevalence, molecular methods for detecting genes, biofilm formation, methods for measuring biofilm. The extracted data were entered into these forms.
Data analysis
For the purpose of data analysis, Comprehensive Meta-Analysis software (version 3.3.070) was employed. The combined prevalence of the targeted variables was estimated by constructing a forest plot utilizing a random-effects model, which provided a 95% confidence interval (CI). Heterogeneity among the data was measured using the Q and I2 indices, with a p value of less than 0.05 denoting statistically significant heterogeneity. To assess potential publication bias, both Egger’s linear regression test and a funnel plot were utilized.
Results
Among the 417 articles retrieved from the different databases, there were 201 duplicates that were deleted. Records were screened. A series of articles were removed and the rest of the articles were evaluated for retrieval. At this stage again, with the exclusion of several articles, the eligibility of other studies was checked, and finally, due to the reasons mentioned in Figure 1, only 15 articles were eligible to be included in this review. Included studies used standard techniques for biofilm evaluation such as microtiter plate (MTP), Tissue culture plate (TCP), and Congo Red Agar (CRA). All the studies included here, used PCR for the detection of ica operon genes (Table 1).
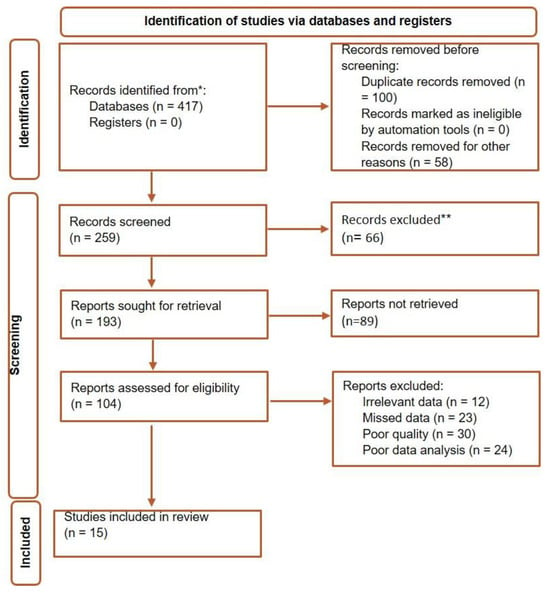
Figure 1.
Flow diagram for selection of studies included in this review.

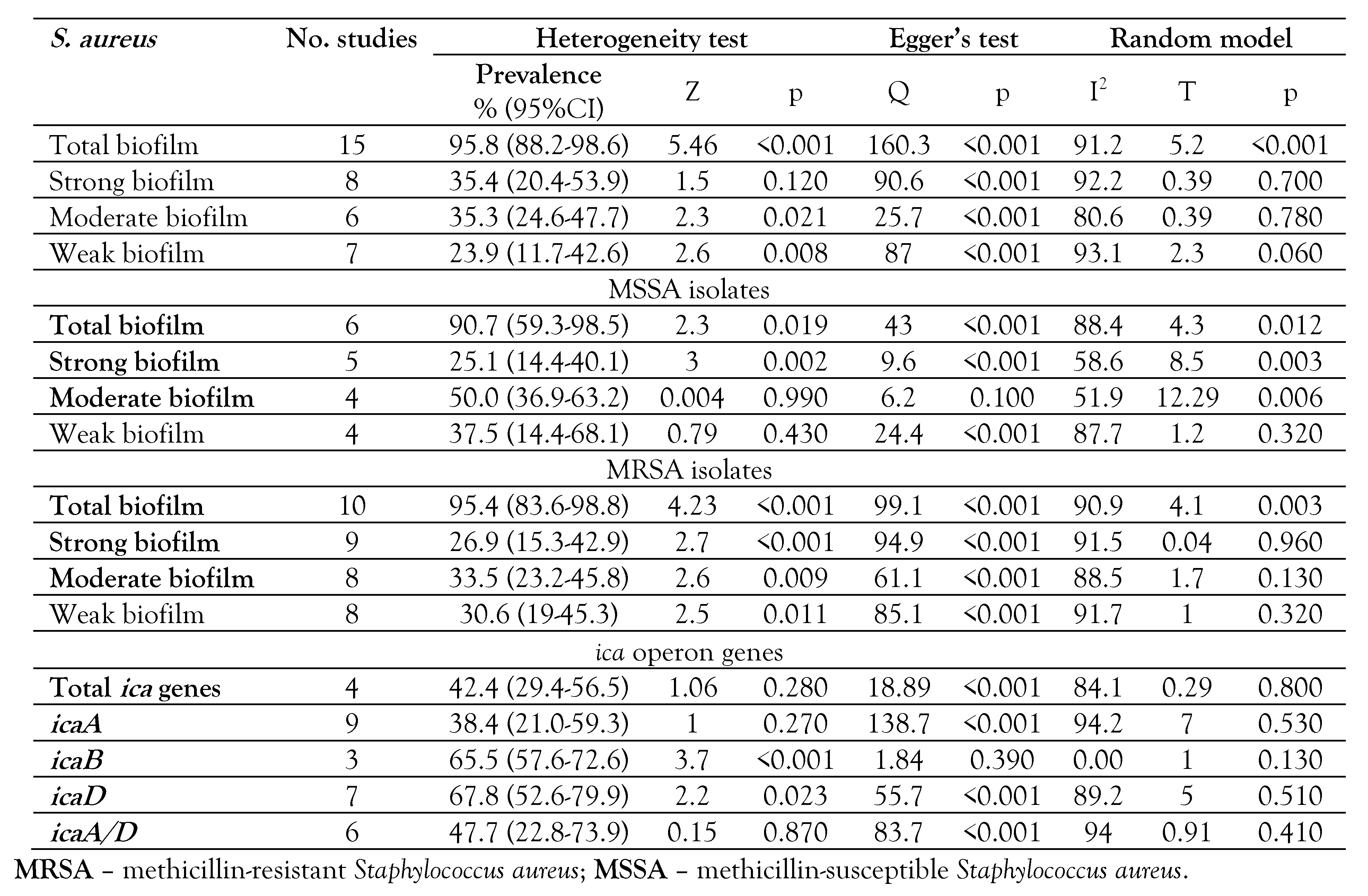
Table 1.
Characteristics of included studies in the present review.
Prevalence of total ica operon genes in S. aureus
The prevalence of ica operon genes varied between 28.0% and 51.5%. The prevalence of total ica operon genes in S. aureus was reported at 42.4% (95%CI: 29.4-56.5), Z=1.06, p=0.280. The frequencies of icaA, icaB, icaD and icaA/D were reported 38.4% (95%CI: 21.0-59.3), 65.5% (95%CI: 57.6-72.6), 67.8% (95%CI: 52.6-79.9), and 47.7% (95%CI: 22.8-73.9), respectively (Supplementary S2a, b, c, d, e and Table 2). The prevalence of icaA gene varied (between 3.1% and 80%), icaB (between 52.0% and 68.8%), icaD (between 38.5% and 91.5%), and icaA/D (between 3.8% and 74.2%).

Table 2.
Subgroups analysis for different variables in the present review.
Total biofilm formation rate in S. aureus strains
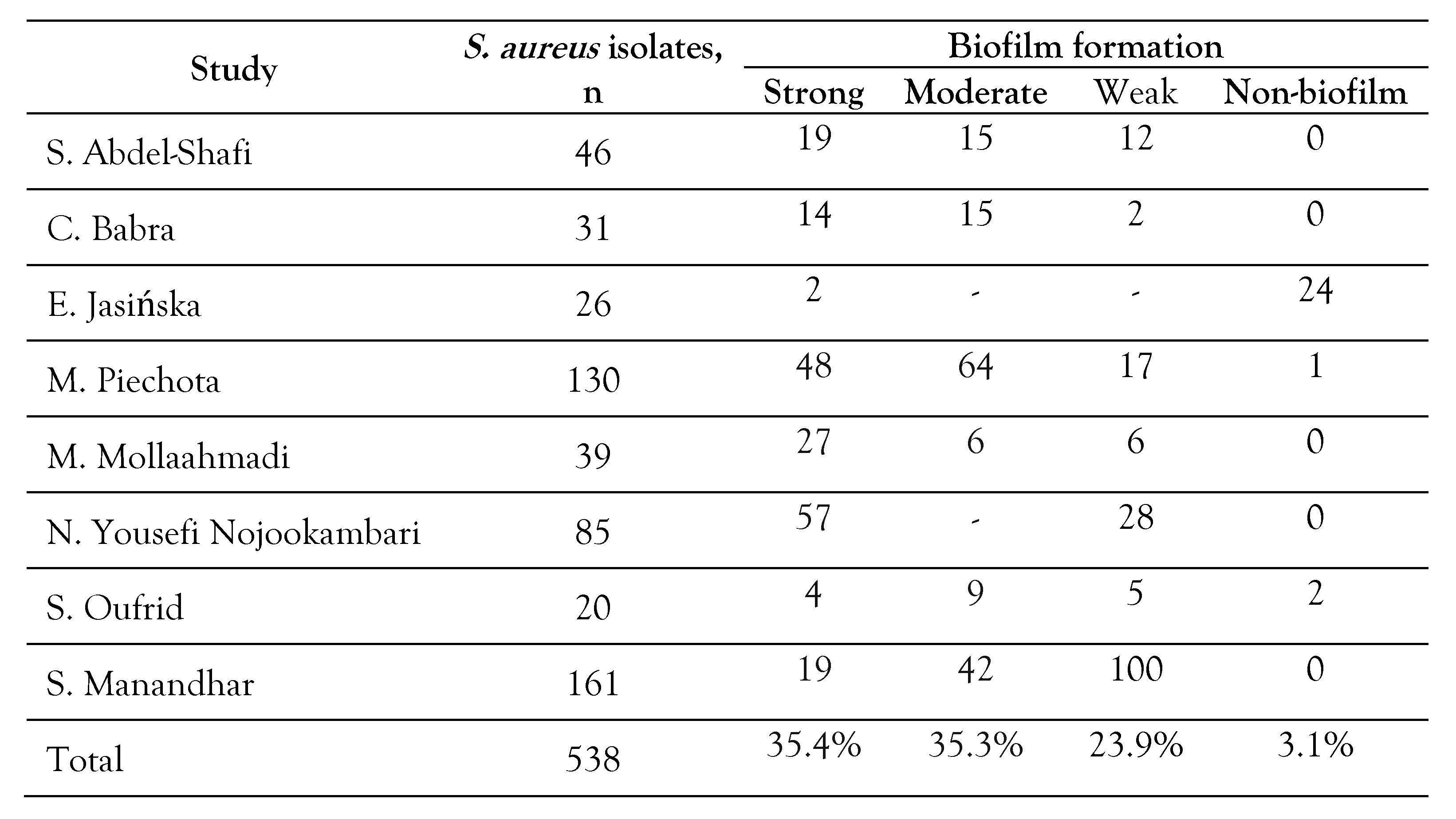
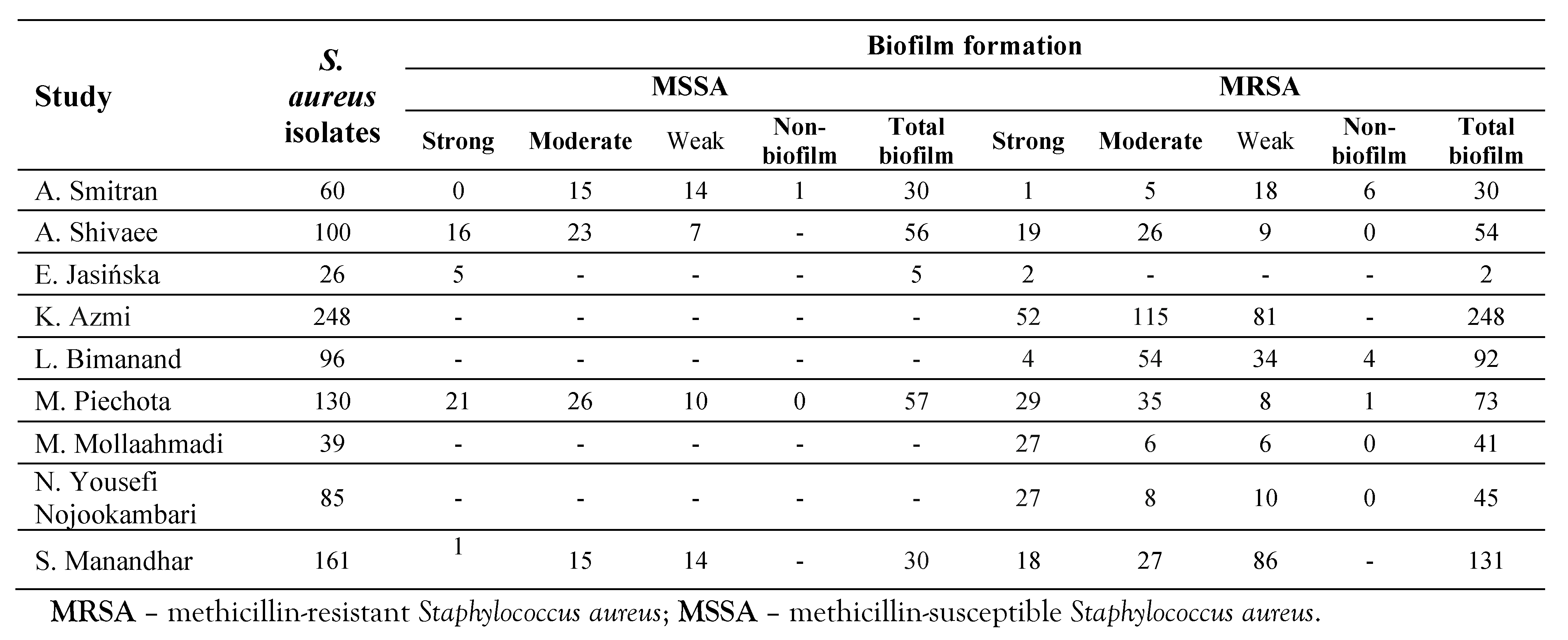
The prevalence of biofilm formation by S. aureus isolates in different studies was reported between 10-100% (Table 3). The rate of total biofilm in S. aureus was 95.8% (95%CI: 88.2-98.6), Z=5.4, p<0.001. The rates of total strong, moderate, and weak biofilm in S. aureus were reported at 35.4% (95%CI: 20.4-53.9), 35.3% (95%CI: 24.6-47.7), and 23.9% (95%CI: 11.7-42.6), respectively (Supplementary S3a, b, c, d and Table 2). The rate of total biofilm in MRSA strains was reported at 95.4% (95%CI: 83.6-98.8), Z=4.23, I2=90.9, p<0.001 (Table 2 and Table 4). The rate of total strong, moderate and weak biofilm in MRSA strains was 26.9% (95%CI: 15.3-42.9), 33.5% (95%CI: 23.2-45.8), and 30.6% (95%CI: 19.0-45.3), respectively (Table 2 and Table 4). The rates of total biofilm in MSSA isolates were reported at 25.1% (95%CI: 14.4-40.1), 50.0% (95%CI: 36.9-63.2), and 37.5% (95%CI: 14.4-68.1), respectively (Table 2 and Table 4).

Table 3.
Total biofilm producing isolates of S. aureus.

Table 4.
Biofilm production in both MSSA and MRSA isolates.
Correlation between the prevalence of these genes with biofilm formation and antibiotic resistance
Most studies encompassed in this review have indicated a connection between ica genes and biofilm, regardless of the extent of biofilm development. Certain studies have established a link between robust biofilm production and increased antibiotic resistance, whereas other investigations have found no such association. They concluded that the ability to form biofilm, regardless of its degree, can increase antibiotic resistance.
Heterogeneity analysis and publication bias among the studies included
The heterogeneity indices reported for total biofilm were as follows: Q2=180.3, I2=91.2, and t=5.2, with a significance level of p<0.001. A visual assessment of the funnel plot concerning ica operon genes indicated the presence of publication bias, which was further corroborated by the Egger regression test yielding p<0.001. Conversely, the funnel plot analysis for ica operon genes also suggested publication bias among the reports (Figure 2a), while the Egger regression test indicated no such bias (p=0.790). Additionally, both the funnel plot and the Egger regression test confirmed the presence of publication bias related to biofilms (Fig. 2b), with a significance level of p<0.001.
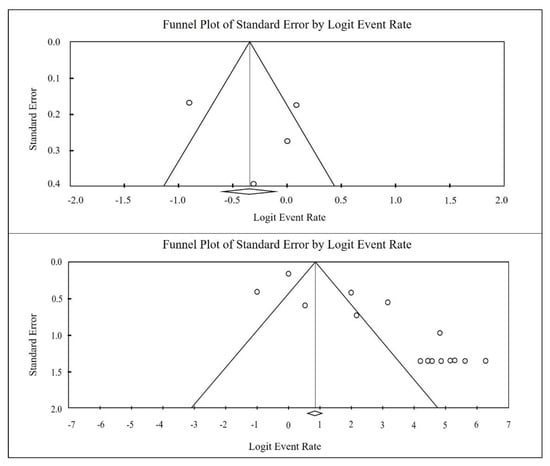
Figure 2.
Funnel plot of meta-analysis on the ica gene prevalence (top image), and biofilm formation (below image) in S. aureus isolated from clinical samples.
Sensitivity analyses
To perform this test, the studies with both the highest and lowest sample sizes were omitted from the analysis, and a new meta-analysis was conducted. The results indicated that the original findings remained consistent.
Discussion
Data obtained from this review showed that the prevalence of ica operon genes varied between 28.0-51.5%, while the prevalence of total ica operon genes in S. aureus was reported at 42.4%. The combined prevalence of around 42.0% of ica genes indicates that the biofilm formed by S. aureus is not only dependent on the presence of these genes but other mechanisms independent of these genes are involved. In line with this fact, Piechota et al. [21] reported that although the prevalence of icaABCD genes was in a small percentage of MSSA isolates, there was no significant correlation between the existence of strong, moderate, poor biofilm types, and the strains that harbored these genes.
Biofilm prevalence of S. aureus isolates in different studies was reported between 10%-100%, while the rate of combined biofilm rate in S. aureus strains was 95.8%. This high rate of combined biofilm production in either MSSA or MRSA strains shows the importance of biofilm in the pathogenesis of S. aureus, even though methicillin-resistant strains use this biological-biofilm layer to prevent antibiotic penetration inside bacteria.
An interesting finding obtained from these studies was that apart from whether the strains were resistant to methicillin (MRSA) or not, the high biofilm formation ability has been reported by most reports except the study conducted by Jasińska et al. [6] It should be mentioned that the reports of some studies indicated a high rate of biofilm production in methicillin-sensitive strains (MSSA) compared to MRSA strains [12,24]. Furthermore, some research findings have shown that there is not a notable difference in biofilm when comparing MSSA and MRSA [8,14,24], but this was the opposite regarding the prevalence of strains containing ica genes, so MRSA strains showed a higher rate [24]. S. Oufrid et al. [23] reported that 100% of the strains with ica genes were also biofilm producers. Similar data has been presented by dos Santos-Goes et al. [18], where using PCR technique, more than 91% of the isolates had icaD gene and the simultaneous presence of icaD and icaA genes was reported in about 59%, which showed the role of these genes for creating biofilm because 100% of them were biofilm producers. Furthermore, findings from a study carried out by Azmi et al. [19] indicated that all strains harboring the icaD/icaA genes were associated with biofilm formation, which indicates a correlation between the existence of these genes and phenotypic biofilm. Manandhar et al. [24] showed that MRSA strains were resistant to most antibiotics, and the biofilm was also higher in these isolates. Meanwhile, strong biofilm producers displayed more drug resistance. Besides, the biofilm-producing strains that harbor icaAD genes recorded higher drug resistance compared to the planktonic counterparts [24]. El-Mahallawy et al. [17] reported that 92% of the ica-gene-positive strains were biofilm positive and 88% of the ica-gene-negatives were biofilm-producing negative.
In contrary, Jasińska et al. [6] have shown that a small number of biofilm producers harbored ica gene, and no noteworthy correlation was observed between ica gene, biofilm, and methicillin resistance, but Bimanand et at [20]. reported a substantial relationship between ica gene and biofilm, but this was not significant between the biofilm and drug resistance. Shivaee et al. [15] pointed out the high expression of icaA/D genes in both MSSA and MRSA. Reports from Shafi et al. [13] and Babra et al. [16] highlight that, apart from the type of biofilm (strong, moderate, and weak), biofilm producers have a higher level of resistance.
Of course, it is worth mentioning that this difference in the prevalence of the ica operon genes and the amount of biofilm formed by the isolates depends on the type of clinical samples (source), geographical region, molecular techniques used for gene detection, phenotypic method of biofilm detection and many other factors.
The analysis of the studies incorporated in this meta-analysis revealed a notable correlation between biofilm-associated genes and both biofilm development and antibiotic resistance. This finding underscores the critical role of biofilms in fostering resistance, as they create a protective biological layer that hinders the effective penetration of antibiotics and disinfectants into the microorganisms. Microorganisms residing within biofilms exhibit distinct characteristics compared to their planktonic forms [25]; they typically demonstrate enhanced resistance against adverse conditions, including exposure to chemical biocides, bacteriophages, antibiotics, and antibodies [26]. Consequently, it is imperative to implement various strategies aimed at inhibiting biofilm formation, such as modifying the properties of abiotic surfaces to deter biofilm establishment, manipulating signaling pathways to suppress biofilm development and promote dispersal, and employing external forces to eliminate existing biofilms [27].
Regardless of the MRSA status of the strains surveyed or their capacity for biofilm production (whether strong, moderate, or weak), biofilm-producing strains consistently exhibited greater antibiotic resistance. Thus, prioritizing the eradication and prevention of biofilm formation is essential in managing clinical microbial infections. Future research should delve into the interplay between biofilm formation, the ica operon, and its related genes across various bacterial species. Additionally, investigations should be conducted to explore the connections between strains harboring these genes and biofilm-forming strains that exhibit resistance to critical antibiotics, such as vancomycin and teicoplanin, which are commonly employed against multidrug-resistant (MDR) and MRSA infections.
Heterogeneity occurs when a collection of studies that are closely related or conceptually aligned on a specific topic produce results that significantly diverge from what would typically be expected due to sampling error. This situation suggests that the observed differences are not simply random variations; rather, they point to fundamental discrepancies in the data, methodologies, participant characteristics, interventions, and outcomes utilized across the studies. In light of this heterogeneity in our study, a random-effects meta-analysis was employed to synthesize the findings. In the context of heterogeneous studies, a random-effects meta-analysis assigns greater weight to smaller studies compared to a fixed-effect meta-analysis. This approach is justified because smaller studies provide more valuable insights into the variability of effects across the research landscape, rather than merely contributing to an assumption of a uniform intervention effect. Consequently, this method allows for a more nuanced understanding of the data.
To further explore the sources of heterogeneity, publication bias was assessed using funnel plots and the Egger regression test, which corroborated the presence of heterogeneity among the studies included in the analysis. To mitigate the impact of this heterogeneity on the results of the meta-analysis, subgroup analyses were conducted, categorizing studies based on the strength of biofilm formation-strong, moderate, and weak. This stratification aimed to clarify the effects observed and enhance the robustness of the findings.
Findings from this review indicated a notable presence of publication bias among the studies included. To assess this further, a visual examination of the funnel plot was conducted alongside the Egger regression test. The existence of publication bias can skew the results of meta-analyses by exaggerating effect sizes, thus highlighting the importance of its identification and correction. Funnel plots and Egger's Test are effective in detecting such biases, while the trim-and-fill method is used for their correction; nonetheless, these methods have inherent limitations, making sensitivity analyses imperative. In order to validate the results, a sensitivity analysis was performed, revealing no significant changes in the findings of this meta-analysis.
Conclusions
Our findings showed that many studies reported a correlation between the high prevalence of ica operon genes, phenotypic biofilm production, and antibiotic resistance. Also, regardless of whether the strains were MRSA or not, the high biofilm formation ability was reported at about 95.8% by most studies. Although some reported that certain MRSA strains exhibit a notably high capacity for biofilm formation, others did not consider such a difference between MRSA and MSSA.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.18683/germs.2024.1448/s1, S1: The Joanna Briggs Institute Prevalence Critical Appraisal Tool. S2: a. Forest plot of the meta-analysis of the combined ica gene prevalence in S. aureus retrieved from clinical samples; b. Forest plot of the meta-analysis of the icaA gene prevalence in S. aureus retrieved from clinical samples; c. Forest plot of the meta-analysis of the icaB gene prevalence in S. aureus retrieved from clinical samples; d. Forest plot of the meta-analysis of the icaD gene prevalence in S. aureus retrieved from clinical samples. S3: a. Forest plot of the meta-analysis of the total biofilms in S. aureus isolated from clinical samples; b. Forest plot of the meta-analysis of the total strong biofilm in S. aureus isolated from clinical samples; c. Forest plot of the meta-analysis of the Total moderate biofilm in S. aureus isolated from clinical samples; d. Forest plot of the meta-analysis of the total weak biofilm in S. aureus isolated from clinical samples.
Author Contributions
KB, MP and RHA undertook the collection and analysis of the data and completed the background literature review for the manuscript. The laboratory experiments, statistical analyses, and manuscript drafting were executed by MRJA, ZSF, ZG, RM and AK. All authors have read and approved the final manuscript version.
Funding
None to declare.
Data Availability Statement
The data supporting the findings of this study are available within the article [Figures/Tables and supplementary materials].
Acknowledgments
The authors would like to thank the Clinical Research Development Unit of Kashan Shahid Beheshti Hospital in Kashan University Medical Sciences.
Conflicts of Interest
All authors – none to declare.
References
- Khademi, F.; Ghanbari, F.; Mellmann, A.; Najafzadeh, M.J.; Khaledi, A. Phylogenetic relationships among Staphylococcus aureus isolated from clinical samples in Mashhad, Iran. J Infect Public Health. 2016, 9, 639–644. [Google Scholar] [CrossRef][Green Version]
- Shakerimoghaddam, A.; Razavi, D.; Rahvar, F.; et al. Evaluate the effect of zinc oxide and silver nanoparticles on biofilm and icaA gene expression in methicillin-resistant Staphylococcus aureus isolated from burn wound infection. J Burn Care Res. 2020, 41, 1253–1259. [Google Scholar] [CrossRef]
- Hosseini, M.; Shapouri Moghaddam, A.; Derakhshan, S.; et al. Correlation between biofilm formation and antibiotic resistance in MRSA and MSSA isolated from clinical samples in Iran: a systematic review and meta-analysis. Microb Drug Resist. 2020, 26, 1071–1080. [Google Scholar] [CrossRef]
- Omidi, M.; Firoozeh, F.; Saffari, M.; Sedaghat, H.; Zibaei, M.; Khaledi, A. Ability of biofilm production and molecular analysis of spa and ica genes among clinical isolates of methicillin-resistant Staphylococcus aureus. BMC Res Notes. 2020, 13, 19. [Google Scholar] [CrossRef]
- Nikmanesh, Y.; Foolady Azarnaminy, A.; Avishan, P.; et al. A Middle East systematic review and meta-analysis of prevalence and antibiotic susceptibility pattern in MRSA Staphylococcus aureus isolated from patients with cystic fibrosis. J Health Popul Nutr. 2022, 41, 26. [Google Scholar] [CrossRef] [PubMed]
- Jasińska, E.; Bogut, A.; Magryś, A.; Olender, A. Evaluation of the role of staphylococci in the pathomechanism of conjunctivitis. Int Ophthalmol. 2021, 41, 2585–2600. [Google Scholar] [CrossRef]
- Speziale, P.; Pietrocola, G.; Foster, T.J.; Geoghegan, J.A. Protein-based biofilm matrices in staphylococci. Front Cell Infect Microbiol. 2014, 4, 171. [Google Scholar] [CrossRef] [PubMed]
- Yousefi Nojookambari, N.; Yazdansetad, S.; Ardebili, A.; Saki, M.; Najjari, E. Detection of intercellular adhesion (ica) genes involved in biofilm and slime formation in clinical isolates of Staphylococcus aureus harboring mecA gene. J Babol Uni Med Sci. 2018, 20, 27–35. [Google Scholar]
- Lin, M.H.; Shu, J.C.; Lin, L.P.; et al. Elucidating the crucial role of poly N-acetylglucosamine from Staphylococcus aureus in cellular adhesion and pathogenesis. PLoS One. 2015, 10, e0124216. [Google Scholar] [CrossRef]
- Rohde, H.; Frankenberger, S.; Zähringer, U.; Mack, D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur J Cell Biol. 2010, 89, 103–111. [Google Scholar] [CrossRef]
- Downes, M.J.; Brennan, M.L.; Williams, H.C.; Dean, R.S. Development of a critical appraisal tool to assess the quality of cross-sectional studies (AXIS). BMJ Open. 2016, 6, e011458. [Google Scholar] [CrossRef]
- Šmitran, A.; Sladojević, Ž.; Božić, L. , et al. Comparison of biofilm production and virulence genes distribution among human and canine isolates of Staphylococcus aureus. Iran J Vet Res. 2023, 24, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Shafi, S.; El-Serwy, H.; El-Zawahry, Y.; Zaki, M.; Sitohy, B.; Sitohy, M. The association between icaA and icaB genes, antibiotic resistance and biofilm formation in clinical isolates of staphylococci spp. Antibiotics. Antibiotics. 2022, 11, 389. [Google Scholar] [CrossRef]
- Manandhar, S.; Shrestha, R.; Tuladhar, R.S.; Lekhak, S. Inducible clindamycin resistance and biofilm production among staphylococci isolated from tertiary care hospitals in Nepal. Infect Dis Rep. 2021, 13, 1043–1052. [Google Scholar] [CrossRef]
- Shivaee, A.; Sadeghi Kalani, B.; Talebi, M.; Darban-Sarokhalil, D. Does biofilm formation have different pathways in Staphylococcus aureus? Iran J Basic Med Sci. 2019, 22, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Babra, C.; Tiwari, J.; Costantino, P.; et al. Human methicillin-sensitive Staphylococcus aureus biofilms: potential associations with antibiotic resistance persistence and surface polysaccharide antigens. J Basic Microbiol. 2014, 54, 721–728. [Google Scholar] [CrossRef] [PubMed]
- El-Mahallawy, H.A.; Loutfy, S.A.; El-Wakil, M.; El-Al, A.K.A.; Morcos, H. Clinical implications of icaA and icaD genes in coagulase negative staphylococci and Staphylococcus aureus bacteremia in febrile neutropenic pediatric cancer patients. Pediatr Blood Cancer. 2009, 52, 824–828. [Google Scholar] [CrossRef]
- dos Santos-Goes, I.C.R.; Romero, L.C.; Turra, A.J.; et al. Prevalence of nasal carriers of methicillin-resistant Staphylococcus aureus in primary health care units in Brazil. Rev Inst Med Trop Sao Paulo. 2021, 63, e14. [Google Scholar] [CrossRef]
- Azmi, K.; Qrei, W.; Abdeen, Z. Screening of genes encoding adhesion factors and biofilm production in methicillin resistant strains of Staphylococcus aureus isolated from Palestinian patients. BMC Genomics. 2019, 20, 578. [Google Scholar] [CrossRef]
- Bimanand, L.; Taherikalani, M.; Jalilian, F.A.; et al. Association between biofilm production, adhesion genes and drugs resistance in different SCCmec types of methicillin resistant Staphylococcus aureus strains isolated from several major hospitals of Iran. Iran J Basic Med Sci. 2018, 21, 400–403. [Google Scholar] [CrossRef]
- Piechota, M.; Kot, B.; Frankowska-Maciejewska, A.; Grużewska, A.; Woźniak-Kosek, A. Biofilm formation by methicillin-resistant and methicillin-sensitive Staphylococcus aureus strains from hospitalized patients in Poland. BioMed Res Int. 2018, 2018, 4657396. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi Mollaahmadi, C.; Anzabi, Y.; Shayegh, J. Comparison of the frequency of biofilm-forming genes (icaABCD) in methicillin-resistant S. aureusstrains isolated from human and livestock. Arch Razi Inst. 2021, 76, 1655–1663. [Google Scholar] [CrossRef]
- Oufrid, S.; Ghazlane, Z.; Jamali, L.; et al. Correlation between staphylococcal biofilm formation in vitro and potential for catheter-related infections. J Infect Dev Ctries. 2015, 9, 368–372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Manandhar, S.; Singh, A.; Varma, A.; Pandey, S.; Shrivastava, N. Biofilm producing clinical Staphylococcus aureus isolates augmented prevalence of antibiotic resistant cases in tertiary care hospitals of Nepal. Front Microbiol. 2018, 9, 2749. [Google Scholar] [CrossRef]
- Hathroubi, S.; Mekni, M.A.; Domenico, P.; Nguyen, D.; Jacques, M. Biofilms: microbial shelters against antibiotics. Microb Drug Resist. 2017, 23, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Müsken, M.; Pawar, V.; Schwebs, T.; et al. Breaking the vicious cycle of antibiotic killing and regrowth of biofilm-residing Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2018, 62, e01635–18. [Google Scholar] [CrossRef]
- Yin, W.; Xu, S.; Wang, Y.; et al. Ways to control harmful biofilms: prevention, inhibition, and eradication. Crit Rev Microbiol. 2021, 47, 57–78. [Google Scholar] [CrossRef]
© GERMS 2025.